Abstract
Hepatocellular carcinoma (HCC) is characterized by poor prognosis and remains a leading cause of cancer mortality worldwide. Advanced HCC is managed with several first-line therapies, including tyrosine kinase inhibitors (TKI) and immunotherapy (mAb-PD-1 and mAb-VEGF). However, the efficacy of HCC therapeutics is often short-lived. Recent studies have demonstrated that the activation of the Nrf2-Bcl-xL pathway contributes to poor prognosis in a subset of HCC patients. Here, we found that dimethyl fumarate (DMF), a drug used for treating psoriasis and multiple sclerosis, regulates the Nrf2-Bcl-xL signaling axis to inhibit HCC growth in a mice xenograft model. Mechanistically, the downregulation of the Nrf2-Bcl-xL axis led to mitochondria stress and apoptosis in vitro and in vivo. Enforced Nrf2 or Bcl-xL expression in HCC cells markedly reversed the antitumor effects of DMF in HCC cells. Importantly, DMF enhanced sorafenib’s antitumor effects. Collectively, our results demonstrate new mechanism insights into the antitumor effects of DMF and that Nrf2-targeted therapy might improve HCC treatment outcomes.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is one of the most aggressive cancers worldwide, with few therapeutic options at advanced stages. About 841,000 new HCC cases and 782,000 deaths were recorded in 20181,2. The incidence is projected to increase owing to the prevalence of metabolic disorders and viral infections3,4. Tyrosine kinase inhibitors (TKIs), such as sorafenib, and Lenvatinib, are usually prescribed for systemic therapy for patients with advanced HCC5,6. However, the therapeutic effects of TKI monotherapy are short-lived and associated with substantial side effects. To meet clinical needs, the combination of anti-PDL1 monoclonal antibody (atezolizumab) and anti-VEGF monoclonal antibody (bevacizumab) was approved as a first-line treatment for advanced HCC treatment in 20207,8. The combination therapy more than doubled the life expectancy of advanced HCC patients9. There is also an ongoing Phase 2 trial for the combination of Lenvatinib (TKI) with nivolumab (anti-PD1) as first-line therapy for HCC (NCT03841201). These recent trends suggest that targeting alternative signaling pathways could improve HCC treatment outcomes.
Oncogenic activation of survival pathways contributes to chemoresistance in HCC10,11. The transcription factor NF-E2-related factor 2 (Nrf2) plays a vital role in detoxifying and mounting antioxidant responses against harmful substances12. However, emerging data suggest that constitutive activation of Nrf2 contributes to the malignant phenotype in HCC13,14. Somatic mutation in Nrf2 or its negative regulator, Keap1, recurs in HCC patients15,16,17 and independently drives rat hepatocarcinogenesis18,19,20. Constitutively active Nrf2 gives tumors a survival advantage by detoxifying chemotherapeutics and upregulating antioxidant defense systems21. Another mode of action by which Nrf2 activation contributes to tumor progression relates to the upregulation of Bcl2 antiapoptotic proteins22,23. It is well known that alterations in the delicate balance of Bcl-2 family proteins influence a plethora of biological functions ranging from survival24, metastasis25, and angiogenesis26,27. Antiapoptotic B-cell lymphoma-extra-large (Bcl-xL) is highly expressed in one-third of HCC patients28,29. In this context, several studies have identified the role of Nrf2 in promoting HCC proliferation and invasion through the Bcl-xL upregulation22,30,31. These findings underscore the potential of Nrf2-Bcl-xL axis to promote tumor growth and provide insight into how targeting this signaling axis might improve HCC treatment.
Discovering new uses for clinically approved drugs with well-defined profiles may be cost-effective and valuable for patients. Dimethyl fumarate (DMF) is an ester of fumaric acid that has demonstrated promising results in psoriasis and multiple sclerosis treatment32. DMF is also examined in an ongoing phase II clinical trial for Cutaneous T Cell Lymphoma (CTCL) treatment (NCT02546440). Previous research has shown that DMF possesses antitumor properties. In breast cancer cells, DMF inhibits the nuclear factor κB pathway33. It also inhibited the growth of melanoma and cervical cancer by promoting cell cycle arrest and apoptosis34,35,36. In human pancreatic cells, DMF could induce cell metabolism dysfunction possibly via targeting the enzyme methylenetetrahydrofolate dehydrogenase 1 (MTHFD1)37. It also induced apoptosis in ovarian carcinoma OVCAR3 cells by regulating Nrf238. However, little is known about the effects and intrinsic molecular mechanisms of DMF in HCC.
In addressing the unmet need for effective HCC therapeutic, we provide an evaluation of DMF’s therapeutic potential on HCC. Treatment with DMF inhibited tumor growth by targeting the Nrf2-Bcl-xL axis and resulted in the accumulation of damaged mitochondria. Furthermore, DMF was as efficient as sorafenib in two xenograft models, and their combination synergistically improved treatment outcomes. Taken together, our study provides evidence to evaluate the potential of DMF, alone or in combination with sorafenib, for HCC treatment.
Materials and methods
Cell lines and cell culture
Human HCC cell lines Huh7 (RRID:CVCL_0336) and HepG2 (RRID:CVCL_0027) were acquired from JCRB Cell Bank, Osaka, Japan, and the American Type Culture Collection (ATCC), respectively. MIHA cell (Immortalized Human Hepatocytes) was obtained from Zhangang Xiao’s lab at the Department of Pharmacology, Southwest Medical University, Luzhou, China. The cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Gibco), 100 IU/ml penicillin, and 100 mg/ml streptomycin. Cells were cultured at 37 °C in a 5% CO₂ incubator and routinely screened for mycoplasma contamination using a sensitive PCR-based method to ensure culture integrity. Human HCC cell lines Huh7 and HepG2 were authenticated yearly by short tandem repeat (STR) profiling by using the PowerPlex16 Cell-ID assay (Promega).
Reagents
Dimethyl fumarate (purity > 99%) and sorafenib (Bay 43-9006, purity > 99%) were purchased from Sigma (St. Louis, MO, USA) and MCE (USA) respectively.
Cell survival and death assays
Cell survival was determined using Cell Counting Kit-8 (CCK-8, Beyotime, Shanghai, China). Briefly, cells were treated with indicated concentrations of DMF for 24 h and 48 h. Following treatment, the optical density of viable cells was measured at 450 nm in a spectrophotometer (Tecan Group Ltd, Männedorf, Switzerland). Cell viability assays were performed in triplicate.
Cell death was assessed using the Annexin V/7-AAD Apoptosis Detection kit (Yeasen, China). In brief, cells were collected and incubated with Annexin V and 7-AAD at room temperature for 15 min in the dark. Subsequently, stained samples were analyzed using a BD LSRFortessa flow cytometer. Annexin V/7-AAD assays were performed in triplicate.
Terminal deoxytransferase-mediated dUTP-biotin nick-end labeling (TUNEL-Alexa 640) assay was used to detect cell apoptosis in GFP-expressing cell lines. Cells were fixed with 4% paraformaldehyde for 20 min. Cell permeabilization and staining procedures were performed according to the manufacturer’s guidelines (Yeasen, China). After TUNEL staining, the nuclei were stained with DAPI, and cells were imaged by fluorescence microscopy (IX70, Olympus, Tokyo, Japan). At least three independent experiments were performed.
Edu staining assay
Ethynyl-2-deoxyuridine incorporation assay was used to determine cell proliferation of huh7 in vitro. 5-Ethynyl-2′-deoxyuridine (EdU), an analog of thymidine, is incorporated into dividing cells during DNA synthesis (Beyotime, Shanghai, China). In brief, cells were incubated with Edu (20 µM) for 1 h after cells were treated with 150 µM DMF or a control vehicle for 24 h. Afterward, cells were fixed with 4% paraformaldehyde in PBS at room temperature for 15 min. After rinsing thrice, cells were permeabilized with 0.3% Triton-X100 in PBS. The incorporated Edu was stained by incubating cells with 1 × Apollo solution for 30 min at room temperature (R.T.) in the dark. Finally, cell nuclei were stained using 1 × Hoechst nuclear dye staining for 30 min and then detected by fluorescence microscopy.
RNA sequencing
Total RNA was extracted from cells treated with vehicle control or DMF for 48 h using TRIzol reagent (Invitrogen), and RNeasy MinElute Cleanup Kit (Qiagen). Each group had three biological replicates (six RNA samples in total). After that, RNA integrity analysis was performed, and the cDNA library was constructed using Illumina TruSeq RNA Sample Preparation Kit. RNA sequencing was performed using Illumina HiSeq 4000 by Novogene (Beijing, China). In brief, the purified RNA samples were used to construct the sequencing libraries with the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (New England Biolabs, USA). The data mining and pathway analysis were performed using the BGI in-house customized data mining system called Dr.Tom (http://report.bgi.com) and the filtering threshold was P value < 0.001 and FC ≥ 1. The raw RNA seq data has been uploaded to GEO (https://ncbi.nlm.nih.gov/sra/PRJNA842764).
Mitochondrial O2- detection
Mitochondrial O2- levels were detected using MitoTracker® Red CM-XRos (Yeasen, China). Cells were washed in PBS and incubated with 150 nM MitoSOXTM Red at 37 C for 30 min. Fluorescence was detected using a BD LSRFortessa flow cytometer according to the manufacturer’s protocol. O2- levels were expressed as mean fluorescence intensity (MFI). Data were analyzed using FlowJo software (version 10.0.7r2; Treestar). Undetached cells were also stained after treatment and imaged using confocal microscopy.
Mitochondrial membrane potential assay
After treatment for 48 h, cells were collected in 0.5 mL medium and incubated with 0.5 mL JC-1 (Yeasen, China) working solution. Cells were maintained at 37 °C in a 5% CO2 incubator for 30 min. After that, the cells were centrifuged at 1000 r/min for 3 min at 4 °C. The cells’ mitochondrial membrane potential (ΔΨm) was detected with flow cytometry according to the manufacturer’s instructions.
Quantitative PCR
RT-qPCR assays were carried out using 1 μg RNA extracted from Huh7 cells. cDNA was synthesized using HiScript® III-RT SuperMix for qPCR kit (Vazyme) following the manufacturer’s protocol. Real-time qPCR was performed using qPCR SYBR Green Master Mix (Q111-02, Vazyme). PCR was performed in the relative quantification of mRNA expression using the comparative cycle threshold (C.T.) method. 18S rRNA expression was used as the endogenous control. The sequence of primers used can be found in Supplementary Table 1.
Adenosine-triphosphate (ATP) measurement
Intracellular ATP was measured using an ATP assay kit (Beyotime, China) according to the manufacturer’s instructions. In brief, cells were washed with PBS, lysed, and centrifuged at 12,000 g for 5 min at 4 °C. The supernatant was collected and mixed with an ATP detection working solution and then measured using a spectrophotometer. The ATP concentration in each sample was calculated using a standard curve normalized to total protein.
Mitochondrial isolation
Mitochondria were isolated from cytoplasmic fraction using a mitochondrial/cytosolic fraction isolation kit (Beyotime, China) following the manufacturer’s instructions. In brief, cells (or animal tissue) were washed with ice-cold PBS, suspended in mitochondria extraction mixed buffer, and lyzed/ homogenized on ice. The cell suspension was then centrifuged at 600 g for 10 min at 4 C. To collect the mitochondrial fraction, the supernatant was centrifuged at 11,000 g for 10 min and the pellet was immediately used or stored at − 80 °C (resuspended in mitochondrial storage solution).
Western blot
Cells were washed with PBS and lysed using ice-cold lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS) supplemented with a Roche protease inhibitor cocktail and phosphatase inhibitor. After centrifugation at 14,000 rpm for 20 min at 4 °C, the supernatant was collected. Protein concentrations were quantified, and samples were separated by 8–12% SDS–polyacrylamide gel electrophoresis before being transferred onto polyvinylidene difluoride (PVDF) membranes (G.E., MA, USA). Membranes were blocked with 5% bovine serum albumin (BSA) in Tris-buffered saline containing 0.1% Tween-20 (TBST) for 1 h and incubated with specific primary antibodies followed by HRP-conjugated secondary antibodies. Protein bands were visualized using the C600 Western blot imaging system (Azure Biosystems). Note: Membranes were cut prior to antibody incubation to enable efficient probing of multiple targets on the same blot and to minimize antibody consumption. The corresponding uncropped blots are provided and clearly documented in the supplementary figure. The following antibodies were used in this study: Beta-actin (sc-517582, Santa Cruz), cytochrome c (cat. no. bs-0013R, Bioss), Bcl-xL (cat. no. 4C12A6, Proteintech), Bak (cat. no. BSM-1284R, Bioss), Bax (cat. no. BSM-0127M, Bioss), Nrf2 (cat. no. BSM-52179R, Bioss), Histone H3 (cat. no. AF0009, Beyotime), AKT (cat. no. BS-50505R, Bioss), p-AKT (cat. no. BSM-33281M, Bioss), Stat3 (cat. no. BSM-52210R, Bioss), p- Stat3 (cat. no. BS-1658R, Bioss).
Transmission and Scanning electron microscopy (TEM/SEM)
Sample preparation for electron microscopy was carried out with aid from the Imaging laboratory at the Guangdong Institute of Agricultural Sciences. In brief, samples for transmission electron microscopy were fixed in 2.5% glutaraldehyde for 30 min at room temperature and then post-fixed with 2% osmium tetroxide for 1 h at 4 °C. This was followed by dehydration in increasing concentration of ethanol (30%-100%) and polymerization by epoxy resin. Ultra-thin sections were obtained and double-stained with 4% uranyl acetate and lead citrate. Finally, the ultrastructure of the cells was observed under a Tecnai G2 F20 S-TWIN transmission electron microscope (FEI). Cellular morphology was studied using SEM. Cells were fixed in 2.5% glutaraldehyde. After washing in phosphate buffer, samples were subjected to post-fixation with 1% OsO4 and dehydrated with an increasing concentration of ethanol. Subsequently, samples were dried and coated with Au–Pd (80:20) using a Polaron E5000 sputter coater. Images were then taken with the S.E. detector.
Transfection
A lentivirus expressing human BCL2L1 (Bcl-xL) was purchased from RiboBio (Guangzhou RiboBio Technology, China). HCC cell lines, Huh7 and HepG2, were transduced with the lentivirus, a multiplicity of infection of 16 along in the presence of 5 μg/mL polybrene for 48 h. Lentivirus-infected cells were screened with an increasing concentration of puromycin (Sigma Aldrich, USA) for 6 days. Finally, healthy colonies were selected and cultured in 6-well plates with a fresh culture medium. NFE2L2 (Nrf2) plasmid was obtained from the same company for trainset Nrf2 expression.
Xenograft models
Animal experiments were performed in compliance with the institutional guidelines and prior approval from the Animal Experimentations Ethics Committee, Southern Medical University, for the care and use of experimental animals. Athymic female mice (NU/NU), aged 4–6 weeks, were subcutaneously inoculated with 1 × 107 huh7 cells. When the tumors became palpable (0.1–0.2 cm3), the mice received an intraperitoneal injection of DMF (30 mg/kg), oral sorafenib (30 mg/kg), or combination once daily for 14 days. All drugs were dissolved in 0.8% methylcellulose daily. The growth tumor curves were determined after measuring the tumor volume using the equation V = (length × width2)/2. The length and width of the tumor were measured using vernier caliper. After the experiment, the mice were euthanized by cervical dislocation. Serum alanine transaminase (ALT), aspartate transaminase (AST), blood urea nitrogen (BUN), and creatinine (ALP) determinations were performed using Beckman Coulter AU5800 automatic biochemical analyzer (Mannheim, Germany), and the respective diagnostic kits (A.U. Chemistry Systems, Beckman Coulter).
Immunofluorescence staining analyses
After treatment, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 for 15 min, blocked with PBS buffer containing 5% bovine serum albumin, and washed with PBS three times. The cells were incubated with primary antibody for 2 h, followed by 1-h incubation with Alexa Fluor-594 conjugated anti-rabbit immunoglobulin antibody. The nuclei were stained using DAPI (4 = 6-diamidino-2-phenylindole) for 1 h. The images were captured on a Zeiss laser scanning microscope (LSM 880 NLO with Airyscan; Germany).
Immunohistochemical staining (IHC)
Firstly, tissues were fixed in formalin. Next, the samples were embedded in paraffin and stained with the following primary antibodies activated caspase-3 (dilution 1:200, Bioss BSM-33199 M), or Bcl-xL (dilution 1:100, Cell signaling technology, USA) and Ki67 (Wanleibio WL03319,). After 1 h of incubation at 37 °C, and corresponding HRP-labelled streptavidin solution was added for 10 min and then stained with diaminobenzidine (DAB). HRP-conjugated Affinipure Goat Anti-Mouse IgG(H + L)(Origene) and HRP-conjugated Affinipure Goat Anti-RabbitIgG(H + L)(Origene) were used as secondary antibodies.
Statistical analysis
The data were analyzed using GraphPad Prism 8 (GraphPad Software, USA). Each experiment had 3 to 5 biological independent samples for each group/condition. Results are reported as the mean value ± s.e.m (unless otherwise specified) of at least 3 independent experiments. An unpaired t-test was used to analyze the differences between 2 groups, one-way analysis of variance (post hoc Tukey’s test) was used to compare the differences of more than 2 groups. P < 0.05 was considered significant.
Results
Dimethyl fumarate (DMF) represses the tumorigenicity of hepatocellular carcinoma cells in vivo
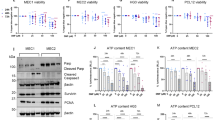
To investigate if DMF displays anticancer effects, eight-week-old female athymic BALB/c nude mice were engrafted with Huh7 cells (Fig. 1A). The mice received DMF (30 mg/kg body weight), sorafenib (30 mg/kg), or combined treatment, once daily, for 14 days. After treatment, tumor volume in all groups significantly reduced compared with the model group (Fig. 1B). DMF and sorafenib both inhibited tumor growth, and the co-treatment of both drugs markedly decreased tumor volumes (Fig. 1C) while all groups showed similar body weight profiles (Fig. 1D). We then performed immunohistochemistry staining of tumor sections. A significant decrease in proliferation marker ki67 while active caspase 3 increased after DMF treatment relative to model group (Fig. 1E, Fig. S1A). Notably, the biochemical results indicated that the co-treatment did not induce renal or hepatotoxicity in the treated mice (Table 1). These results suggested that DMF possesses an antitumor effect in vivo.
Dimethyl fumarate (DMF) represses the tumorigenicity of hepatocellular carcinoma cells in vivo. (A) A schematic of the Huh7 cell line-derived xenograft tumor model exposed to drug treatment for 14 days. 1 × 107 Huh7 cells were injected once into the mice to establish the xenograft model. (B) The xenograft mice were randomly divided into four groups and treated with vehicle control (0.8% methylcellulose, oral), DMF (30 mg/kg body weight i.p.), sorafenib (30 mg/kg/day, oral), or both daily for 14 days. The image shows tumours harvested from each treatment group (C, D) Tumour volume and body weight in all groups were recorded. The fourth tumour in the DMF + sora group was too small to be measured accurately. The S.D for the DMF + sora group does not include the fourth tumour. Results are mean ± SD. **P ≤ 0.01. n = 4 biologically independent samples) mean ± s.d. unless otherwise specified (E) Representative images of IHC for Ki67 and cleaved-caspase 3 in control (0.8% methylcellulose), and DMF ((30 mg/kg body weight) treatment groups of mice were detected by immunohistochemistry. Quantification of the IHC images can be found in the supplementary figure. Bar scale 20 μm (F) Total RNA was isolated from five mice of each group (DMF, 30 mg/kg body weight i.p. or 0.8% methylcellulose, oral) and the expression of target genes were analysed by qPCR (n = 5 biologically independent samples) mean ± s.e.m. *P < .05, **P < 0.01.
Information about the effects of DMF on gene transcription in HCC cells remains unknown. To address this knowledge gap, an analysis of transcriptional pathways was carried out on Huh7 cells treated with DMF or a control vehicle for 48 h. Compared with gene expression in the model group cells, 2031 upregulated genes and 1891 downregulated genes were found in the cells treated with DMF (|log2FC|≥ 1 and P-value ≤ 0.001, Fig. 1F). Interestingly, we observed the modulation of many genes involved in cell cycle, apoptosis, metabolic pathways, and protein processing in the endoplasmic reticulum in DMF-treated cells following KEGG pathway analysis. Based on our RNA-seq data, we selected several dysregulated genes involved in these pathways for further investigation. Their mRNA expression levels were evaluated using RT-qPCR. The qPCR analysis confirmed significant alterations in the expression of these genes in two cell lines (Huh7 and HepG2) as well as in tumor tissues from our animal study. Notably, cell cycle regulators CDC6 and MCM7, which are dysregulated in HCC, were consistently downregulated in the DMF-treated groups (Fig. S1B–D). The transcriptomic results suggested that DMF exerts regulatory effects on vital pathways involved in oncogenesis and tumor progression. Taken together, these data demonstrated that DMF treatment significantly inhibited tumor growth in vivo and that a variety of pathways were involved in the inhibition of tumorigenicity. it highlights key genes of interest that were dysregulated according to our RNA-seq data—specifically those involved in cell cycle regulation, apoptosis, metabolic pathways, and protein processing.
DMF represses cell proliferation and induces apoptosis
Based on the signaling pathways from the above bioinformatics study, we then performed in vitro experiments to confirm the antiproliferation effects of DMF on HCC cells using cell counting kit-8 (CCK-8) assay. Incubation of Huh7 and hepG2 cells with DMF resulted in reduced cell viability in a time and dose-dependent manner (Fig. 2A). In contrast, MIHA cells were less sensitive to DMF treatment (Fig. S2A). Notably, we observed that co-treatment of DMF and sorafenib repressed HCC cell growth compared to the single treatment. Our data showed that co-treatment of DMF and sorafenib reduced huh7 cell growth and enhanced the decrease of AKT and STAT3 phosphorylation, two markers linked to a good prognosis of HCC (Fig. S2B–D). Next, EdU incorporation assay was performed to assess cell division after DMF treatment. We observed that DMF treatment reduced the number of dividing cells in the S phase of the cell cycle compared to the model group. (Fig. 2B). To clarify whether DMF-induced cell growth inhibition is associated with apoptosis, annexin V/7AAD staining was performed. We observed that DMF treatment caused a time-dependent increase of apoptotic (annexin V+) cells compared to model group cells (Fig. 2C, D). Furthermore, the incubation of HCC cells with the pan-caspase inhibitor Z-VAD-FMK significantly reduced DMF-induced apoptosis (Fig. 1E). We also observed increased DNA damage and altered cell morphological features, as indicated by the increase of p-H2A.X (Fig. 2F, G). In summary, DMF inhibited hepatoma cell growth in vitro by repressing DNA synthesis and activating apoptosis.
Dimethyl fumarate represses cell proliferation and induces apoptosis. (A) Huh7 and hepG2 hepatocellular carcinoma cell lines were treated with different concentrations of DMF for 24 and 48 h. Cell survival was measured by CCK-8 assay with each point representing the mean of n = 5 biologically independent samples. mean ± s.e.m. (B) EdU staining was used to assess the effect of DMF on the proliferation of HCC cells. Cells were exposed to DMF (150 μM) or vehicle control for 24 h and stained using EdU assay. Figure showing the percentage of EdU-positive cells in each group. Mean was calculated using the percentage of EdU-positive cells from 3 biologically independent samples. mean ± s.d (C, D) Huh7 and HepG2 cells were treated with DMF in a time-dependent manner (0H, 12H, 24H and 48H). Cell death was analyzed using Annexin V-FITC/7-AAD staining. n = 3 biologically independent samples. mean ± s.e.m. (E) Huh7 cells were treated with DMF (150 μM) with or without Z-VAD-MFK for 48H. Cell death was analyzed using Annexin V-FITC/7-AAD staining. n = 3 biologically independent samples mean ± s.e.m. (F) Western blots of DNA damage marker, p-H2A.X, in vehicle control and DMF-treated cells. Cells were treated with vehicle control or DMF (150 μM) for the indicated time points (0H, 12H, 24H and 48H) (G) Scanning electron microscopy images of Huh7 cells treated with DMF (150 μM) or vehicle control for 24H. Control cells displayed normal cellular membranes, while DMF-treated cells had an altered membrane structure after 24 h of treatment (arrows pointing at the altered membrane) *, #P < .05, **P < 0.01.
DMF induces mitochondrial dysfunction and ATP depletion
Fumarates are readily transported into the mitochondria and influence the citric acid cycle through their metabolites39,40. Next, we mapped the morphological effects of DMF on various cellular organelles. Huh7 cells were treated and analyzed with transmission electron microscopy (TEM). The untreated Huh-7 cells showed intact organelles ultrastructures such as nucleus membrane, mitochondria consisting of well-defined cristae, and electron-dense mitochondrial matrix (Fig. 3A). In contrast, the DMF-treated cells exhibited disrupted and engulfed mitochondria, swollen morphology with a visible accumulation of vacuoles in the cytosol, and severely swollen endoplasmic reticulum. The presence of disrupted mitochondria and extensive mitophagy suggested DMF-induced mitochondrial impairment. Next, we investigated the effect of DMF exposure on mitochondrial membrane potential. Consistently, after treatment with DMF for 48 h, a significant loss of mitochondrial membrane potential (Fig. 3B) was observed in DMF-treated cells. In contrast, we observed only a slight mitochondrial damage in MIHA cells (immortalized Human Hepatocytes) following DMF treatment for 48 h (Fig. S3A). As previously reported, compromised mitochondria are often associated with amplified superoxide production and ATP depletion. Therefore, we further examined the mitochondrial superoxide (MitoSOX) production level upon DMF treatment. Our results from flow cytometric analyses showed that DMF triggered an increase in MitoSOX compared to control (Fig. 3C). Similarly, we observed a marked reduction of ATP level in DMF-treated cells (Fig. 3D) and tumor samples (Fig. S3B), further suggesting that mitochondrial dysfunction and membrane integrity damage were induced by DMF exposure. Together, our data demonstrated that DMF triggered mitochondria dysfunction and that mitochondria seemed to be involved in DMF-induced cell death.
Dimethyl fumarate induces mitochondrial dysfunction and ATP depletion. (A) Representative electron microscopic images of huh7 cells treated with 150 μM DMF or vehicle control for 24H. Electron micrograph showing cristae and electron-dense mitochondrial matrix in vehicle-treated cells. Damage mitochondria were observed after DMF treatment. Arrows in the images point at the normal and damaged mitochondria) (B) JC-1 staining by flow cytometry was used to assess mitochondrial membrane potential (MMP) after 150 μM DMF treatment or vehicle control for 48H. Histograms indicate the red/green ratio of JC-1 fluorescence. n = 3 biologically independent samples mean ± s.e.m. (C) MitoSOX fluorescence images of HCC cells treated with 150 μM DMF or vehicle control for 12H. mtROS level was measured by staining with mitoSOx (150 nM) for 0.5 h. Flow cytometry results were calculated and represented as the relative value to the control vehicle (*p < 0.05). n = 3 biologically independent samples mean ± s.e.m. (D) Cellular ATP levels were measured in huh7 treated with 150 μM DMF or vehicle control for 48 h. n = 3 biologically independent samples. mean ± s.e.m. Scale bar, 20 µm. *P < 0.05, **P < 0.01, **P < 0.001.
DMF induces apoptosis through downregulating Bcl-xL
Cell apoptosis is mainly initiated through the extrinsic or intrinsic pathway. Changes in Bcl-2 family proteins expression in the mitochondria regulate the activation of the intrinsic apoptosis pathway. Our earlier results suggested that DMF induced caspase-dependent apoptosis and extensive mitochondrial stress. Next, we assessed the role of mitochondrial in DMF-induced apoptosis. The expression of several Bcl-2 family proteins was assessed using immunoblot assays.
Notably, we observed an increased expression of the pro-apoptotic proteins Bax and Bak, accompanied by a significant downregulation of the anti-apoptotic protein Bcl-xL in both western blot and immunofluorescence analyses of cells treated with or without DMF (Fig. 4A–C, Fig. S4A, B). Additionally, tumor tissues from model mice treated with DMF showed a significant reduction in Bcl-xL expression compared to untreated model mice group (Fig. 4D). Unlike other antiapoptotic Bcl-2 proteins, Bcl-xL is highly expressed in one-third of HCC patients and contributes to therapeutic resistance and low survival rates28,29.
DMF induces apoptosis through downregulating Bcl-xL. (A) Cells were treated with 150 μM DMF or vehicle control for 48 h, and then the expression of mitochondria apoptosis-related proteins was analysed by western blotting. (B, C) Huh7 and HepG2 cells were treated with 150 μM DMF or vehicle control for 48H. The cells were fixed, and the gene expression was assessed by immunofluorescence assay. (D) The expression levels of Bcl-xL in control (0.8% methylcellulose), and DMF ((30 mg/kg body weight) of mice were detected by immunohistochemistry. (E) Bcl-xl overexpression represses DMF-induced apoptosis in Huh7 cells. Representative immunofluorescence images of TUNEL-positive (green) in Huh7 cells treated with 150 μM DMF or vehicle control for 48H. Histogram indicating the percentages of TUNEL-positive cells relative to the total number of cells. Data are presented as mean ± s.e.m. (n = 3 biologically independent samples. (F) Bcl-xl overexpression suppresses DMF-induced increase in cytosolic cytochrome c in huh7 cells treated with 150 μM DMF or vehicle control for 48H. *P < 0.05, **P < 0.01.
Based on this, we next examined whether DMF inhibits HCC progression by downregulating Bcl-xL. Firstly, we stably infected HCC cells with empty or Bcl-xL-expressing vectors and treated them with vehicle control or DMF (Fig. S4C). TUNEL staining assay revealed that Bcl-xL-overexpression significantly reduced apoptosis rate in DMF-treated cells (Fig. 4E Fig. S4D). Additionally, as Bcl-xL is known to regulate mitochondria integrity, we tested whether Bcl-xL overexpression could modulate cytosolic cytochrome c levels in DMF-treated cells. Interestingly, immunoblot assay revealed that DMF-induced cytosolic cytochrome c was significantly decreased in Bcl-xL overexpressing cells (Fig. 4F). Together, these findings suggest that Bcl-xL downregulation by DMF contributed to cytochrome c release, caspase activation, and ultimately apoptosis. Together, these findings showed that Bcl-xL downregulation is vital in DMF-induced tumor growth in vitro and in vivo.
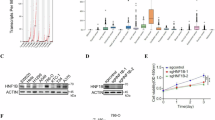
DMF downregulates Bcl-xL through Nrf2 regulation
The Bcl-xL decrease can be caused by either less production or increased protein degradation. Next, we investigated how Bcl-xL was downregulated in DMF-treated cells. Protein degradation is dependent on the proteasome or lysosomal degradation pathway. To clarify the role of proteolysis, we examined the effect of MG132 (ubiquitin–proteasome inhibitor) or the lysosome inhibitor NH4Cl on DMF-induced Bcl-xL reduction. Treatment with either MG132 or NH4Cl could not significantly reverse DMF-induced Bcl-xL downregulation or cell death (Fig. 5A,B). Then we examined the Bcl-xL mRNA level in DMF-treated HCC cells and observed significant downregulation (Fig. 5C, Fig. S2A). Nrf2 is a redox-sensitive transcriptional factor of Bcl-xL22 and has been reported as a target of DMF41,42. We carried out immunoblotting analyses and observed Nrf2 reduction in the DMF-treated cells (Fig. 5D, E, Fig. S5A). Next, we examined whether Bcl-xL downregulation induced by DMF was related to Nrf2 depletion by overexpressing Nrf2 in HCC cells. To do this, cells transfected with Nrf2 or control plasmid were treated with DMF and subjected to immunoblot analysis. Interestingly, the inhibitory effect of DMF on the protein expression of Bcl-xL was reversed by Nrf2 overexpression (Fig. 5F, Fig. S5B). Similarly, we observed reduced apoptosis in Nrf2-overexpressed HCC cells (Fig. 5G, Fig. S5C). Taken together, our results suggest that Nrf2 is an important upstream regulator in DMF-induced Bcl-xL downregulation.
DMF downregulates Bcl-xL through NRF2 regulation. (A) HCC cells were pre-treated with NH4Cl (5 mM), or MG132 (10 µM) for 1 h, followed by DMF treatment (150 µM) for 24 h, and the protein level of Bcl-xl was detected. (B) Cells were pre-treated with NH4Cl (5 mM), or MG132 (10 µM) for 1 h, followed by DMF treatment (150 µM) for 48 h. Cell survival was measured by CCK-8 assay with each point representing the mean of n = 5 biologically independent samples. mean ± s.e.m. (C) Bcl-xl mRNA was extracted from huh7 and quantified using qRT-PCR after 150 μM DMF treatment or vehicle control for 12H, and 24H. mean ± s.e.m. (n = 4 biologically independent samples). (D) Huh7 and hepG2 cells were treated with 150 μM DMF or vehicle control for 12H and expression of Nrf2 was assessed using western blot. (E) Immunofluorescence staining for Nrf2 in huh7 cells treated with 150 μM DMF or vehicle control for 12H treatment (F) Nrf2 overexpression suppresses DMF-induced Bcl-xl and N-cadherin downregulation in huh7 and hepG2 cells. Cells were exposed to with 150 μM DMF or vehicle control for 48H. Cell lysate were collected and subjected to western blot (G) Cells transfected with Nrf2 or control plasmid were treated with DMF or vehicle control for 48H. The apoptosis rate was analyzed with 7-AAD/Annexin V-FITC stain using flow cytometry. Each point representing the mean of n = 3 biologically independent samples. mean ± s.e.m. *P < 0.05, **P < 0.01.
Discussion
In the present study, we provided the most comprehensive preclinical assessment of DMF in HCC to date. Ultrastructural and molecular analysis revealed that DMF regulated the NRF2-Bcl-xL axis in HCC cells, leading to cell division arrest, mitochondria dysfunction, and, ultimately, growth inhibition. Notably, DMF functioned synergistically with sorafenib in vitro and in vivo to suppress tumor growth.
DMF has demonstrated promising efficacy in the treatment of psoriasis and multiple sclerosis treatment43,44,45. A review of the literature indicated that DMF exerts multiple pharmacological effects, including anti-inflammatory46,47, immunomodulatory48,49, and anticancer effects33,37,50,51,52,53,54,55,56,57,58,59 in different cell types. Despite these promising results, the mechanisms underlying the anticancer action of DMF are not fully understood, and its cellular targets in HCC cells remain unidentified. Firstly, we confirmed the antitumor effects of DMF by using mice tumor xenograft models. Fewer Ki-67-positive cells were observed in the tumor treated with DMF compared to the untreated group, indicating that DMF could inhibit tumor growth in vivo. By combining electron microscopy, deep sequencing, and protein profile analysis, we showed that the mechanism of action of DMF centers around the mitochondria and is dependent on the downregulation of the Nrf2-Bcl-xL signaling axis. Nrf2 is typically considered a protective mechanism that helps maintain cellular homeostasis under stressful conditions12. Under the physiological state, low Nrf2 expression is maintained through a negative feedback system controlled by E3 ubiquitin ligases60,61. However, Nrf2 is persistently activated in many cancer types due to somatic mutations of Nrf2 or other oncogene activations62,63,64,65. Such mutations make tumor cells chemoresistant and resilient to stress. In HCC, Nrf2 gain-of-function mutation is associated with poor prognosis15,16,17 and independently drives hepatocarcinogenesis in animal models18,19,20. Aside from its cytoprotective functions, Nrf2 also increases the threshold for caspase activation by upregulating antiapoptotic Bcl2 proteins’ expression22,23. Nrf2-dependent Bcl-xL upregulation promotes HCC tumor growth30. Unlike other antiapoptotic Bcl-2 proteins, Bcl-xL is highly expressed in one-third of HCC patients and contributes to therapeutic resistance and low survival rates28,29. We showed that DMF could regulate the Nrf2/Bcl-xL signaling axis. In addition, the overexpression of Nrf2 or Bcl-xL rendered HCC cells refractory to the DMF antitumor effects, suggesting the functional relevance of the Nrf2/Bcl-xL signaling axis. This is consistent with existing literature that DMF can downregulate Nrf2 in cancer cells at a cytotoxic concentration38,66. Our data lend evidence that DMF could be used to target Nrf2-Bcl-xL in HCC and other cancer types. The doses used for in vivo and in vitro experiments are similar to previous studies. DMF (144.127 g/mol) has a relatively lower molecular weight and requires higher molar concentration for in vitro studies compared to sorafenib (464.825 g/mol) or other cases, rapamycin (914.172 g/mol).
The short-lived and poor therapeutic response of HCC has been partly attributed to intra-tumor and inter-tumor molecular heterogeneity67,68. Given this observation, the combination of sorafenib with other chemical drugs with alternated mechanisms of action could amplify its anticancer activity and improve patients’ prognoses. Interestingly, DMF combined with sorafenib treatment exhibited greater antitumor efficacy than DMF or sorafenib treatment alone, as indicated by the marked inhibition of tumor growth in the mice xenograft model. Notably, the treatment was well tolerated in both xenograft models. Furthermore, our results showed that the co-treatment enhanced the decrease of AKT and STAT3 phosphorylation, two markers linked to a good prognosis of HCC. A recent report also showed that DMF suppressed HCC progression by activating the SOCS3/JAK1/STAT3 signaling pathway53. Previous studies have shown that modest doses of DMF can result in high local concentrations of its primary metabolite, MMF. For instance, doses as low as 20 mg/kg in rats produced MMF levels ranging from 450 to 474 µM in the portal vein69. Based on this, our intraperitoneal administration of DMF at 30 mg/kg is likely sufficient to achieve or exceed the 100 µM threshold in local hepatic tissues, as further supported by our immunohistochemistry findings (e.g., Ki-67 and caspase-3). Despite these encouraging outcomes, a notable limitation of this study lies in its reliance on tumor cell-derived xenografts, which do not fully capture the heterogeneity of HCC. Although our data suggest that DMF modulates mitochondrial redox balance and integrity, the precise upstream signaling events remain incompletely understood, underscoring the need for more detailed mechanistic analyses. Future work employing patient-derived xenograft models will be crucial for gaining deeper insights into DMF’s antitumor potential in HCC. Moreover, DMF has been shown to downregulate Nrf2 by promoting the degradation of its stabilizer protein, DJ-138,66. Drugs that directly target Nrf2 might be more effective at addressing Nrf2 overexpression in cancer cells.
In conclusion, our present study revealed important details regarding the anticancer effects of DMF in HCC. Mechanistically, DMF inhibits the Nrf2-Bcl-xL axis, causing mitochondria dysfunction and apoptosis in HCC cells. Although we focused on the Nrf2-Bcl-xL axis, DMF could regulate other pathways, for example, NFKB, to repress HCC growth. Besides, any potential clinical application of DMF for cancer treatment must consider previously reported DMF’s side effects, including gastrointestinal tract irritation, flushing, and lymphocytopenia70. Our findings support further evaluation of targeting the Nrf2-Bcl-xL axis for HCC treatment as a single therapy or in combination with sorafenib. Additional studies might be needed to identify the patient subgroup(s) that will benefit from DMF treatment. This study provides insight into developing a new therapeutic strategy for HCC.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. The raw RNA seq data has been uploaded to GEO (https://ncbi.nlm.nih.gov/sra/PRJNA842764).
Abbreviations
- HCC:
-
Heptocellular carcinoma
- Nrf2:
-
Nuclear factor erythroid 2-related factor 2
- Bcl-xL:
-
B-cell lymphoma extra-large
- PARP-1:
-
Poly(ADP-ribose) polymerase-1
- CDC6:
-
Cell division cycle 6
- MCM7:
-
Minichromosome maintenance complex component 7
- SSBP1:
-
Small acid-soluble spore protein B
- XBP1:
-
X-box binding protein 1
- RNF185:
-
Ring finger protein 185
- FLCN:
-
Folliculin
- CARS2:
-
Cysteinyl-TRNA synthetase 2
- LAMB1:
-
Laminin subunit beta 1
- MitoSOX:
-
Mitochondrial superoxide
- ALT:
-
Alanine aminotransferase
- AST:
-
Aspartate aminotransferase
References
Forner, A., Reig, M. & Bruix, J. Hepatocellular carcinoma. Lancet 391(10127), 1301–1314 (2018).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68(6), 394–424 (2018).
Liu, Z. et al. Global incidence trends in primary liver cancer by age at diagnosis, sex, region, and etiology, 1990–2017. Cancer 126(10), 2267–2278 (2020).
Petrick, J. L. et al. International trends in hepatocellular carcinoma incidence, 1978–2012. Int. J. Cancer 147(2), 317–330 (2020).
Raoul, J. L. et al. Systemic therapy for intermediate and advanced hepatocellular carcinoma: Sorafenib and beyond. Cancer Treat. Rev 68, 16–24 (2018).
Rimassa, L., Pressiani, T. & Merle, P. Systemic treatment options in hepatocellular carcinoma. Liver Cancer 8(6), 427–446 (2019).
Finn, R. S. et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 382(20), 1894–1905 (2020).
Casak, S. J. et al. FDA approval summary: Atezolizumab plus bevacizumab for the treatment of patients with advanced unresectable or metastatic hepatocellular carcinoma. Clin. Cancer Res. 27(7), 1836–1841 (2021).
Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 7(1), 6 (2021).
Chow, A. K., Yau, S. W. & Ng, L. Novel molecular targets in hepatocellular carcinoma. World J. Clin. Oncol. 11(8), 589–605 (2020).
Ma, S. et al. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene 27(12), 1749–1758 (2008).
Kaspar, J. W., Niture, S. K. & Jaiswal, A. K. Nrf 2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 47(9), 1304–1309 (2009).
Menegon, S., Columbano, A. & Giordano, S. The dual roles of NRF2 in cancer. Trends Mol. Med. 22(7), 578–593 (2016).
Wu, S., Lu, H. & Bai, Y. Nrf2 in cancers: A double-edged sword. Cancer Med. 8(5), 2252–2267 (2019).
Guichard, C. et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 44(6), 694–698 (2012).
Cleary, S. P. et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 58(5), 1693–1702 (2013).
Sanghvi, V. R. et al. The oncogenic action of NRF2 depends on De-glycation by fructosamine-3-kinase. Cell 178(4), 807–819 (2019).
Zavattari, P. et al. Nrf2, but not beta-catenin, mutation represents an early event in rat hepatocarcinogenesis. Hepatology 62(3), 851–862 (2015).
Orru, C. et al. Genetic inactivation of Nrf2 prevents clonal expansion of initiated cells in a nutritional model of rat hepatocarcinogenesis. J. Hepatol. 69(3), 635–643 (2018).
Orru, C. et al. Distinct mechanisms are responsible for Nrf2-keap1 pathway activation at different stages of rat hepatocarcinogenesis. Cancers (Basel) 12(8), 2305 (2020).
Kitamura, H. & Motohashi, H. NRF2 addiction in cancer cells. Cancer Sci. 109(4), 900–911 (2018).
Niture, S. K. & Jaiswal, A. K. Nrf2-induced antiapoptotic Bcl-xL protein enhances cell survival and drug resistance. Free Radic. Biol. Med. 57, 119–131 (2013).
Niture, S. K. & Jaiswal, A. K. Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J. Biol. Chem. 287(13), 9873–9886 (2012).
Cherbonnel-Lasserre, C. & Dosanjh, M. K. Suppression of apoptosis by overexpression of Bcl-2 or Bcl-xL promotes survival and mutagenesis after oxidative damage. Biochimie 79(9–10), 613–617 (1997).
Choi, S. et al. Bcl-xL promotes metastasis independent of its anti-apoptotic activity. Nat. Commun. 7, 10384 (2016).
Giorgini, S. et al. Modulation of bcl-xL in tumor cells regulates angiogenesis through CXCL8 expression. Mol. Cancer Res. 5(8), 761–771 (2007).
Gabellini, C. et al. Interleukin 8 mediates bcl-xL-induced enhancement of human melanoma cell dissemination and angiogenesis in a zebrafish xenograft model. Int. J. Cancer 142(3), 584–596 (2018).
Takehara, T. et al. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology 34(1), 55–61 (2001).
Watanabe, J. et al. Prognostic significance of Bcl-xL in human hepatocellular carcinoma. Surgery 135(6), 604–612 (2004).
Zhang, M. et al. Nrf2 is a potential prognostic marker and promotes proliferation and invasion in human hepatocellular carcinoma. BMC Cancer 15, 531 (2015).
Gan, N., Sun, X. & Song, L. Activation of Nrf2 by microcystin-LR provides advantages for liver cancer cell growth. Chem. Res. Toxicol. 23(9), 1477–1484 (2010).
Meissner, M. et al. Dimethyl fumarate-only an anti-psoriatic medication?. J. Dtsch Dermatol. Ges. 10(11), 793–801 (2012).
Kastrati, I. et al. Dimethyl fumarate inhibits the nuclear factor kappaB pathway in breast cancer cells by covalent modification of p65 protein. J. Biol. Chem. 291(7), 3639–3647 (2016).
Loewe, R. et al. Dimethylfumarate impairs melanoma growth and metastasis. Cancer Res. 66(24), 11888–11896 (2006).
Yamazoe, Y. et al. Dimethylfumarate inhibits tumor cell invasion and metastasis by suppressing the expression and activities of matrix metalloproteinases in melanoma cells. Cell Biol. Int. 33(10), 1087–1094 (2009).
Han, G. & Zhou, Q. Dimethylfumarate induces cell cycle arrest and apoptosis via regulating intracellular redox systems in HeLa cells. In Vitro Cell Dev. Biol. Anim. 52(10), 1034–1041 (2016).
Chen, K. et al. Dimethyl fumarate induces metabolic crisie to suppress pancreatic carcinoma. Front Pharmacol. 12, 617714 (2021).
Saidu, N. E. et al. Dimethyl fumarate controls the NRF2/DJ-1 axis in cancer cells: Therapeutic applications. Mol. Cancer Ther. 16(3), 529–539 (2017).
Passarella, S. et al. Anion transport in rat brain mitochondria: Fumarate uptake via the dicarboxylate carrier. Neurochem. Res. 12(3), 255–264 (1987).
Center for Drug Evaluation and Research. Medical review: Dimethyl fumarate. http://www.accessdata.fda.gov/drugsatfda_docs/nda/2013/204063Orig1s000MedR.pdf
Linker, R. A. et al. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain 134(Pt 3), 678–692 (2011).
Gopal, S. et al. Evidence of activation of the Nrf2 pathway in multiple sclerosis patients treated with delayed-release dimethyl fumarate in the phase 3 DEFINE and CONFIRM studies. Mult. Scler. 23(14), 1875–1883 (2017).
Mrowietz, U. & Asadullah, K. Dimethylfumarate for psoriasis: More than a dietary curiosity. Trends Mol. Med. 11(1), 43–48 (2005).
Mills, E. A. et al. Emerging understanding of the mechanism of action for dimethyl fumarate in the treatment of multiple sclerosis. Front Neurol. 9, 5 (2018).
Xu, Z. et al. Dimethyl fumarate for multiple sclerosis. Cochrane Database Systemat. Rev. 4, 3 (2015).
Lin, S. X. et al. The anti-inflammatory effects of dimethyl fumarate in astrocytes involve glutathione and haem oxygenase-1. ASN Neuro 3(2), 2010000 (2011).
Li, R. et al. Dimethyl fumarate treatment mediates an anti-inflammatory shift in B cell subsets of patients with multiple sclerosis. J. Immunol. 198(2), 691–698 (2017).
Albrecht, P. et al. Effects of dimethyl fumarate on neuroprotection and immunomodulation. J. Neuroinflamm. 9, 163 (2012).
Bombeiro, A. L. et al. Immunomodulation by dimethyl fumarate treatment improves mouse sciatic nerve regeneration. Brain Res. Bull. 160, 24–32 (2020).
Tsubaki, M. et al. Dimethyl fumarate induces apoptosis of hematopoietic tumor cells via inhibition of NF-kappaB nuclear translocation and down-regulation of Bcl-xL and XIAP. Biomed. Pharmacother. 68(8), 999–1005 (2014).
Xie, X. et al. Dimethyl fumarate induces necroptosis in colon cancer cells through GSH depletion/ROS increase/MAPKs activation pathway. Br. J. Pharmacol. 172(15), 3929–3943 (2015).
Takeda, T. et al. Dimethyl fumarate suppresses metastasis and growth of melanoma cells by inhibiting the nuclear translocation of NF-kappaB. J. Dermatol. Sci. 99(3), 168–176 (2020).
Liu, H. et al. Dimethyl fumarate suppresses hepatocellular carcinoma progression via activating SOCS3/JAK1/STAT3 signaling pathway. Am. J. Transl. Res. 11(8), 4713–4725 (2019).
Nicolay, J. P. et al. Dimethyl fumarate restores apoptosis sensitivity and inhibits tumor growth and metastasis in CTCL by targeting NF-kappaB. Blood 128(6), 805–815 (2016).
Kaluzki, I. et al. Dimethylfumarate inhibits melanoma cell proliferation via p21 and p53 induction and bcl-2 and cyclin B1 downregulation. Tumour Biol. 37(10), 13627–13635 (2016).
Kastrati, I. et al. Dimethyl fumarate inhibits the nuclear factor B pathway in breast cancer cells by covalent modification of p65 protein. J. Biol. Chem. 291(7), 3639–3647 (2016).
Booth, L. et al. Regulation of dimethyl-fumarate toxicity by proteasome inhibitors. Cancer Biol. Ther. 15(12), 1646–1657 (2014).
Saidu, N. E. B. et al. Dimethyl fumarate, a two-edged drug: Current status and future directions. Med. Res. Rev. 39(5), 1923–1952 (2019).
Kastrati, I. et al. Dimethyl fumarate impairs breast cancer growth and inhibits the nuclear factor kappa B pathway in breast cancer cells by covalent modification of p65. Cancer Res. 76, 4716 (2016).
Baird, L. & Dinkova-Kostova, A. T. The cytoprotective role of the Keap1-Nrf2 pathway. Arch. Toxicol. 85(4), 241–272 (2011).
Baird, L. & Yamamoto, M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 40(13), 93–100 (2020).
Yoo, N. J. et al. Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology 60(6), 943–952 (2012).
Konstantinopoulos, P. A. et al. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res. 71(15), 5081–5089 (2011).
Shibata, T. et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 135(4), 1358–1368 (2008).
Padmanabhan, B. et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21(5), 689–700 (2006).
Bennett Saidu, N. E. et al. Dimethyl fumarate is highly cytotoxic in KRAS mutated cancer cells but spares non-tumorigenic cells. Oncotarget 9(10), 9088–9099 (2018).
Nguyen, P. H. D. et al. Intratumoural immune heterogeneity as a hallmark of tumour evolution and progression in hepatocellular carcinoma. Nat. Commun. 12(1), 227 (2021).
Liu, J., Dang, H. & Wang, X. W. The significance of intertumor and intratumor heterogeneity in liver cancer. Exp. Mol. Med. 50(1), e416 (2018).
Dibbert, S. et al. Detection of fumarate-glutathione adducts in the portal vein blood of rats: Evidence for rapid dimethylfumarate metabolism. Arch. Dermatol. Res. 305(5), 447–451 (2013).
Linker, R. A. & Gold, R. Dimethyl fumarate for treatment of multiple sclerosis: Mechanism of action, effectiveness, and side effects. Curr. Neurol. Neurosci. Rep. 13(11), 394 (2013).
Acknowledgements
The work was partially supported by grants from Guangdong Basic and Applied Basic Research Foundation, China (No. 2021A1515010713). Zengcheng Technology Innovation Support Fund, Guangzhou, China (2024ZCKJ08).
Author information
Authors and Affiliations
Contributions
X.L, W.F and M.H. designed the study. O.F, and M.A. mainly performed the experiments; J.L. and M.A. performed the cell experiments; Y.Z. and S.D. performed the animal experiments. L.H performed the animal serum biochemical assay, Q.L.and G.W. did the pathology slides and IHC staining; O.F and X.L analyzed data, X.L, W.F and M.H. supervised the study; O.F, X.L and M.H. wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Animal experiments were performed in compliance with the institutional guidelines and prior approval from the Animal Experimentation Ethics Committee, Southern Medical University, for the care and use of experimental animals. The study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Faleti, O.D., Alsaadawe, M., Long, J. et al. Dimethyl fumarate abrogates hepatocellular carcinoma growth by inhibiting Nrf2/Bcl-xL axis and enhances sorafenib’s efficacy. Sci Rep 15, 16724 (2025). https://doi.org/10.1038/s41598-025-00832-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-00832-7