Abstract
The complexity and variability of biological data has promoted the increased use of machine learning methods to understand processes and predict outcomes. These same features complicate reliable, reproducible, interpretable, and responsible use of such methods, resulting in questionable relevance of the derived. outcomes. Here we systematically explore challenges associated with applying machine learning to predict and understand biological processes using a well- characterized in vitro experimental system. We evaluated factors that vary while applying machine learning classifers: (1) type of biochemical signature (transcripts vs. proteins), (2) data curation methods (pre- and post-processing), and (3) choice of machine learning classifier. Using accuracy, generalizability, interpretability, and reproducibility as metrics, we found that the above factors significantly mod- ulate outcomes even within a simple model system. Our results caution against the unregulated use of machine learning methods in the biological sciences, and strongly advocate the need for data standards and validation tool-kits for such studies.
Similar content being viewed by others
Introduction
Machine learning (ML) can arguably be defined as the development of statistical algorithms that confer the ability to learn and adapt without following explicit instructions. 90% of the world’s data has been generated in the last five years1 resulting in the rapid expansion of the use of ML methods. In biology, ML has largely been used to infer functional relationships from data without the need to define them apriori, i.e., predict responses and processes without a strong understanding of underlying systems. Indeed, classifiers such as random forest (RF)2, support vector machine (SVM) 3,4,5, and neural network (NN)6 have been applied to diverse fields such as genomics and immunology. Predictive classifiers have been constructed to gain mechanistic insights into cardiovascular disease7, acute ischemic stroke8, bacterial and viral peritonitis9, bacterial sepsis10, and latent tuberculosis11. Several studies outline processes and considerations that can enable the effective use of data science methods in biology12,13. Whereas the ability to predict outcomes where mechanisms are unknown or insufficiently defined is the ideal end state for use of ML in biology, we argue that at present, without standardization and validation tools, the physiological relevance of outcomes derived from such studies are questionable. There are various reasons for this: (1) Biological datasets, as they currently exist, are inherently small; and may not be enough to derive reliable ML-driven interpretations. (2) Biological data is intrinsically complex – various factors such as the choice of the system from which the data is generated (e.g., in vitro vs. in vivo), conditions of data collection (e.g., instrument, experimentalist, other), type of signatures being measured (e.g., RNA vs. protein), and other factors influence output. (3) Understanding a biological system typically requires the integration of various data sources14,15. Although the ability to integrate complex raw data has been simplified with the use of methods such as deep neural networks16,17, when combined with the intrinsic variability in biological data streams, our ability to derive reliable systems-level understanding from such studies remains uncertain. (4) Choice of the ML classifier could introduce a significant bias in deriving the outcomes. Indeed various ML classifiers – Bayesian18, tree- based19,20, kernel21, network-based fusion methods22, matrix factorization models23, and a range of deep neural networks6,24,25 – can be utilized to capture complexity of biological systems, but conditional application of these methods is not fully understood. 5) Design of such studies involves critical decisions, both with regards to experimental design and data curation and analyses, which can potentially impact accuracy, reliability, and reproducibility. For instance, necessary data preparation in the form of cleaning, normalization, scaling and standardization; training/testing data split; the extent of hyperparameter tuning; implementation of feature selection; and visualization and interpretation of the results are (as yet) non-standardized decision points in the analysis pipeline that can influence outcomes.
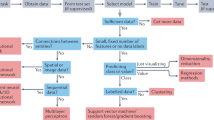
In this manuscript, for the first time, we begin to explore the impact of the above critical factors on outcomes, using Lipopolysaccharide (LPS)-mediated toll-like receptor (TLR)-4 signaling as a model system (Fig. 1). LPS is an essential lipoglycan in the cell walls of Gram-negative bacteria and a pathogen-associated molecular pattern recognized by the human TLR-4, resulting in innate immune stimulation26,27,28. The resulting cytokine and chemokine responses have been measured at both the RNA transcript and protein levels29,30, and as such, it is a well-characterized signal transduction system. In this manuscript, we systematically evaluated the impact of three distinctive aspects of study design on predicting LPS exposure using the cytokine and chemokine expression profiles as outcomes: (1) choice of biochemical signature (tran- scripts vs. proteins), (2) data curation decisions (pre- and post-processing methods such as parameter optimization, data normalization, scaling, feature selection and others), and (3) choice of ML classifier (five “off-the-shelf” algorithms that have been routinely used in exploring biological systems7,9,10,11,31). We explored model building with “fat” biological data, i.e., datasets with few observations relative to the number of parameters, which is typical in biological systems32,33. We evaluated outcomes on the basis of accuracy, interpretability, and identification of signals previously reported from experimental analysis (ground-truth)29,30. Our work demonstrates the impact of these design decisions, and reiterates need for standardization and benchmarking to derive reliable, physiologically valid outcomes from such studies.
Results
We compared five classifier types – single layer neural net (NN), random forest (RF), elastic-net regularized generalized linear model (GLM), support vector machine (SVM), and na¨ıve bayes (NB) – for their ability to accurately predict LPS stimulation or lack thereof and identify features critical to prediction accuracy. The criteria for our non-exhaustive classifier selection were (1) previous use for the interrogation of other/similar biological processes7,9,10,11,31 and (2) ready availability (of-the-shelf) for use by investigators. While more complex classifiers exist, e.g. multi-layer neural networks34, auto-encoders35, and gradient methods36, we found that, for our dataset, the additional complexity of multi-layer neural networks did not demonstrably improve classification performance (see Fig. S8). Variable importance was similarly unchanged. This may be an indication that this dataset is more linearly separable and thus does not require the non-linearity capabilities afforded by multi-layer neural nets37. This may not be true of all datasets, so one must assess if the complexity is necessary for each dataset individually. Furthermore, training of these more complicated models is time-intensive and less straight-forward for a non-expert user, with additional hyper-parameters not tuned with the caret package. Therefore, for the remainder of the analysis, we use a single-layer NN classifier when comparing to the other classifiers evaluated.
Impact of training data proportion on accuracy
With overall classification accuracy as a metric (Eq. 1), we evaluated performance of classifiers against varying train/test proportions (Fig. 2a, Fig. S1). As fraction of data designated to training increased, accuracy on the test set also increased across all classifiers, as might be anticipated. Across training sets, RF and GLM outperformed other classifiers for the transcripts data, whereas GLM and NN outperformed on protein data. For transcripts data, the fraction of RF classifiers that achieved 100% accuracy against the test set (500 data assortments) rapidly increased as training set size increased, followed closely by GLM. For the proteins, GLM and NN had the highest proportion of 100% testing accuracy, followed by SVM. RF was notably less accurate for the proteins, with < 50% of classifiers achieving 100% accuracy even when trained on over 80% of the data. NB was consistently less accurate than other classifiers for both datasets, reach- ing 50% classifiers with 100% accuracy at 86% training data for the transcripts data,
Type and construction of classifier impacts prediction accuracy and predictor importance. (A) Proportion of models trained on 500 different random training samples that yielded 100% accuracy on the test set. (B) Average rank (less than or equal to 15) of each transcript/protein for the entire panel of cytokines and chemokines for repetitions of five-fold cross validations performed on all classifiers for both datasets. Gray tiles: cytokines/chemokines present, but lacking average rank ≤ 15. Blank tiles: cytokine/chemokine present in only one of the datasets. (C) average accuracy of each model type against the reserved fold of the normalized training set at a range of values for parameters controlling the model structure for transcripts (top) and protein (bottom) for each of the five classifiers.
and no classifiers with 100% accuracy until 76% of the data for training for the pro- tein data. Thus, classifier accuracy changes dramatically based on the proportion of training data and the random sample in the split. Based on the performance of the various models at differing train/test proportions, we identified a 70%/30% (transcripts), and 64%/36% (proteins) as split percentages for use in the rest of the analyses in our study. The proteins dataset is smaller than the transcripts dataset, simply as a factor of the number of analytes measured. Thus, we had to choose different train/test percentages for the two datasets, minimizing exclusion of significant data, ensuring high accuracy and approximately equal-sized test data to compare between datasets. We contend that the decision of train/test ratio, as determined by such factors, could add bias to the outcomes, which further substantiates need for standardization metrics for such analyses.
Impact of hyperparameterization
Hyperparameters are factors that dictate how each classifier is constructed. We evaluated accuracy of each classifier across a range of hyperparameter values. Figure 2c shows average accuracy against the reserved fold testing data, over 250 random assortments of training data, and demonstrates that the effect of hyperparameter tuning on accuracy can vary greatly by classifier type. Hyperparameter optimization affects accuracy of GLM, SVM, and NB significantly. In general, effects are more pronounced with protein data than transcript data. This is possibly due to the smaller size, increased standard deviations29, and “missingness” of this data, especially when compared to transcripts (Fig. S2). For RF and NN, most combinations of hyperparameters do not significantly impact average classifier accuracy with the transcript data. For GLM, SVM, and NB, hyperparameters governing method of variable reduction and exclusion can impact performance, particularly for small datasets likely because they tend to use fewer variables in the final model, whereas RF and NN tend to use many more variables. GLM, SVM, and NB exhibit ranges of hyperparameter values with dramatically lower accuracy than optimal with both datasets.
Assessing classifier predictor selection
Identification of key features is a primary purpose of using ML methods, whether it is to enable foundational understanding or drive applied bioscience. Figure 2b depicts features with highest average rank for each classifier for both datasets. All five classifiers selected similar small clusters (6 to 11) of features as most important (lowest rank), but some interesting patterns were observed. For example, NN consistently ranked only two variables as most important (CXCL1 and CCL5 for transcripts, CXCL8 and CXCL3 for proteins), while assigning a broad array of ranks across the remaining cytokines/chemokines (Fig. S3). GLM highly weighted up to three variables (CCL5, CXCL2, and LTB for transcripts, CXCL8 and IL6R for proteins), and consistently excluded others (Fig. S3). GLM distributes middle tier ranks among a third group of features, but does not incorporate all cytokines/chemokines. Differences between SVM and NB are negligible likely because importance is calculated in a model-agnostic manner with these classifiers. For the transcripts, we observed eight features with average ranks ≤ 15 with every classifier- CCL5, CXCL1, CXCL2, LTB, CCL20, IL-6, CXCL8, CXCL5- albeit asso- ciated with varying orders of importance. Two additional features, CCL2 and CSF3, showed average rank ≤ 15 in all classifiers except NN. Thus, important features for RF, GLM, NB, and SVM are similar, if not perfectly aligned. However, there are significant differences in outcomes among selected classifiers, especially with regards to prediction accuracy. We note that the most highly ranked features overlap with known cytokines/chemokines significantly upregulated in LPS-exposed cells, as determined by ground-truth experimental data29. NN also selected many of the same variables, but the importance of CCL2 and CSF3 and others were obscured, potentially because of wide variability in rankings across training repeats. With the protein data, all five classifiers identified IL-6, CXCL8, CXCL5, and CXCL3 as top predictors of LPS exposure, again a refreshing consistency in outcome, albeit with varying order of importance. This is largely consistent with experimental studies, which reported IL-6, CXCL8, and CXCL3 to be significantly upregulated in LPS exposure29. CCL2 was identified as one of the top predictors by four of the five classifiers (all but GLM), while CSF3 was identified by three. Interestingly, NN and GLM, which are the most accurate protein classifiers, do not include CSF3 in the top 15 predictors. In all, consistency among the top 15 predictors is lower for proteins, compared to transcripts. SVM, NB, and RF share eight predictors, including all five selected by NB. The most accurate classifiers, GLM and NN, share six top predictors, including IL6 and CD147(BSG) which were not selected by the other three classifiers. Thus, while there is some consistency in outcomes, there is significant variability based on the choice of dataset (proteins or transcripts), classifier, and hyperparameters used in the analysis.
Comparison of transcripts vs. proteins datasets
Overall, the following cytokines and chemokines were selected as top predictors in both the transcripts and proteins datasets: IL-6, CCL2, CXCL8, CXCL5, and CSF3. However, the top five predictors for transcripts– CCL5, CXCL1, CXCL2, LTB, and CCL20 – were not measured in the protein panel. It is encouraging that when measured in both panels, predictors with high importance in classification with one data type retain their importance with the other data type. Yet, there remains significant variability in top predictors, highlighting the need for validation standards before deriving physiological relevance from such studies.
Changes in importance distributions between data types
Fig. 3b demonstrates distribution of scaled importance scores for seven (of a total 12) overlapping features, with each of the five classifiers, for both data types. The other five overlapping cytokines/chemokines were not identified as important features by any classifier. The importance distributions between data types (transcripts vs. proteins) differ between classifier types, altering the outcome of the study. For RF, overlapping features are less important for transcripts data and more important for protein data. For instance, only CXCL5 had a similar distribution of importance scores for both transcripts and proteins. RF was seen to attribute high importance to IL6 with much greater consistency in the proteins data than it does for the transcripts, suggesting the classifier distributes predictive power differently with the two data types. Indeed, RF is much less accurate for the protein data (Fig. 3a). The other four classifiers differ significantly in the impact of data type on distributions of scaled importance. For NN, distributions are generally similar between the two data types, with the notable exception of CXCL8. Indeed, CXCL8 is consistently more important for all classifier types with the use of protein data, as compared to transcripts. Some of the cytokines/chemokines ranked with higher importance in the transcripts panel do not have corresponding signatures in the proteins panel (the latter being a smaller array, as noted earlier), which may be one of the reasons for this discrepancy. For GLM, CCL2 and CSF3 are largely excluded from importance in the protein data, despite having moderate importance in the transcript data. Despite using fewer predictors, GLM achieves similar testing accuracy on the protein data as it does with transcripts. This may be because of the limited feature selection using this classifier38, and shows that other predictors can compensate. Indeed, CCL2 and CSF3 are highly correlated with heavily weighted features such as CXCL5, CXCL8, and IL-6 (Fig. S4). Because importance for SVM and NB is calculated in a model agnostic manner, they are less informative; with identical importance distributions between the two both on transcripts and proteins, although it is noted that SVM is significantly more accurate than NB for both data types. Thus, relative importance of predictors differs across classifiers and data types, complicating accurate biological interpretation.
Impacts of data type on model accuracy and feature importance between classifiers. (A) Testing accuracy of five classifiers over 500 random training and testing data splits for the transcript (blue) and protein (yellow) data. Solid line: mean accuracy on the hold out test set for the 500 training interactions, Ribbons: one standard deviation above and below the mean. (B) Scaled importance values closest to 1 indicate highest importance for a given classifier. Widest portions of the distribution indicate the most frequent scaled importance across the fifty repetitions of five-fold cross validation feature selection for each of the models.
Impact of data curation and classifier construction decisions on accuracy
We determined that the choice of raw (not normalized) vs. normalized data impacts the optimal hyperparameter values and accuracy. Raw data has different magnitudes for each predictor. For GLM, NB, and SVM, use of raw data did not impact the choice of hyperparameter values, or corresponding classification accuracy; though importance order was affected with the use of GLM. Most significantly, data normalization changes optimal hyperparameter values for NN (Fig. 4a). Indeed normalizing data affects both average accuracy and optimal hyperparameter ranges for both types of data with this classifier. The decay parameter values, which produced the most accurate NNs shifted from lower decay values with normalized data, to higher ones with the use of raw data. The change in accuracy between NN classifiers trained on normalized and raw data was more pronounced with protein data. For RF, using raw data negatively impacted overall accuracy on transcripts data (Fig. S5), narrowing optimal hyperparameter range. Yet, it had minimal effect on accuracy of protein data (Fig. S6).
Impact of data curation on classifier performance (A) Impact of Feature Scaling on Accuracy: For NN we show the average accuracy on the reserved fold of the training set for the transcript (left) and protein (right) datasets for the scaled (top) and un-scaled (bottom) data at a range of values for parameters controlling the model structure.(B) Impact of Feature Set Size on Accuracy: The average accuracy (solid line) and +/- one standard deviation (ribbons) are shown for model sizes ranging from 1–84 for the transcripts data for the five classifiers. These cross validated accuracies were generated via recursive feature selection for NB, NN, SVM, and RF, while GLM elastic net performs feature selection via regularization parameterization. (C) Biological Variation has an outsized impact on model accuracy for certain model configurations. GLM parameter optimization across the transcript dataset shows that structural parameter values cause the GLM to perform very differently against the reserved fold (a) vs. the test set (b) vs. the validation set (c) (the data gathered at a later date and by a different experimentalist). Note that against the reserved fold, there are highly accurate models even at high λ and high α values, which corresponds to models with very few predictors ( 2–4). However, as these models are tested against unseen data in the test and particularly in the validation set, the very minimal models begin to fail because of variations in the primary predictor, CCL5. In order to perform better across data batches, the classifer needs to preserve more predictors (lower α and/or lower λ values).
Impact of number of features used in the classifier
Because of the large number of predictors in the transcripts data, and high accuracy among all five classifiers, we evaluated the effect of eliminating predictors on accuracy. Figure 4b shows average accuracy of each classifier over all folds and repetitions for different feature set sizes. Accuracy of RF and NN are least impacted by feature set size, and the classifiers retain high accuracy across most sizes. Still, accuracy drops precipitously at the small- est sizes evaluated (< 2–3 features). SVM is also minimally impacted by changes in size above a minimum threshold of 2 features. NN demonstrates small, but noticeable increases in accuracy as more features are included. In contrast, NB shows gradual increase in accuracy as number of features drops until it reaches its optimal size of 10 features, wherein the accuracy degrades with less features used. Similarly, GLM performed slightly better at smaller sizes, achieving highest accuracy at 6 predictors. Down-selection of correlated features was evident in optimal sizes for the transcripts data, wherein optimal sizes were consistently smaller than the largest cluster of cor- related features (Fig. S7). In general, different classifiers perform better with varying feature set sizes. Thus, feature reduction is not consistent across classifiers.
Assessing batch effects
We evaluated prediction accuracy of each classifier against dataset(s) generated by different researcher(s), with assured use of the same proto- cols, materials, instruments, and all experimental parameters. Thus, the change in the investigator is the primary source of variation in the data, although it can be argued that the change in the date of performance might also be a factor. We noted that this difference – a single point of variation- has a pronounced effect on those classifiers that rely heavily on only a few predictors. For example, GLMs were 100% accurate on the transcripts training set and nearly 100% accurate on the test set with as few as two variables (this size corresponding to high values of hyperparameters α and/or λ). Yet, GLMs performed poorly against transcripts validation set (Fig. 4c) run by a different investigator. CCL5, for instance, was not upregulated to the same extent in the transcript validation set as it was in the testing set. Therefore, smaller GLMs, which consistently relied primarily on CCL5 for classification, failed to predict LPS exposure in all cases, despite it being the same experimental system.
Discussion
ML is based on pattern recognition, and consequently, improves performance/yields from a given task with experience39,40. Experience, as pertains to ML, is derived from previously collected data relevant to the task being studied. Thus, availability of large and reliable datasets is critical to the success of ML in any discipline. Yet, intrinsic variability and diversity of biological data and the relatively smaller sizes of datasets complicates application of ML in this discipline. In addition, lack of standardization with regards to design of the ML workflow further contributes to the ineffective use of data science in biology. In this study, we systematically decoded the impact of distinctive inputs that can influence the ability of a ML classifier to unravel a simple immunological signal transduction event in an in vitro cell system (Fig. 1). Our results strongly state that achieving reliable, interpretable, reproducible use of ML in biology requires standardization, controls and validation datasets.
First, the experimental design, which includes data type (transcripts vs. proteins) and data processing and curation choices, significantly modulate the outcome of a study. Even with a controlled in vitro experimental design, the use of multi-omic data, differences in data size, distribution, choice of classifier and data curation methods and other factors modulated accuracy of predictions and ranking of significant features in our study. Given that current sources of biological data are seldom large enough to allow for reliable uncertainty quantification, the standard practice of integrating information from different sources, and deriving salient conclusions from associated analysis, while understandable, can be associated with questionable reliability, reproducibility and physiological relevance. Indeed, our results show that while highly accurate ML classifiers could be generated with each of the five classifiers tested for both data types, differences in accuracy and interpretability urge caution in both data preparation and interpretation.
Additionally, inherent differences in classifier types may result in differing importance structures, with some classifiers (e.g., NB and GLM) relying more heavily on a limited number of predictors while others spread the importance over multiple related predictors. It should be noted that importance information for all classifiers indicated strong biologically relevant signals which mirrored the previously reported experimental analysis of this data29 and supported by literature connections to LPS and TLR-4 signaling41,42,43. However, particularly with the protein data, different classifier structures tended to select different “important” signals outside a small core group, suggesting that looking at an ensemble of classifier types may be most informative when identifying biologically relevant signals. For example, the choice of using RF with the protein data would highlight CXCL10 as a significant biomarker, in contrast with choosing NN or GLM, which would not align with that selection. Our study raises an important question – how can we confirm biological significance of the outcomes of such studies? Taken together, our results motivate meticulous standardization, rigorous comparison, and communication of data preparation for ML classifiers applied to biological data (particularly “fat” data). Without prior knowledge, and when relying on a single classifier type, a researcher may have difficulty identifying such biases in importance. How we design the combination of data curation and classifier depends on the desired use of the model and the intended generalizability of the system. While the application of AI/ML in biological sciences has extensive potential, we are not yet entirely ready for the apriori use of these tools to unravel unknown or poorly known processes; and some degree of mechanistic understanding is recommended to ensure physiological relevance of the analysis. Effective use of ML in biological sciences necessitates careful both careful data preparation and benchmarking against known biological mechanisms and multiple classifier types, and the development of standards and synthetic data sets to assess validity of outcomes.
Methods
Source data sets, normalization, missingness and standardization for ML analysis
We previously reported on comprehensive transcript and protein profiles of LPS- mediated induction of cytokines, which were assembled into a database and utilized here29,30. Therein, LPS from Pseudomonas aeruginosa, was used to stimulate A549 lung epithelial cells, which is known to express a variety of TLRs including TLR-444. Following stimulation, cells were harvested, lysed, and lysate harvested for protein analysis and RNA extracted for transcript analysis. This database30 comprises of 110 paired samples from twelve different cell culture lineages that under- went 5 different passages, before being designated as control or treated with LPS. The transcript profiles of 84 cytokines and chemokines, standardized to a housekeeper cytokine/chemokine for normalization was determined. The final 10 samples were generated 6 months after the first 100 by another researcher to introduce an additional layer of data variability. These 10 samples are referred to as the “validation data”. For 78 of the 110 samples a protein panel of 69 cytokines and chemokines was also assessed to generate a matching protein dataset. The protein data was transformed from the raw fluorescence signal to pg/mL using the standard curve.
Missing data values were treated differently in the two data sets. For the transcripts data, missingness is assumed to be due to an insufficient number of cycles in the PCR process, and therefore missing data were represented by setting the CT value to 41, one cycle greater than the number used in the protocol, according to the standard practice in the field. While there are more advanced methods for handling non-detects in qPCR data45, the relatively low number of missing values, 4% of all observations, made this unnecessary. For the protein data, missing values were one of two types (1) NP (no particles detected in flow cytometry), representing a lack of signal in the fluorescence data, (2) signal higher/lower than that of the highest/lowest dilution of the standard curve. The values that were outside the range of the standard curve were assigned to the min or max dilution value of standard curve, as is standard practice29. This type of missingness represented 64% of the data. The NP values, which represented 0.5% of the observations, were filled by the mean value for the data, stratified by status of the LPS-mediated stimulation46. Due to the high proportion of missing observations in the protein data, additional steps were taken to minimize the impact of filled values on the modeling by pruning features in the data that contained more than 70% missing observations. This eliminated 39 cytokine/chemokine features from the protein data leaving 30 remaining predictors above the 70% missingness cutoff. Fig. S2 shows the distribution of percentages of the data is missing for the entire panel of cytokines and chemokines.Except for where the impact of data normalization was being explicitly evaluated, classifiers were run with normalized data. Both types of data were normalized to a [0,1] scale using min/max normalization with the minimum and maximum values for each particular cytokine/chemokine in the training set. The datasets analysed during the current study are available from the corresponding author on reasonable request, and are included in the publication by Jacobsen et al.29.
Machine learning classifers
ML classifiers were all run in R Statistical Software (version 4.2.2)47. Classifiers were constructed using the following R packages: the RandomForest48 package for RF models; neuralnet49, nnet50, and RSNNS51 for NN models; glmnet52,53 for GLM models; e107154 for SVM models; and naivebayes55 for NB models. Addi- tional details about methodology and specific implementation of each model type and analysis can be found in Development and Evaluation of Machine Learning Classifiers and Implementation Details in the Supplementary Information. The code developed during the current study are available from the corresponding author on reasonable request. Performance of the classifiers was evaluated primarily based on the overall classification accuracy:
We also calculated other accuracy metrics such as Sensitivity (Eq. S2), Specificity (Eq. S3), Precision (Eq. S4), and Area under the ROC curve (AUC)56,57, however for this particular dataset, the results did not differ significantly from that of overall accuracy. The performance of the classifiers based on these other metrics can be seen in Figs. S9 and S10 and Tables S4 and S5.
Determining proportion of training data
To evaluate the impact of training set size, 500 random balanced subsets of the data were generated for different training percentages ranging from 10 to 90% of the data assigned as training, with the remainder assigned as testing. Average overall classification accuracy against the remaining testing data was calculated for each classifier at each training percentage using default hyperparameter values either hard-coded into the functions used or determined in preliminary analyses. The values used are described in Table S2. For the remainder of the analyses, a training set size of 70% was used for transcripts data and a training set size of 64% was used for protein data to preserve accuracy without over-fitting. Different percentages of training data were used to have a closer to equal number of testing samples (30 transcript and 28 protein) for both datasets while maintaining a robust training dataset size.
Hyperparameter tunning
Hyperparemeter tuning was performed using the caret package58, running across a predefined set of 50 repeats, each with 5-fold cross-validation for a total of 250 runs per classifier. The hyperparameters for which tuning was performed are detailed in Table 1. For most classifiers, a tuning grid was set up spanning values for the one to three hyperparameters in each model structure and the “train” function from the caret package was used to tune over all hyperparameters simultaneously, and accuracy against the reserved fold – the random 20% of the training data left out of the training for each run – was evaluated at each hyperparameter combination. The exception is RF, where the “train” function did not accommodate training the number of trees (ntrees). For this classifier, then, the “train” function was applied manually at each tested value of ntrees. The “method” option was set to “rf” for RF, “glment” for GLM, “nnet” for NN, “mlpML” for three layer neural net (NN3), “svmLinear” for SVM, and “na¨ıve bayes” for NB. The tuned parameters (used for feature importance determination) are recorded in Table S3.
Feature selection methods
The impact of the number of features that the model is trained on was assessed by tuning the hyperparameters across the range of model sizes due to the interdependence of the number of features and the hyperparamter configuration. For RF, NN, SVM, and NB, recursive feature elimination (RFE) was used concurrently to perform feature selection. We used the “rfe” function from the caret package58, to perform 50 repetitions of 5-fold cross validated RFE. This demonstrates change in accuracy as the least important feature is sequentially eliminated. For the GLM we used elastic net regularized regression to perform feature selection. When smaller values of hyperparameters α and λ are used, the GLM elastic-net regularization shares predictive weight amongst correlated variables, as opposed to down-selecting as does the default LASSO-regularized classifier60 (default α = 1). We suggest that this allows the classifier to be more robust to noise or batch effects affecting a small number of predictors, again highlighting the variability in performance across hyperparameters when applying to new data.
Model interpretability via feature importance
Feature importance was evaluated on classifiers of normalized data, such that the coefficient values or weights were not biased by the differences in concentration magnitudes of the various signaling molecules. For RF, GLM, and NN, importance was calculated using the “varImp” function from the caret package. For SVM and NB, which do not have a default feature importance measure, caret defaults to using a filter importance method via a receiver operating characteristic (ROC)58,61 analysis to characterize feature importance.
Data availability
The datasets analysed during the current study are available from the corresponding author on reasonable request, and are included in the publication by Jacobsen et al. [29].
References
Machine learning: the power and promise of computers that learn by example. The Royal Society. https://royalsociety.org/-/media/policy/ projects/machine-learning/publications/machine-learning-report.pdf (2017)
Chen, X. & Ishwaran, H. Random forests for genomic data analysis. Genomics 99, 323–329 URL (2012). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3387489/
Yang, Z. R. Biological applications of support vector machines. Brief. Bioinform. 5, 328–338 (2004).
Gholami, R. & Fakhari, N. in Chap. 27 - Support Vector Machine: Principles, Parameters, and Applications (eds Samui, P., Sekhar, S. & Balas, V. E.) Handbook of Neural Computation 515–535 Academic Press, (2017). https://www.sciencedirect.com/science/article/pii/B9780128113189000272
Lai, K., Twine, N., O’Brien, A., Guo, Y. & Bauer, D. in Artificial Intelligence and Machine Learning in Bioinformatics (eds Ranganathan, S., Gribskov, M., Nakai, K. & Sch¨onbach, C.) Encyclopedia of Bioinformatics and Computational Biology 272–286 Academic Press, Oxford, (2019). https://www.sciencedirect.com/science/article/pii/B9780128096338203257
Maslova, A. et al. Deep learning of immune cell differentiation. Proceedings of the National Academy of Sciences 117, 25655–25666 (2020). https://www.pnas.org/doi/10.1073/pnas.2011795117. Publisher: Proceedings of the National Academy of Sciences.
Jiang, Y. et al. Cardiovascular disease prediction by machine learning algorithms based on cytokines in Kazakhs of China. Clin. Epidemiol. 13, 417 (2021).
Martha, S. R. et al. Expression of Cytokines and Chemokines as predictors of stroke outcomes in acute ischemic stroke. Front. Neurol. 10 URL https://www.frontiersin.org/articles/ (2020). https://doi.org/10.3389/fneur.2019.01391
Zhang, J. et al. Machine-learning algorithms define pathogen-specific local immune fingerprints in peritoneal dialysis patients with bacterial infections. Kidney Int. 92, 179–191 (2017).
Lamping, F. et al. Development and validation of a diagnostic model for early differentiation of sepsis and non-infectious Sirs in critically ill children-a data- driven approach using machine-learning algorithms. BMC Pediatr. 18, 1–11 (2018).
Robison, H. M. et al. Precision Immunoprofiling to reveal diagnostic signatures for latent tuberculosis infection and reactivation risk stratification. Integr. Biology. 11, 16–25 (2019).
Chicco, D. Ten quick tips for machine learning in computational biology. BioData Mining 10, 35 (2017). https://doi.org/10.1186/s13040-017-0155-3
G. Greener, J., Kandathil, S. M., Moffat, L. & Jones, D. T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 23, 40–55 (2022). https://www.nature.com/articles/s41580-021-00407-0 Number: 1 Publisher: Nature Publishing Group.
Jakhar, S., Bitzer, A. A., Stromberg, L. R. & Mukundan, H. Pediatric Tuberculo- sis: The iImpact of omics on diagnostics development. Int. J. Mol. Sci. 21, 6979 (2020). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7582311/
Chen, C. et al. Applications of multi-omics analysis in human diseases. MedComm 4, e315 (2023). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10390758/
Fortelny, N. & Bock, C. Knowledge-primed neural networks enable biologically interpretable deep learning on single-cell sequencing data. Genome Biol. 21 (190). https://doi.org/10.1186/s13059-020-02100-5 (2020).
Muzio, G., O’Bray, L. & Borgwardt, K. Biological network analysis with deep learning. Brief. Bioinform. 22, 1515–1530. https://doi.org/10.1093/bib/bbaa257 (2021).
Bartosik, A. & Whittingham, H. in Chap. 7 - Evaluating safety and toxicity (ed.Ashenden, S. K.) The Era of Artificial Intelligence, Machine Learning, and Data Science in the Pharmaceutical Industry 119–137 (Academic Press) https://www.sciencedirect.com/science/article/pii/ B9780128200452000088 (2021).
Pisano, F. et al. Decision trees for early prediction of inadequate immune response to coronavirus infections: a pilot study on COVID-19. Front. Med. 10 URL https://www.frontiersin.org/articles/ (2023). https://doi.org/10.3389/fmed.2023.1230733
Qi, Y. in Random Forest for Bioinformatics (eds Zhang, C. & Ma, Y.) Ensemble Machine Learning: Methods and Applications 307–323 (Springer, New York, NY, 2012).
Oz¸sen, S., Gu¨ne¸s, S., Kara, S. & Latifo˘glu, F. Use of kernel functions in artificial immune systems for the nonlinear classification problems. IEEE Trans. Inform. Technol. Biomedicine: Publication IEEE Eng. Med. Biology Soc. 13, 621–628 (2009).
Chierici, M. et al. Integrative network fusion: A multi-omics approach in molecular profiling. Front. Oncol. 10 URL https://www.frontiersin.org/journals/oncology/articles/ (2020). https://doi.org/10.3389/fonc.2020.01065
Tang, D., Park, S. & Zhao, H. N. I. T. U. M. I. D. Nonnegative matrix factorization-based immune-tumor microenvironment deconvolution. Bioinformatics 36, 1344– 1350 URL (2019). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8215918/
Li, G., Iyer, B., Prasath, V. B. S., Ni, Y. & Salomonis, N. DeepImmuno: deep learning-empowered prediction and generation of Immunogenic peptides for T-cell immunity. Brief. Bioinform. 22, bbab160 (2021).
Kang, Y., Vijay, S. & Gujral, T. S. Deep neural network modeling identi- Fies biomarkers of response to immune-checkpoint therapy. iScience 25, 104228 (2022).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 124, 783–801 (2006). https://www.cell.com/cell/abstract/ S0092-8674(06)00190-5. Publisher: Elsevier.
Lu, Y. C., Yeh, W. C. & Ohashi, S. P. LPS/TLR4 signal transduction path- way. Cytokine 42, 145–151 (2008). https://www.sciencedirect.com/science/article/pii/S1043466608000070
Stromberg, L. R. et al. Presentation matters: Impact of association of amphiphilic LPS with serum carrier proteins on innate immune signaling. PloS ONE 13, e0198531 (2018). https://doi.org/10.1371/journal.pone.0198531
Jacobsen, D. E. et al. Correlating transcription and protein expression profiles of immune biomarkers following lipopolysaccharide exposure in lung epithelial cells. PLoS One (2024).
Kubicek-Sutherland, J. Expression profiles of immune biomarkers following LPS exposure (2023).
Dhaubhadel, S. et al. High dimensional predictions of suicide risk in 4.2 million Us veterans Using ensemble transfer learning. Sci. Rep. 14, 1793 (2024).
Maronna, R. A. & Yohai, V. J. Correcting MM estimates for fat data sets. Comput. Stat. Data Anal. 54, 3168–3173 (2010). https://www.sciencedirect.com/science/article/pii/S0167947309003314
Demir-Kavuk, O., Kamada, M., Akutsu, T. & Knapp, E. W. Prediction using step-wise L1, L2 regularization and feature selection for small data sets with large number of features. BMC Bioinformatics 12, 412 (2011). https://doi.org/10.1186/1471-2105-12-412
Cetin, O. & Temurtas, F. A comparative study on classification of magnetoen- cephalography signals using probabilistic neural network and multilayer neural network. Soft. Comput. 25, 2267–2275 (2021).
Li, P., Pei, Y. & Li, J. A comprehensive survey on design and application of autoencoder in deep learning. Appl. Soft Comput. 138, 110176 (2023). https://www.sciencedirect.com/science/article/pii/S1568494623001941
Cai, J., Wang, H. & Zhou, D. X. Gradient learning in a classification setting by gradient descent. J. Approximation Theory. 161, 674–692 (2009). https://www.sciencedirect.com/science/article/pii/S0021904508002608
Ghiasi-Shirazi, K. Competitive cross-entropy loss: A study on training single- layer neural networks for solving nonlinearly separable classification problems. Neural Process. Lett. 50, 1115–1122 (2019).
Parmar, A. et al. (eds) Baig, Z. A Review on Random Forest: An Ensemble Classifier. (eds Hemanth, J., Fernando, X., Lafata, P. & Baig, Z.) International Conference on Intelligent Data Communication Technologies and Internet of Things (ICICI) 2018, Lecture Notes on Data Engineering and Communications Technologies, 758–763 Springer International Publishing, Cham, (2019).
Xu, C. & Jackson, S. A. Machine learning and complex biological data. Genome Biology 20, 76 (2019). https://doi.org/10.1186/s13059-019-1689-0
Rahmani, A. M. et al. Machine learning (ML) in Medicine: review, applications, and challenges. Mathematics 9, 2970 URL (2021). https://www.mdpi.com/2227-7390/9/22/2970. Number: 22 Publisher: Multidisciplinary Digital Publishing Institute.
Sochal, M., Ditmer, M., Gabryelska, A. & Bia-lasiewicz, P. The role of Brain-Derived neurotrophic factor in Immune-Related diseases: A narrative review. J. Clin. Med. 11, 6023 (2022). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9604720/
Yao, C. et al. Lipopolysaccharide induces inflammatory microglial activation through CD147-mediated matrix metalloproteinase expression. Environ. Sci. Pollut. Res. 30, 35352–35365 (2023). https://doi.org/10.1007/s11356-022-24292-y
Wu, D. et al. Micro-concentration Lipopolysaccharide as a Novel Stimulator of Megakaryocytopoiesis that Synergizes with IL-6 for Platelet Production. Sci. Rep. 5, 13748 (2015). https://www.nature.com/articles/srep13748. Number: 1 Publisher: Nature Publishing Group.
MacRedmond, R., Greene, C., Taggart, C. C., McElvaney, N. & O’Neill, S. Respi- Ratory epithelial cells require toll-like receptor 4 for induction of human β-defensin 2 by lipopolysaccharide. Respir. Res. 6, 1–11 (2005).
McCall, M. N., McMurray, H. R., Land, H. & Almudevar, A. On non-detects in qPCR data. Bioinformatics 30, 2310–2316 (2014). https://doi.org/10.1093/bioinformatics/btu239
Kwak, S. K. & Kim, J. H. Statistical data preparation: management of missing values and outliers. Korean J. Anesthesiol. 70, 407–411 (2017). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5548942/
R Core Team. R: A Language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. (2023). https://www.R-project.org/
Liaw, A. & Wiener, M. Classification and regression by randomforest. R News. 2, 18–22 (2002). https://CRAN.R-project.org/doc/Rnews/
Fritsch, S., Guenther, F. & Wright, M. N. neuralnet: Training of Neural Net- works https://CRAN.R-project.org/package=neuralnet. R package version 1.44.2. (2019)
Venables, W. N. & Ripley, B. D. Modern applied statistics with S fourth ednSpringer, New York https://www.stats.ox.ac.uk/pub/MASS4/. ISBN 0-387-95457-0. (2002)
Bergmeir, C. & Ben´ıtez, J. M. Neural networks in R using the Stuttgart neural network simulator: RSNNS. J. Stat. Softw. 46, 1–26 (2012).
Friedman, J., Tibshirani, R. & Hastie, T. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010).
Tay, J. K., Narasimhan, B. & Hastie, T. Elastic net regularization paths for all generalized linear models. J. Stat. Softw. 106, 1–31 (2023).
Meyer, D., Dimitriadou, E., Hornik, K., Weingessel, A. & Leisch, F. e1071 Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien https://CRAN.R-project.org/package=e1071. R package version 1.7–13. (2023)
Majka, M. Naivebayes: high performance implementation of the naive bayes algorithm in R URL (2019). https://CRAN.R-project.org/package=naivebayes. R package version 0.9.7.
Foody, G. M. Challenges in the real world use of classification accuracy metrics: From recall and precision to the matthews correlation coefficient. PLoS ONE 18, 1–27 (2023). https://doi.org/10.1371/journal.pone.0291908
Bradley, A. P. The use of the area under the Roc curve in the evaluation of machine learning algorithms. Pattern Recogn. 30, 1145–1159 (1997). https://www.sciencedirect.com/science/article/pii/S0031320396001422
Kuhn & Max. Building predictive models in r using the caret package. J. Statistical Software 28, 1–26. https://www.jstatsoft.org/index.php/ jss/article/view/v028i05 (2008).
Andriushchenko, M., D’Angelo, F., Varre, A. & Flammarion, N. Why do we need weight decay in modern deep learning? Preprint at https://.org/abs:2310.04415 (2023).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1 (2010).
FawcettT. An introduction to ROC analysis. Pattern Recognit. Lett. 27, 861–874 (2006). https://www.sciencedirect.com/science/article/pii/S016786550500303X
Acknowledgements
This work was performed at the Los Alamos Nation Laboratory, operated by Triad National Security LLC, for the National Nuclear Security Administration of the United States Department of Energy and at the Lawrence Berkeley National Laboratory, operated by the University of California for the United States Department of Energy. Funding for this work was provided by the Defense Threat Reduction Agency, Joint science and technology office, Chemical and Biological Technologies program (Principal Investigators Carrie Manore and Harshini Mukundan). The views expressed in this article are those of the authors and do not reflect the official policy or position of the U.S. Department of Defense or the U.S. Government.
Author information
Authors and Affiliations
Contributions
KMM contributed to the design, development and execution of data integration, machine learning analysis, authoring the manuscript and editing. KW contributed to the integration, analysis and machine learning; and authoring and editing the manuscript. TRL contributed to data integration. DEJ, MMM, with JZK contributed to the experimental data used for this effort; and editing of the manuscript, including figures. JZK lead experimental data generation, and contributed to the authoring and editing of the manuscript. SB contributed to the development of the concept, authoring and editing. CM lead the machine learning analysis, helped conceive the ML strategy, and co-lead the effort. HM conceived and developed the concept, lead the effort, procured funding; and contributed to the experimental design and analysis; authoring and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Martinez, K.M., Wilding, K., Llewellyn, T.R. et al. Evaluating the factors influencing accuracy, interpretability, and reproducibility in the use of machine learning classifiers in biology to enable standardization. Sci Rep 15, 16651 (2025). https://doi.org/10.1038/s41598-025-00245-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-00245-6