Abstract
Although reduced levels of global 5-hydroxymethylcytosine (5hmC) have been observed in medulloblastomas (MBs), it remains unclear whether immunohistochemical (IHC) evaluation of 5hmC can serve as a prognostic biomarker for patients with MB. We performed IHC staining using a 5hmC antibody on a cohort of 114 pediatric MBs, including 69 non-WNT/non-SHH MBs. The 5hmC staining score was evaluated using a 9-point scale based on both the staining intensity and the percentage positive cells. We found that a low 5hmC staining score (< 5 points) was associated with poor outcomes in patients with non-WNT/non-SHH MB (both P < 0.001). Multivariate Cox regression analyses demonstrated the 5hmC staining score was an independent prognostic predictor for progression-free survival (P < 0.001) and overall survival (P = 0.004) in patients with non-WNT/non-SHH MB. Both the receiver operating characteristic curves and calibration curves demonstrated the excellent performance of the nomogram models established based on the Cox regression models. The high predictive accuracy of the nomogram models was confirmed in a validation cohort comprising 32 patients with non-WNT/non-SHH MB. In conclusion, IHC evaluation of 5hmC may serve as a cost-effective and readily accessible approach for the prognostic stratification of patients with non-WNT/non-SHH MB.
Similar content being viewed by others
Introduction
Medulloblastoma (MB) is the most common malignant tumor of the central nervous system (CNS) in children1. A recent consensus has been reached that there are at least four molecular subgroups of MBs (WNT, SHH, Group 3 [G3], and Group 4 [G4]) with distinctive genetic, transcriptional, and epigenetic features2,3,4. Given the overlap between G3- and G4-MBs, the 2016 World Health Organization classification of CNS tumors designates these MBs as non-WNT/non-SHH MBs5,6. Non-WNT/non-SHH MBs account for nearly 65% of all MB cases and exhibit heterogeneous molecular characteristics and outcomes7,8,9,10. Several recent studies have demonstrated that the survival rate for this group of patients exceeds 50%3,10; however, some patients still exhibit extremely poor outcomes, highlighting the need for more precise prognostic biomarkers and treatment strategies.
5-Hydroxymethylcytosine (5hmC) is a stable epigenetic modification that is particularly enriched in the CNS11. Recent studies have shown that 5hmC plays crucial roles in neuronal development and differentiation12,13. The dysregulation of 5hmC has been implicated in various neurological disorders, including neurodevelopmental and neurodegenerative diseases14,15,16,17. Furthermore, it is widely recognized that methylation aberrations, such as promoter hypermethylation and genome-wide DNA hypomethylation are key features in cancer18. Given the importance of 5hmC-mediated demethylation cycle in the active removal of 5-methylcytosine (5mC)19, abnormal 5hmC modifications may play the crucial role in cancer development. Interestingly, several studies on brain tumors have demonstrated that loss of 5hmC associated with high proliferation and invasiveness, as well as poor outcomes20,21,22,23, suggesting that 5hmC may serve as a potential prognostic biomarker for brain tumors.
Our previous study demonstrated that a reduction in global 5hmC levels in MBs correlated with poor outcomes, as assessed by ultra-high-performance liquid chromatography–tandem mass spectrometry (UHPLC-MS/MS)24. In this study, we hypothesized that immunohistochemical (IHC) staining using a 5hmC antibody could serve as a potential prognostic biomarker for pediatric patients with MB.
Results
Patient cohort
This study included 114 pediatric patients with MB. Detailed data are presented in Supplementary Table 1. The mean patient age at surgery was 7.4 years (range, 2–18 years). The male-to-female ratio was 2.6:1. Molecular analysis revealed 10 WNT tumors (8.8%), 35 SHH tumors (30.7%), 23 G3 tumors (20.2%), and 46 G4 tumors (40.3%). The discovery cohort comprised 69 patients with non-WNT/non-SHH MB (Table 1). The average age was 7.3 years (range: 2–16 years). The male-to-female ratio was 3.9:1. The follow-up period ranged from 4 to 166 months. During the follow-up period, 25 patients (36.2%) died due to disease progression.
IHC analysis
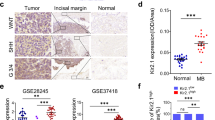
IHC analysis showed that peritumoral brain tissues exhibited significantly higher levels of 5hmC staining than tumor tissues (287.0 ± 12.3 vs. 139.3 ± 68.15; P < 0.001; Fig. 1a, b). The staining level of 5hmC positively correlated with global 5hmC levels in the MBs (r = 0.63, P < 0.001; Fig. 1c). Given the significant inter-tumoral variation in 5hmC staining levels in the MBs, we constructed a 5hmC staining score system using a 9-point scale (Fig. 1d). Using the optimal cut-off value of 5 points, the 114 MBs were divided into high-score (n = 71) and low-score (n = 43) groups. Most large cell/anaplastic MBs were categorized into the low-score group (Fig. 1e). A higher proportion of low-score cases was observed in G3-MBs compared to the other MB subgroups (Fig. 1f). Among non-WNT/non-SHH MBs, there were no significant differences in the consistenty of the assessment of 5hmC staining scores between theassessors, nor in the distribution between the high- and low-score groups (Supplementary Fig. 1a, b).
5hmC immunohistochemical staining in medulloblastomas (MBs). (a) Representative image of 5hmC staining in the peritumoral brain tissues (PBTs) and MBs. (b) Box plot showing the difference in 5hmC staining level between PBT and MB. 5hmC staining level was calculated by multiplying the staining intensity by the percentage of positive tumor cells. ***P < 0.001, by unpaired Student’s t-test. Scale bars represent 50 μm. (c) Scatter plot illustrating the significant correlation between 5hmC staining level and global 5hmC level (n = 30). Global 5hmC level was calculated by by UHPLC-MS/MS analysis. Pearson’s correlation coefficient was used to assess the association. (d) Representative images of MB with different 5hmC immunohistochemical staining scores. Nine-point scale of 5hmC staining score based on staining intensity (no staining = 0, weak staining = 1, moderate staining = 2, strong staining = 3) and the percentage of positive cells; (< 5% = 0, 5–30% = 1, 30–60% = 2, > 60% = 3). Scale bars represent 100 μm. (e) The proportion of high and low 5hmC scores by pathological subtypes. (f) The proportion of high (> 5) and low (< 5) 5hmC scores by molecular subgroups. *P < 0.05, **P < 0.01, by Chi-square test. CMB, classic medulloblastoma; DNMB, desmoplastic nodular medulloblastoma; LC/AMB, large cell/anaplastic medulloblastoma.
Survival analysis
In the discovery cohort of 69 patients with non-WNT/non-SHH MB, tumors with a low 5hmC score showed a poorer prognosis than those with a high 5hmC score (5-year PFS: 27.9% ± 9.4% vs. 83.1 ± 6.4%, P < 0.001; 5-year OS: 26.2% ± 9.2% vs. 85.0% ± 6.4%, P < 0.001. Figure 2a, b). No significant association between 5hmC score and prognosis was observed in patients with WNT- or SHH-MBs. Univariate and multivariate Cox regression analyses identified the 5hmC score (low vs. high) as an independent prognostic predictor in patients with non-WNT/non-SHH MB (PFS: HR = 4.901, 95% CI = 1.909–12.586, P < 0.001; OS: HR = 4.905, 95% CI = 1.639–14.674, P = 0.004; Fig. 3; Supplementary Tables 2, 3).
MBs with a low 5hmC score are correlated with poor prognosis. (a, b) Kaplan-Meier plots of estimated PFS (a) and OS ( b) time distributions stratified by 5hmC score (low 5hmC score vs. high 5hmC score) were analyzed in the discovery cohort (n = 69). Survival differences were calculated using continuous log-rank tests. The numbers below the x-axis indicate the number of people at risk at each time point. OS, overall survival; PFS, progression-free survival.
Nomogram models
Nomogram models were established based on the multivariate Cox regression models (Fig. 4a). The ROC curves for the 3-, 5-, and 10-year PFS and OS demonstrated excellent predictive accuracy for prognosis, with area under the curve (AUC) values ranging from 0.855 to 0.908 (Fig. 4b, c). The calibration plots showed acceptable agreement between the nomogram-based predictions and actual observations (Supplementary Fig. 2a, b). The C-index confirmed the good performance of the nomogram models (range: 0.807–0.814).
Nomogram models for prognostic prediction in patients with non-WNT/non-SHH MB. (a) Nomograms were generated based on the multivariable Cox regression models. Each variable was translated into a risk score based on its contribution to the outcome of the model, and the scores for each case were then summed to calculate the total score. The sum of the scores from each variable was used to calculate the 3-year, 5-year, or 10-year PFS and OS probabilities. (b, c) Time-dependent ROC curves of 3-, 5-, and 10-year PFS (b) and OS (c) for predictive accuracy of nomograms. The prognostic predictive accuracies were analyzed using the area under the ROC curves (AUCs) at 3, 5, and 10 years. GTR, gross total resection; STR, subtotal resection; OS, overall survival; PFS, progression-free survival; ROC, receiver operating characteristic.
Validation cohort
A total of 32 non-WNT/non-SHH MBs were included in the validation cohort (Table 1), comprising 14 cases with high 5hmC score and 18 cases with low 5hmC score. No significant differences in demographic, clinical, or histopathological features were observed between the discovery and validation cohorts. In this cohort, MBs with low 5hmC staining score also showed poorer OS and PFS rates compared to those with high 5hmC staining score (5-year PFS: 41.7% ± 12.0% vs. 92.9% ± 6.9%, P = 0.012; 5-year OS: 50.4% ± 12.5% vs.92.9% ± 6.9%, P = 0.034; Supplementary Fig. 3a, b). The ROC analysis substantiated the good performance of the nomogram models in predicting PFS and OS (AUC: range from 0.759 to 0.821; Supplementary Fig. 4a, b).
Discussion
Identifying reliable biomarkers is crucial for tailoring individual treatment strategies for patients with MB. However, there remains a paucity of effective biomarkers or indicators for predicting prognosis in clinical practice, particularly among patients with non-WNT/non-SHH MBs. In this study, we identified that IHC detection of 5hmC can be a rapid and reliable method for assessing global 5hmC levels in MBs. We revealed that the 5hmC staining score is a potential valuable independent prognostic biomarker for patients with non-WNT/non-SHH MB. Importantly, our data demonstrated the good performance of 5hmC-based nomogram models for predicting outcomes in patients with non-WNT/non-SHH MB.
Through IHC and LC-MS/MS analyses, several studies have shown the relation between 5hmC levels and outcomes in multiple types of cancers23,25,26,27,28. In this study, we demonstrated that a low level of 5hmC staining was strongly correlated with poor prognosis in patients with non-WNT/non-SHH MB. Non-WNT/non-SHH MBs account for a dominant proportion of MBs and display relatively poor outcomes. Recent studies have shown that non-WNT/non-SHH MBs exhibit significant inter-tumoral heterogeneity10,29. However, valuable biomarkers for distinguishing tumor heterogeneity in non-WNT/non-SHH MBs are still lacking. We have previously identified 5hmC as a valuable prognostic marker for MBs24, however, its application is limited owing to the relative complexity and instability of LC-MS/MS methods30,31. In contrast, the determination of 5hmC levels by IHC can be easily applied in clinical practice and does not require additional equipment. Notably, 5hmC holds the potential to be the first epigenetic marker utilized in immunohistochemical analysis of MB. By integrating the 5hmC staining scores, our nomogram models exhibited high precision and reliability in predicting long-term OS and PFS rates. This offers a novel and practical tool for clinicians to identify high-risk patients with non-WNT/non-SHH MBs, which can potentially enable more proactive interventions. Thus, future clinical trials of non-WNT/non-SHH MBs should consider the application of IHC detection of 5hmC for the development of individual treatment strategies.
Our data provide novel evidence that the loss of 5hmC may contribute to the tumorigenesis and tumor development in non-WNT/non-SHH MBs. 5hmC is abundant in the mammalian brain and plays an important role in brain development32. 5hmC accumulates in human brains throughout the lifespan, especially in most neuronal populations of adult and pediatric brains23,33. As embryonal residual tumors, MBs exhibit an extremely low mutational burden2. This indicates that other mechanisms, such as epigenetic dysregulation, may play an important role in the pathogenesis of MBs. 5hmC modifications co-localize with gene bodies, and enhancers are known to mark transcriptional activation34,35. Although the molecular mechanisms underlying the action of 5hmC in MBs remain unclear, the strong relationship between 5hmC levels and prognosis in non-WNT/non-SHH MBs suggest that the mechanisms responsible for regulating 5hmC modification may represent potential future therapeutic targets.
Our study has several limitations. First, this is a retrospective, single-institution study with a small sample size. Potential selection and recall biases are inevitable. Large-scale multicenter prospective studies are required to fully assess the potential clinical applications of the 5hmC staining score system. Additionally, several molecular factors of MB, such as MYC or MYCN amplification and TP53 mutations, were not included in our multiple Cox regression models. Future studies should take into account the integration of 5hmC alongside these molecular factors to achieve a more comprehensive prognostic assessment.
Conclusion
In summary, our study demonstrated that IHC detection of 5hmC may be a potential prognostic biomarker for patients with non-WNT/non-SHH MBs. Future studies should investigate 5hmC as a prognostic biomarker for risk stratification and the development of personalized treatment strategies.
Materials and methods
Ethical approval, consent, and human sample collection
This study was performed in line with the principles of the Declaration of Helsinki and was approved by the Ethics Committee of Beijing Tiantan Hospital, Capital Medical University (no. KY2018-020-01 and KY2020-086-02). Patients aged < 18 years who underwent initial MB resection at Beijing Tiantan Hospital between 2010 and 2019 were included. Tumor location, size, and intracranial or spinal metastases were assessed using perioperative craniospinal magnetic resonance imaging scans. Tumor samples were collected from patients after obtaining informed consent from their parents or legal guardians. Tumors that received radiation or any other adjuvant treatment before surgery were excluded. Using the same inclusion criteria, an independent cohort of patients who underwent surgery between 2020 and 2024 at the same hospital formed the validation cohort.
Immunohistochemistry
IHC staining was performed on formalin-fixed paraffin-embedded (FFPE) samples using a Leica Bond RXm automated staining processor (Leica Biosystems, Buffalo Grove, IL, USA). Briefly, sections were incubated with primary antibodies against 5hmC (1:500; ab214728; Abcam, Cambridge, UK) and Ki-67 (1:300; 9449 S; Cell Signaling Technology, Danvers, MA, USA) at 4℃ overnight. After a 1-h incubation with a secondary antibody (Dako, Glostrup, Denmark) at 37 °C, the signal was visualized using a DAB Chromogenic Kit (Dako).
For each tumor sample, five random tumor fields of view at 40× magnification were captured using the Aperio ImageScope viewing program (Leica Biosystems). Subsequently, two neuropathologists independently quantified calculated the percentage of 5hmC-positive positive cells using Image-Pro Plus image analysis software. The staining intensity was also assessed by the two neuropathologists according to the following criterion: 1 (no or weak staining), 2 (moderate staining), or 3 (strong staining). For discordant data between the two neuropathologists, a third neuropathologists would review the cases and a consensus would be reached among the three neuropathologists at the end. All processes were performed by neuropathologists in a blinded manner (unaware of all clinical, prognostic and molecular data).
We then employed two approaches to evaluate the results of 5hmC staining in MBs: (a) The 5hmC staining level was determined using a semi-quantitative system by multiplying the staining intensity by the percentage (0–100%) of positive tumor cells, as previously described23; These continuous variables were used to evaluate the correlations between 5hmC staining and global 5hmC level detected by UHPLC-MS/MS, as well as to evaluate the differences in 5hmC staining between peritumoral brain tissue (PBT) and tumors. (b) The 5hmC staining score (9-point scale) was calculated by multiplying the staining intensity by the score corresponding to the percentage of positively stained tumor cells (< 5% = 0, 5–30% = 1, 30–60% = 2, > 60% = 3), as previously described36,37. This score was used in the survival analysis and the establishment of risk classification models. Additionally, the areas of tumor tissues and PBT were distingwished by cell types (tumor cells vs. glial cells and neurons), cell density, and expression of Ki-67 antibody (Supplementary Fig. 5). All 5hmC staining level and score were caculated based on the image views of tumor-specific regions.
Molecular subgrouping analysis
The molecular classification of the FFPE samples was determined using the NanoString nCounter system (NanoString Technologies, Seattle, Washington)3. Immunochemistry with antibodies against β-catenin, SFRP1, NPR3, and KCNA1 was used to validate the molecular subgrouping, as described previously38.
UHPLC-MS/MS analysis
UHPLC-MS/MS was used to measure the global 5hmC level, as we previously described24,39. Briefly, DNA extraction was performed using the Wizard® Genomic DNA Purification Kit (A1620; Promega, Madison, WI, USA) according to the manufacturer’s protocol. Nucleosides were separated by UHPLC on a T3 column (Waters, Milford, MA, USA; 186003538) and detected using a triple-4 quadrupole tandem MS spectrometer (Waters, ACQUITY UPLC XEVO TQ-S). The mass/change (m/e) transitions of 228.4 to 112.2 (cytosine), 242.3 to 126.1 (mC), and 258.2 to 124.2 (hmC) were monitored and recorded. Global 5hmC level was calculated using the following formula: 5hmC% = M (5hmC)/(M [cytosine] + M [5mC] + M [5hmC]) × 100.
Statistical analysis
All statistical analyses were performed using GraphPad Prism V9.0 (Graphpad Software, San Diego, CA, USA). Binary and categorical variables were compared using chi-square or Fisher’s exact tests. Continuous variables are presented as the mean ± standard deviation and compared using Student’s t-tests. Kaplan-Meier analysis was used to determine overall survival (OS) and progression-free survival (PFS). For survival analyses, OS was defined as the time from diagnosis to death, and PFS was defined as the time from the date of target tumor resection to the date of tumor progression. Significant differences between survival curves were determined using the log-rank test. For prognostic analysis, we divided the 5hmC score into high-score and low-score groups, and the Ki-67 index into high and low groups, both based on the optimal cut-off values. The optimal cut-off values for the 5hmC score (< 5 and ≥ 5) and Ki-67 index (< 30% and ≥ 30%) were determined as the points with the most statistically significant splits in the cohort using Cutoff Finder (http://molpath.charite.de/cutoff/index.jsp)40. Univariate and multivariate Cox proportional hazards regression models were used to assess the impact of covariates on PFS and OS, with hazard ratios (HR) and 95% confidence intervals (CIs) reported. A backward stepwise selection method was used to construct a multivariate model. A nomogram was developed using the “rms” package in R (version 4.3.1) based on the Cox model to predict 3-, 5-, and 10-year PFS and OS rates in the discovery cohort. Calibration curves were generated to compare the associations between observed and predicted outcomes. Time-dependent receiver operating characteristic (ROC) curve analysis was used to evaluate the discriminative ability of the nomogram (“time ROC” package in R). Statistical significance was defined as a P value < 0.05.
Data availability
The dataset and material are available from the corresponding authors on reasonable request.
Change history
26 June 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-08165-1
References
Sheng, H. et al. Heterogeneity and tumoral origin of Medulloblastoma in the single-cell era. Oncogene 43, 839–850. https://doi.org/10.1038/s41388-024-02967-9 (2024).
Northcott, P. A. et al. The whole-genome landscape of Medulloblastoma subtypes. Nature 547, 311–317. https://doi.org/10.1038/nature22973 (2017).
Kool, M. et al. Molecular subgroups of Medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, group 3, and group 4 Medulloblastomas. Acta Neuropathol. 123, 473–484. https://doi.org/10.1007/s00401-012-0958-8 (2012).
Taylor, M. D. et al. Molecular subgroups of Medulloblastoma: The current consensus. Acta Neuropathol. 123, 465–472. https://doi.org/10.1007/s00401-011-0922-z (2012).
Louis, D. N. et al. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 131, 803–820. https://doi.org/10.1007/s00401-016-1545-1 (2016).
Louis, D. N. et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 23, 1231–1251. https://doi.org/10.1093/neuonc/noab106 (2021).
Cavalli, F. M. G. et al. Intertumoral heterogeneity within Medulloblastoma subgroups. Cancer Cell. 31, 737–754. https://doi.org/10.1016/j.ccell.2017.05.005 (2017).
Goschzik, T. et al. Prognostic effect of whole chromosomal aberration signatures in standard-risk, non-WNT/non-SHH Medulloblastoma: A retrospective, molecular analysis of the HIT-SIOP PNET 4 trial. Lancet Oncol. 19, 1602–1616. https://doi.org/10.1016/S1470-2045(18)30532-1 (2018).
Sharma, T. et al. Second-generation molecular subgrouping of Medulloblastoma: an international meta-analysis of group 3 and group 4 subtypes. Acta Neuropathol. 138, 309–326. https://doi.org/10.1007/s00401-019-02020-0 (2019).
Korshunov, A. et al. Transcriptome analysis stratifies second-generation non-WNT/non-SHH Medulloblastoma subgroups into clinically tractable subtypes. Acta Neuropathol. 145, 829–842. https://doi.org/10.1007/s00401-023-02575-z (2023).
Globisch, D. et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5, e15367. https://doi.org/10.1371/journal.pone.0015367 (2010).
Takai, H. et al. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell. Rep. 9, 48–60. https://doi.org/10.1016/j.celrep.2014.08.071 (2014).
Azizgolshani, N. et al. DNA 5-hydroxymethylcytosine in pediatric central nervous system tumors May impact tumor classification and is a positive prognostic marker. Clin. Epigenetics. 13, 176. https://doi.org/10.1186/s13148-021-01156-9 (2021).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in purkinje neurons and the brain. Science 324, 929–930. https://doi.org/10.1126/science.1169786 (2009).
Al-Mahdawi, S., Virmouni, S. A. & Pook, M. A. The emerging role of 5-hydroxymethylcytosine in neurodegenerative diseases. Front. Neurosci. 8, 397. https://doi.org/10.3389/fnins.2014.00397 (2014).
Zhang, Y. et al. Selective loss of 5hmC promotes neurodegeneration in the mouse model of Alzheimer’s disease. Faseb J. 34, 16364–16382. https://doi.org/10.1096/fj.202001271R (2020).
Godler, D. E. & Amor, D. J. DNA methylation analysis for screening and diagnostic testing in neurodevelopmental disorders. Essays Biochem. 63, 785–795. https://doi.org/10.1042/ebc20190056 (2019).
Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159. https://doi.org/10.1056/NEJMra072067 (2008).
He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. https://doi.org/10.1126/science.1210944 (2011).
Jin, S. G. et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 71, 7360–7365. https://doi.org/10.1158/0008-5472.Can-11-2023 (2011).
Johnson, K. C. et al. 5-Hydroxymethylcytosine localizes to enhancer elements and is associated with survival in glioblastoma patients. Nat. Commun. 7, 13177. https://doi.org/10.1038/ncomms13177 (2016).
Szabó, B. et al. Demethylation status of somatic DNA extracted from pituitary neuroendocrine tumors indicates proliferative behavior. J. Clin. Endocrinol. Metab. https://doi.org/10.1210/clinem/dgaa156 (2020).
Orr, B. A., Haffner, M. C., Nelson, W. G., Yegnasubramanian, S. & Eberhart, C. G. Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS One 7, e41036. https://doi.org/10.1371/journal.pone.0041036 (2012).
Zhao, F. et al. Loss of 5-Hydroxymethylcytosine as an epigenetic signature that correlates with poor outcomes in patients with Medulloblastoma. Front. Oncol.. https://doi.org/10.3389/fonc.2021.603686 (2021).
Chen, K. et al. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell. Res. 26, 103–118. https://doi.org/10.1038/cr.2015.150 (2016).
Qi, J. et al. Regional gain and global loss of 5-hydroxymethylcytosine coexist in genitourinary cancers and regulate different oncogenic pathways. Clin. Epigenetics 14, 117. https://doi.org/10.1186/s13148-022-01333-4 (2022).
Canberk, S. et al. The role of 5-Hydroxymethylcytosine as a potential epigenetic biomarker in a large series of thyroid neoplasms. Endocr. Pathol. 35, 25–39. https://doi.org/10.1007/s12022-024-09800-9 (2024).
Mason, E. F. & Hornick, J. L. Succinate dehydrogenase deficiency is associated with decreased 5-hydroxymethylcytosine production in Gastrointestinal stromal tumors: Implications for mechanisms of tumorigenesis. Mod. Pathol. 26, 1492–1497. https://doi.org/10.1038/modpathol.2013.86 (2013).
Pfister, S. M. et al. A summary of the inaugural WHO classification of pediatric tumors: Transitioning from the optical into the molecular era. Cancer Discov. 12, 331–355. https://doi.org/10.1158/2159-8290.Cd-21-1094 (2022).
Vogeser, M. & Parhofer, K. G. Liquid chromatography tandem-mass spectrometry (LC-MS/MS)--technique and applications in endocrinology. Exp. Clin. Endocrinol. Diabetes 115, 559–570. https://doi.org/10.1055/s-2007-981458 (2007).
Shen, Q. et al. Graphene based pipette tip solid phase extraction of marine toxins in shellfish muscle followed by UPLC-MS/MS analysis. Talanta 116, 770–775. https://doi.org/10.1016/j.talanta.2013.07.042 (2013).
Hahn, M. A. et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell. Rep. 3, 291–300. https://doi.org/10.1016/j.celrep.2013.01.011 (2013).
Szulwach, K. E. et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 14, 1607–1616. https://doi.org/10.1038/nn.2959 (2011).
Song, C. X. et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 29, 68–72. https://doi.org/10.1038/nbt.1732 (2011).
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380. https://doi.org/10.1016/j.cell.2012.04.027 (2012).
Hristov, A. C. et al. HMGA1 correlates with advanced tumor grade and decreased survival in pancreatic ductal adenocarcinoma. Mod. Pathol. 23, 98–104. https://doi.org/10.1038/modpathol.2009.139 (2010).
Yang, B. et al. Evaluation of global DNA hypomethylation in human prostate cancer and prostatic intraepithelial neoplasm tissues by immunohistochemistry. Urol. Oncol. 31, 628–634. https://doi.org/10.1016/j.urolonc.2011.05.009 (2013).
Zhao, F. et al. Molecular subgroups of adult Medulloblastoma: A long-term single-institution study. Neuro Oncol. 18, 982–990. https://doi.org/10.1093/neuonc/now050 (2016).
Wu, T. et al. Characterization of global 5-hydroxymethylcytosine in pediatric posterior fossa ependymoma. Clin. Epigenetics 12, 19. https://doi.org/10.1186/s13148-020-0809-8 (2020).
Zhao, F. et al. Prognostic value of Ki-67 index in adult Medulloblastoma after accounting for molecular subgroup: A retrospective clinical and molecular analysis. J. Neurooncol. 139, 333–340. https://doi.org/10.1007/s11060-018-2865-x (2018).
Acknowledgements
We thank the patients and their families who provided the tissues for this study. We gratefully thank Miss Jing Zhang for her excellent technical assistance. This study was supported by the National Natural Science Foundation of China under grant number 81902862 to F.Z..
Author information
Authors and Affiliations
Contributions
F.Z. conceived and designed the study. L.W., and F.Z. developed the methodology. H.Z., S.R., Z.L., and W.Z. acquired the data. L.W., H.Z., S.R., W. Z., and Z.L. analyzed and interpreted the data. All authors contributed to the writing, review, and revision of the manuscript. F.Z. provided administrative, technical, and material support. F.Z. and Y.T. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent to participate
Written informed consent was obtained from the parent and/or legal guardian of all minor participants involved in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: In the original version of this Article, Yong-ji Tian was omitted as a corresponding author. Correspondence and requests for materials should also be addressed to tianyongji@bjtth.org.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, HG., Ren, Sk., Zhou, WT. et al. Immunohistochemical detection of 5-hydroxymethylcytosine as a prognostic biomarker in non-WNT/Non-SHH medulloblastomas. Sci Rep 15, 15633 (2025). https://doi.org/10.1038/s41598-025-00052-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-00052-z