Abstract
Coronary artery disease (CAD) remains a leading global cause of mortality. The expression of small ubiquitin-like modifier 1 (SUMO-1) is reduced in heart failure. However, the mechanisms underlying its modification in CAD remain underexplored. This study sought to identify SUMOylation-related biomarkers and elucidate the potential mechanisms in CAD pathogenesis. This study analyzed three CAD datasets (GSE42148, GSE23561, and GSE121893) alongside 187 SUMOylation-related genes (SRGs). The overlap between differentially expressed genes (DEGs) and SRGs was used to identify differentially expressed SUMOylation-related genes (DE-SRGs). Biomarkers were validated through expression profiling and receiver operating characteristic (ROC) curve analysis. Enrichment and immune infiltration analyses were performed to explore the molecular mechanisms by which these biomarkers influence CAD. A drug-gene interaction network was constructed using the Drug-Gene Interaction database (DGIdb). Single-cell analysis was conducted to identify key cellular players and validate the differential expression of biomarkers across cell types. A total of 12 DE-SRGs were identified in CAD. Among them, SUMO1 and PPARG were validated as biomarkers, with their expression significantly elevated in the CAD group compared to the control group. Single-sample gene set enrichment analysis (ssGSEA) revealed distinct immune cell distributions in CAD, with central memory CD4+ T cells and memory B cells positively correlated with the biomarkers. Gene set enrichment analysis (GSEA) linked these biomarkers to ribosomal activity, olfactory transduction, and other pathways. Single-cell analysis confirmed the expression of SUMO1 and PPARG in endothelial cells, particularly in the CAD group. Additionally, SUMO1 was differentially expressed in cardiomyocytes, exhibiting higher expression in controls. SUMO1 and PPARG were identified as novel SUMOylation-related biomarkers in CAD, suggesting new therapeutic avenues for CAD management.
Similar content being viewed by others
Introduction
Coronary artery disease (CAD), also known as coronary heart disease (CHD), is a leading cause of increasing cardiovascular disease prevalence and mortality globally1. An estimated 330 million individuals are affected by cardiovascular disease, with 11.39 million specifically suffering from CAD2. Key risk factors for CAD include age, dyslipidemia, smoking, and heredity. The pathogenesis of CAD involves multiple factors, including the development of coronary atherosclerosis and coronary artery stenosis, but the precise mechanisms remain unclear3. Currently, patients with CAD are primarily treated with conventional drugs or surgical interventions, though many experience poor prognoses. Drug therapies are hindered by issues of dependence and patient adherence, while percutaneous coronary intervention (PCI) carries risks of restenosis and other complications. Surgical approaches involve significant trauma and have uncertain long-term outcomes4. As such, the discovery of novel biomarkers and an understanding of the underlying molecular mechanisms could enhance our understanding of CAD’s pathogenesis, offering potential targets for clinical diagnosis and treatment5.
Small ubiquitin-like modifier (SUMO) is a highly conserved small protein (approximately 15 kDa) widely present in eukaryotic cells. Its structure and reaction mode are similar to ubiquitin, but its functions differ. SUMOylation, the process by which SUMO is covalently attached to proteins, regulates several biological processes, including cell metabolism, DNA replication and repair, cell division, and signal transduction6. Reduced SUMO-1 expression has been observed in failing hearts, and SUMO-1 knockdown in mice leads to heart failure, suggesting a critical role in cardiac function7. However, limited research has focused on the role of SUMOylation modification in CAD’s development and progression. Identifying biomarkers linked to SUMOylation modification is therefore vital for advancing CAD treatment strategies.
This study employs bioinformatics to identify SUMOylation-related genes (SRGs) as key biomarkers in CAD development. The biological functions and potential mechanisms of these biomarkers are explored, providing novel strategies for CAD treatment. The findings may offer valuable insights into the pathogenesis of CAD and contribute to the development of targeted therapeutic approaches.
Materials and methods
Data source
The CAD datasets GSE121893, GSE42148, and GSE23561 were sourced from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). A total of 187 SUMOylation-related genes (SRGs) were obtained from the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb) using the keyword ‘REACTOME_SUMOYLATION’. The GSE42148 dataset includes transcriptome microarray sequencing data from 13 peripheral blood samples of patients with CAD and 11 normal control peripheral blood samples, using the GPL13607 platform5. The GSE23561 dataset consists of transcriptome microarray data from peripheral blood samples of six patients with CAD and 9 normal control peripheral blood samples, utilizing the GPL10775 platform8. The single-cell GSE121893 dataset encompasses sequencing data from 2 normal samples and 2 CAD samples using the GPL18573 Illumina NextSeq 500 platform9.
Differential expression analysis
Differential expression analysis was conducted using the ‘limma’ 3.56.2 R package10 to identify differentially expressed genes (DEGs) between CAD and control samples from the GSE42148 dataset. DEGs were defined with |log2 fold change (FC)| ≥ 0.5 and adjusted P-value (adj.P) < 0.05. The results were visualized in a volcano plot using the “ggVolcano” 0.0.2 R package11. Additionally, a circular heatmap of the top 20 upregulated and downregulated DEGs was generated using the ‘circlize’ package12.
Identification of DE-SRGs and functional enrichment analysis
The DEGs were intersected with SRGs using the ‘VennDiagram’ 1.6.20 R package13 to derive differentially expressed SUMOylation-related genes (DE-SRGs). Gene Ontology (GO) enrichment analysis, covering biological processes (BP), cellular components (CC), and molecular functions (MF), as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, were performed on the DE-SRGs using the ‘org.Hs.eg.db’ 3.17.0 R package as the background gene set and the ‘clusterProfiler’ 4.8.2 R package14. Enriched entries for BP, CC, MF, and KEGG were displayed using the ‘ggplot2’ 3.4.4 R package15 (P < 0.05).
PPI network construction
To explore protein-protein interactions (PPIs) of the DE-SRGs, these genes were uploaded to the STRING database (http://www.string-db.org/), with a confidence score threshold set at ≥ 0.4. A PPI network was then constructed using Cytoscape 3.7.1 software16. Significant modules within the PPI network were identified using the MCODE plugin in Cytoscape (degree = 2, node score = 0.2). The genes within these significant modules were designated as module genes.
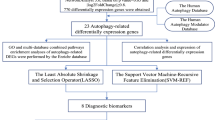
Machine learning
The module genes were analyzed using the ‘Boruta’ 8.0.0 R package (DOI: https://doi.org/10.18637/jss.v036.i11), resulting in the identification of candidate genes 1. In the process of feature selection using Boruta, the random seed was set to 81 to ensure reproducibility. The feature selection was performed with the formula ‘group ~ .’, using the Boruta function on the Bor dataset. Subsequently, the ‘e1071’ 1.7.13 R package (https://rdrr.io/rforge/e1071/) was used for support vector machine-recursive feature elimination (SVM-RFE) to assess the significance of the module genes, leading to the identification of candidate genes 2. In the process of feature selection using Support Vector Machine (SVM), the random seed was initially set to 438 to ensure the results could be reproduced. Feature selection was performed using the svmRFE function, with k set to 10, indicating 10-fold cross-validation. Additionally, when the number of features exceeded 100, halve.above = 100 was set. The number of folds for external cross-validation was set to 8. Finally, an SVM model was trained using a linear kernel (kernel = “linear”). Finally, the intersection of these two gene sets yielded the key genes.
Identification of biomarkers
The ‘ggpubr’ 0.6.0 R package (https://rpkgs.datanovia.com/ggpubr/) was employed to visualize the expression levels of key genes in the GSE42148 and GSE23561 datasets using the Wilcoxon method (P < 0.05). Genes exhibiting consistent expression trends and significant differences were selected for further analysis. Receiver operating characteristic (ROC) curves for these key genes were generated using the ‘pROC’ 1.18.4 R package17, with genes showing an area under the curve (AUC) > 0.7 in both datasets considered potential biomarkers.
Artificial neural network (ANN) analysis of biomarkers
An ANN model was developed for the biomarkers in the GSE42148 dataset with the ‘neuralnet’ 1.44.2 R package16. The ROC curve (AUC > 0.7) was subsequently plotted using the ‘pROC’ 1.18.4 R package to assess model accuracy17. The confusion matrix was constructed using the ‘Caret’ 6.0–94 R package (DOI:https://doi.org/10.18637/jss.v028.i05) to evaluate the model’s predictive power, and generalization weights were visualized via the ‘gwplot’ function.
Functional annotation of biomarkers
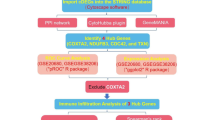
Each biomarker’s expression value served as a phenotype file, and the correlation coefficients between each gene and all other genes in the GSE42148 dataset were ranked. Gene set enrichment analysis (GSEA) for each biomarker was conducted using KEGG gene sets (c2.cp.kegg.v7.5.1.entrez.gmt) via the ‘clusterProfiler’ 4.8.2 R package14. In the GSE42148 dataset, Spearman’s correlation analysis (cor > 0.6, P < 0.05) was performed to examine the expression correlations between biomarkers, and the results were visualized using a heatmap with the ‘ggplot2’ 3.4.4 R package (http://groups.google.com/group/ggplot2). To investigate interactions between biomarkers and other functionally related genes, a co-expression network was constructed. Gene function predictions were made through GeneMANIA, based on this network analysis (http://www.genemania.org/). The Human Protein Atlas (HPA) database (https://www.proteinatlas.org/) was used to assess the expression of biomarkers across various tissues. To explore the relationships between biomarkers and CAD-related diseases, the DisGeNET database (https://www.disgenet.org/) was queried, focusing on the top 10 diseases filtered by Score_gda to predict biomarkers associated with CAD.
Immune infiltration analysis
The proportions of 28 immune cell types in all samples of the GSE42148 dataset were calculated using the ‘GSEABase’ 1.62.0 R package (DOI: https://doi.org/10.18129/B9.bioc.GSEABase) and the immune cell mmc3.gmt gene set. Immune cell correlations across all samples were analyzed using Spearman’s correlation method through the ‘psych’ 2.3.12 R package (https://personality-project.org/r/psych/). The abundance of immune cell infiltration between disease and normal control samples in the training dataset was compared using the Wilcoxon test. Immune cells exhibiting significant differences (P < 0.05) between disease and normal control samples were identified. The correlation between these differential immune cells and biomarkers was then assessed using Spearman’s correlation method.
Molecular regulatory network
Transcription factors (TFs) targeting biomarkers were retrieved from the JASPAR database within NetworkAnalyst (https://www.networkanalyst.ca/), and regulatory networks of TFs and biomarkers were subsequently constructed. Using the NetworkAnalyst platform (https://www.networkanalyst.ca/), microRNAs (miRNAs) regulating biomarkers were predicted through the TarBase 8.0 (http://mirtarbase.mbc.nctu.edu.tw/) and ENCORI (https://rnasysu.com/encori/) databases. Predictions from both databases were compared to identify miRNAs targeting the biomarkers. The miRNet and ENCORI databases were used to predict lncRNAs targeted by these miRNAs, and the intersection of results from both databases was analyzed to identify key lncRNAs.
Drug prediction and molecular docking
Biomarker-targeted drugs were identified using the DGIdb database. The predicted drugs were ranked according to their prediction scores in DGIdb, and the top 10 drugs (or all drugs sharing the highest score) were selected. The protein structures of biomarkers were obtained from the Protein Data Bank (PDB) (https://www.rcsb.org/). Small molecules and water were removed, followed by hydrogenation and charge calculations using AutoDock Tools (version 1.5.6)18. The structures of the top-scoring drugs (or randomly selected from ties in scores) were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and evaluated for charge balance and rotatable bonds using AutoDock Tools. The docking boxes were placed based on the receptor’s active site location, and the structure with the lowest binding free energy (indicating the highest binding affinity) was selected.
Single-cell analysis
Based on the dataset GSE121893, single-cell analysis was performed using the ‘Seurat’ 5.0.1 R package19. A Seurat object was generated for each sample with the parameters min.cells = 3 and min.features = 200. The samples were then integrated and filtered using the subset function, with data exclusion criteria including nCount_RNA < 30,000, nFeature_RNA > 200, and nFeature_RNA < 8,000. The filtered data were normalized using the LogNormalize method. To isolate genes with high variability, normalization was carried out via the NormalizeData function in the ‘Seurat’ R package. This procedure involved scaling the expression level of each gene in a cell by the cell’s total gene expression, multiplying by 10,000, followed by a logarithmic transformation to standardize the data. The FindVariableFeatures function was then applied to identify the top 2,000 genes with variable expression levels. Principal component analysis (PCA) was performed on these variable genes, and significant principal components were determined using the Elbowplot function. Unsupervised clustering of the refined cells was conducted with the FindClusters and FindNeighbors functions, using a resolution parameter of 0.5. To annotate cell types in the different clusters, relevant marker genes were retrieved as references9. Biomarker expression levels in different cell types were visualized using the DotPlot function. Additionally, the expression levels of biomarkers across various sample types were analyzed using the rank-sum test for both disease and control samples.
Animals and treatment
Eight-week-old Apolipoprotein E knock-out (ApoE–/–) mice, with an equal gender distribution (50% male and 50% female), were purchased from SLACCAS (Hunan, China). The mice were housed at 25 °C under standard light conditions. All animals were randomly divided into two groups: the experimental group was fed a high-fat diet (HFD) for 16 weeks, consisting of 78.85% regular mouse maintenance feed, 21% fat, and 0.15% cholesterol, to induce atherosclerosis. The control group was provided a standard diet. Animal care and experimental procedures adhered to ethical guidelines for animal welfare and were approved by the Ethics Committee of the Experimental Animal Center at Central South University (Hunan, China) (202408136). All the methods are performed in accordance with ARRIVE guidelines.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Peripheral blood was collected from mice, and 2 µg of total RNA was extracted using TRIZOL reagent (Invitrogen, CA, US). First-strand cDNA synthesis was performed following the manufacturer’s instructions (Takara Biotechnology, Shiga, Japan). qRT-PCR was conducted using the SYBR Green system (Takara Biotechnology). Primer sequences (Table 1) were synthesized by Sangon Biotechnology Co., Ltd. (Shanghai, China). mRNA levels were quantified using the 2−△△Ct method.
Statistical analysis
Statistical analysis was performed using R 4.2.2 software, and differences between groups were assessed using the Wilcoxon rank-sum test. A P-value of less than 0.05 was considered statistically significant.
Results
Identification of DE-SRGs
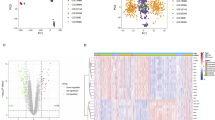
A total of 2,031 DEGs were identified between CAD and normal samples in the GSE42148 dataset, comprising 1,124 upregulated and 907 downregulated genes (Fig. 1a). The heatmap depicting gene expression of the top 20 upregulated and downregulated DEGs, sorted by log2FC, is shown in Fig. 1. Next, 12 DE-SRGs in CAD were identified by intersecting the DEGs with 187 SRGs (Fig. 1c). These DE-SRGs were significantly enriched across 63 GO processes (P < 0.05) (Fig. 1d). Notably, DE-SRGs were primarily enriched in the BP category, including protein minor ubiquitination, miRNA metabolic processes, and intracellular signaling pathways. In terms of CC, the DE-SRGs were predominantly enriched in nuclear speckles, PRC1 complexes, and PcG protein complexes. The MFs involved included SUMO ligase activity, nuclear retinoid X receptor binding, and ubiquitin-like protein binding. KEGG pathway analysis revealed four significantly enriched pathways (P < 0.05), specifically polycomb repressive complex, transcriptional dysregulation in cancer, aldosterone-regulated sodium reabsorption, and thyroid cancer (Fig. 1e).
Identification of DE-SRGs. (a) Distribution of DEGs in CAD and normal samples. (b) Heatmap of expression levels of the top 20 DEGs categorized by upregulation and downregulation. (c) Identification of DE-SRGs specific to CAD. (d) Enrichment of the identified DE-SRGs in biological processes, cellular components, and molecular functions via GO analysis. (e) Pathways enriched by DE-SRGs in KEGG pathway analysis.
SUMO1 and PPARG were identified as biomarkers
To investigate the protein interactions of DE-SRGs, a PPI network was constructed (Fig. 2a). Key modules playing significant roles in these interactions were identified, with one crucial module containing six genes: DDX17, SUMO1, PPARG, PHC2, CBX4, and DDX5 (Fig. 2b). These six genes were further analyzed to identify key genes using two machine learning algorithms within the GSE42148 dataset. Boruta analysis identified three candidate key genes (SUMO1, PPARG, and DDX5) (Fig. 2c), while the SVM-RFE algorithm yielded six candidate genes (DDX17, SUMO1, PPARG, PHC2, CBX4, and DDX5) (Fig. 2d). The intersection of key genes from both algorithms revealed three genes (SUMO1, PPARG, and DDX5) as the key genes in this study (Fig. 2e). Notably, SUMO1 and PPARG showed differential expression (P < 0.05) between CAD and control groups in both datasets, consistently exhibiting the same expression trends, with higher expression levels in disease samples compared to normal samples (Fig. 2f). To evaluate their diagnostic potential, ROC curves were plotted for SUMO1 and PPARG in both datasets, revealing AUC values greater than 0.7, thus identifying them as biomarkers (Fig. 2g).
Identification of SUMO1 and PPARG as biomarkers. (a) Construction of a PPI network for DE-SRGs. (b) Identification of important modules in the PPI network. (c) Boruta analysis identifying three candidate key genes (SUMO1, PPARG, and DDX5). (d) SVM-RFE algorithm identifying six candidate key genes (DDX17, SUMO1, PPARG, PHC2, CBX4, and DDX5). (e) Intersection of key genes identified by both algorithms, resulting in three key genes (SUMO1, PPARG, and DDX5). (f) Differential expression of SUMO1 and PPARG between CAD and control groups in two datasets. (g) ROC curves of SUMO1 and PPARG in the two datasets.
ANN analysis
An ANN model was constructed based on these biomarkers in the GSE42148 dataset (Fig. 3a). ROC curves of the ANN model were plotted in both the GSE42148 and GSE23561 datasets to assess model accuracy. AUC values greater than 0.7 in both datasets confirmed the reliability of the model (Fig. 3b). Generalization values presented in Fig. 3c-d were non-zero, indicating the nonlinear impact of biomarkers (SUMO1 and PPARG) on classification outcomes.
Artificial neural network analysis. (a) Architecture diagram of the ANN model based on biomarkers in the GSE42148 dataset. (b) ROC curves of the ANN model in the GSE42148 and GSE98583 datasets. (c-d) Graphs illustrating the nonlinear effects of biomarkers (SUMO1 and PPARG) on classification outcomes.
Functional enrichment analysis
Gene correlation analysis revealed a positive correlation between the expression levels of SUMO1 and PPARG, with a correlation coefficient of 0.76 and a P-value of 2.3e-05 (Fig. 4a). Co-expression network analysis via GeneMANIA identified several associated genes, including PIAS1, NR2F2, and UBE21, involved in functions such as protein ubiquitination, regulation of protein polymerization, and monocarboxylate transport (Fig. 4b). GSEA demonstrated distinct enrichment patterns: PPARG was enriched in ribosomes, olfactory transduction, lysosomes, neuroactive ligand-receptor interactions, and Fcγ R-mediated phagocytosis, while SUMO1 showed enrichment in olfactory transduction, ribosomes, neurotrophic signaling pathways, Leishmania infection, and chronic myeloid leukemia (Fig. 4c, d). According to the HPA database, PPARG exhibited the highest expression in adipose tissue, whereas SUMO1 was predominantly expressed in the liver (Fig. 4e).
Functional enrichment analysis. (a) Gene correlation analysis between SUMO1 and PPARG expression levels. (b) GeneMANIA co-expression network illustrating interactions among genes, such as PIAS1, NR2F2, and UBE21. (c-d) GSEA results highlighting enrichment pathways associated with PPARG and SUMO1. (e) Expression distribution map from the HPA database across various tissues.
Immune microenvironment analysis
Figure 5a presents immune cell infiltration across all 28 samples. Correlation analysis among immune cells revealed several significant relationships. Specifically, activated B cells and immature B cells exhibited a positive correlation, as did myeloid-derived suppressor cells (MDSCs) and central memory CD4+ T cells (Fig. 5b). The Wilcoxon rank-sum test revealed significant differences in central memory CD4+ T cells, eosinophils, mast cells, and memory B cells between the two groups (P < 0.05) (Fig. 5c). Further correlation analysis between differential immune cells and biomarkers revealed a positive correlation between PPARG and central memory CD4+ T cells and memory B cells (P < 0.05). SUMO1 exhibited a positive correlation with central memory CD4+ T cells, eosinophils, and memory B cells (P < 0.05) (Fig. 5d).
Immune microenvironment analysis. (a) Overview of immune cell infiltration across 28 samples. (b) Correlation analysis of immune cells in all samples. (c) Wilcoxon rank-sum test showing significant differences in central memory CD4+ T cells, eosinophils, mast cells, and memory B cells between groups (P < 0.05). (d) Correlation analysis between differential immune cells and biomarkers.
Regulatory network analysis
To elucidate the molecular regulatory mechanisms of biomarkers, TF-biomarker regulatory networks were established. A total of 25 TFs were predicted, with PPARG and SUMO1 sharing common TFs, including E2F1, SREBF1, and HINFP (Fig. 6a). A lncRNA-miRNA-mRNA regulatory network was constructed to further explore these mechanisms, consisting of 81 nodes (63 lncRNAs, 16 miRNAs, and 2 marker genes) and 177 edges (Fig. 6b). A biomarker-targeted drug interaction network was also created (Fig. 6c), involving 34 edges and 36 nodes. The drug with the highest score predicted by SUMO1 was CHEMBL588143 (N-(4-acetamidophenyl)-3-(2-hydroxy-4-methylphenyl)-1 H-pyrazole-5-carboxamide), while 23 drugs, including ELATOSIDE D and MBX-2044, were predicted by PPARG, with consistent scoring. CHEMBL588143 and CHEMBL483838 were selected for molecular docking targeting SUMO1, while ECHINATIN and ELATOSIDE D were selected for molecular docking targeting PPARG (Supplementary Table 1). The docking binding energies of these four drugs were all below − 5 kcal/mol. Notably, PPARG showed the highest affinity for ECHINATIN, with a docking binding energy of − 8.4 kcal/mol (Fig. 6d). Disease association analysis revealed a network with 33 edges and 35 nodes. SUMO1 predicted diseases such as hypodontia and oligodontia, while PPARG was associated with diseases such as obesity, familial partial lipodystrophy type 3, and non-insulin-dependent diabetes (Fig. 6e).
Regulatory network analysis. (a) Transcription factors associated with biomarkers and their regulatory network. (b) lncRNA-miRNA-mRNA regulatory network diagram. (c) Network diagram illustrating interactions between biomarkers and targeted drugs. (d) Results of drug molecule docking. (e) Disease association analysis chart.
Single-cell analysis
After ensuring the quality of single-cell sequencing data for CAD (Supplementary Fig. 1a), a total of 1,630 cells and 2,153 genes were obtained. From these, 2,000 highly variable genes were selected (Supplementary Fig. 1b). PCA was performed, and the first 30 principal components were selected for unsupervised clustering (Fig. 7a). Cells were then classified into 6 distinct clusters using the UMAP clustering approach (Fig. 7b). Indicative genes for sectors 0 and 3 included TTN, MYH7, MYH6, and TNNT2. Cluster 1 was characterized by marker genes such as VWF, IFI27, and PECAM1. Cluster 2 was marked by genes including DCN, C7, LUM, FBLN1, and COL1A2. Genes in cluster 5 included PTPRC, CD163, CCL4, and CXCL8, while cluster 4 featured ACTA2, CALD1, and MYH11 (Fig. 7c). The final annotation revealed five cell types: macrophages, fibroblasts, endothelial cells, smooth muscle cells, and cardiomyocytes (Fig. 7d). Furthermore, SUMO1 and PPARG were predominantly expressed in endothelial cells in the disease group. In cardiomyocytes, SUMO1 exhibited differential expression between the disease and control groups, with higher expression in the control group (Fig. 7e–g).
Single-cell analysis. (a) PCA results. (b) UMAP clustering plot categorizing cells. (c) Indicative genes in each cluster. (d) Annotated depiction of the final five cell types: myocardial cells, endothelial cells, fibroblasts, smooth muscle cells, and macrophages. (e-g) Cell type-specific expression of SUMO1 and PPARG in the disease and control groups.
Verification of the results
Six genes (DDX17, SUMO1, PPARG, PHC2, CBX4, and DDX5) were identified in MCODE for subsequent machine learning analysis. The expression levels of these genes in the peripheral blood of CAD mice were validated, showing elevated transcription levels for all six genes following CAD modeling (Fig. 8).
Discussion
This study employed a comprehensive approach, including transcriptome data analysis, machine learning, and PPI network analysis, to identify SUMO1 and PPARG as biomarkers associated with SUMOylation modifications in CAD development. These findings offer a novel perspective on CAD biomarker research and may establish a theoretical foundation for the development of new therapeutic strategies.
In this study, differential analysis of samples from the GSE42148 dataset identified 2,031 DEGs, which were subsequently intersected with SRGs to yield 12 DE-SRGs. Following MCODE gene screening and Boruta and SVM_REF analysis, three key genes—SUMO1, PPARG, and DDX5—were identified. Expression levels of SUMO1 and PPARG showed consistent trends across both datasets. Differential gene analysis and machine learning techniques emphasized the potential involvement of SUMO1 and PPARG in CAD pathogenesis. Elevated SUMO1 expression was closely associated with the onset of heart failure, while PPARG was implicated in metabolic regulation, particularly in lipid metabolism and insulin sensitivity. These findings suggest that SUMOylation modifications may drive CAD progression by altering cardiovascular metabolic pathways. SUMO1 exerts its effects primarily through interactions with signaling molecules in Wnt and TGF-β pathways, playing a pivotal role in cell proliferation, migration, and transformation, thereby influencing cell cycle regulation and apoptosis. These interactions are also linked to tumorigenesis20,21,22. Previous studies have demonstrated that SUMO1 upregulation enhances protein stability and activity, maintains cellular homeostasis, and regulates processes such as the cell cycle and apoptosis23. While there is no consensus on SUMO1 expression levels in patients with CAD, existing studies suggest that CAD may influence SUMO1 expression; however, the underlying mechanisms remain unclear24,25.However, SUMO1 plays a critical role in various diseases, including cancer, immune disorders, and neurodegenerative diseases. In cancer, SUMO1 regulates the cell cycle and DNA repair, promoting tumorigenesis and drug resistance. By activating the NF-κB signaling pathway, it facilitates the formation of a tumor-inflammatory microenvironment, thereby accelerating tumor progression26,27. Simultaneously, SUMO1 helps tumors evade immune surveillance by inhibiting tumor antigen presentation and the expression of immune checkpoint molecules. Inhibiting SUMO1 can enhance T-cell activity28. Additionally, SUMO1 is closely associated with immune infiltration and prognosis, potentially serving as a prognostic biomarker. In neurodegenerative diseases, SUMO1 affects neuronal survival by modulating protein stability, and its abnormal expression may serve as an early diagnostic marker for Alzheimer’s disease and progressive supranuclear palsy29,30. Targeting SUMO1 offers a potential strategy for treating a variety of diseases31. PPARG, a nuclear receptor transcription factor, primarily functions by binding to ligands, forming PPARG-ligand complexes that enter the nucleus, bind target genes, and regulate their transcriptional activity, thereby affecting cell function. Additionally, PPARG participates in signal transduction by forming heterodimers with other nuclear receptors, such as RXR, and regulates lipid metabolism, glucose metabolism, angiogenesis, inflammation, and other physiological processes32,33. Additionally, PPARG expression is modulated in patients with CAD34,35. Moreover, PPARG holds significant clinical importance in regulating blood lipids, adiponectin, and insulin sensitivity, particularly in individuals with increased cardiometabolic risk. The interaction between dietary fatty acid choices and PPARG genotypes may provide a basis for personalized dietary interventions and the management of metabolic diseases36.
SUMO1 may regulate PPARG activity through SUMOylation, thereby influencing its function in CAD. For example, SUMO1 may enhance PPARG stability through SUMOylation, promoting its regulatory role in lipid metabolism and inflammation24. This interaction may affect coronary artery lesions by modulating lipid metabolism pathways, immune responses, or inflammatory processes. Additionally, other SUMOylation-related genes, such as DDX5 and DDX17, may interact with SUMO1 and PPARG through the SUMOylation pathway, collectively regulating CAD progression. SUMOylation may also influence cardiovascular diseases by regulating key factors like PPARG, NF-κB, and p53. Enhancing PPARG stability through SUMOylation could mitigate atherosclerosis by promoting anti-inflammatory and lipid-lowering effects6. Future studies should further explore the interactions among these genes and their specific mechanisms in CAD, as well as develop therapeutic strategies targeting SUMO1-PPARG interactions.
GSEA revealed the enrichment of SUMO1 and PPARG in several biological processes, including ribosome function, olfactory conduction, and lysosome activity, reflecting the intricate pathophysiology of CAD. Notably, the involvement of these pathways suggests that CAD development is not solely a consequence of vascular disease but also encompasses extensive alterations in cellular metabolism, immune response, and signal transduction. Prior research has identified various mechanisms underlying CAD, such as vascular endothelial dysfunction, inflammation, lipid metabolism disturbances, oxidative stress, and the proliferation and migration of smooth muscle cells25,37,38,39,40. These mechanisms interact to drive the formation and progression of atherosclerotic plaques, ultimately leading to myocardial infarction or ischemia. Studies have shown that SUMO1 expression is diminished in failing hearts, and transferring the SUMO1 gene significantly improved cardiac function in a mouse model of pressure overload-induced heart failure7.
In the present study, immune infiltration analysis emphasized the role of immune cells in CAD, highlighting dynamic changes in different immune cell populations under both normal and disease conditions. SUMO1 and PPARG were found to be correlated with several differential immune cells, suggesting that immunomodulation may play a significant role in CAD development and providing a solid foundation for future studies targeting immune pathways. Previous research has established that immune regulation is pivotal in CAD progression41. The immune system contributes to coronary artery atherosclerosis through various cell types and molecular mechanisms, promoting both inflammation and anti-inflammatory repair processes. For instance, blood monocytes can infiltrate the arterial wall and differentiate into macrophages, which engulf oxidized low-density lipoprotein (oxLDL) to form foam cells, contributing to plaque formation and growth. Macrophages also secrete pro-inflammatory cytokines that perpetuate and intensify the inflammatory response42. Certain T cell subsets, including Th1 and CD8 + cells, contribute to arterial inflammation, further activating macrophages and promoting cytotoxicity through cytokine release, such as interferon-gamma (IFN-γ)43. B cells can produce antibodies, some of which may target self-lipoproteins, influencing plaque stability and growth. However, B cells may also play a protective role by producing antibodies against oxLDL, promoting immune clearance44. Neutrophils participate in plaque formation and instability in conjunction with CD4 + T cells, accelerating LDL oxidation through the secretion of enzymes like myeloperoxidase. Additionally, the release of free radicals and enzymes from neutrophils exacerbates cellular and matrix damage45. Peripheral blood serves as a crucial subject for studying CAD, providing key insights into the mechanisms of CAD, plaque progression, and thrombus formation, such as LDL levels, inflammatory factors, and immune cells46. Transcriptomic analysis of peripheral blood in patients with CAD has revealed gene modules associated with atherosclerotic plaque formation (e.g., oxidative stress and lipid metabolism), which are highly consistent with the pathological processes at disease sites47. Peripheral blood gene expression profiles can provide insights into the molecular changes at disease sites, such as coronary arteries, particularly in the context of immune cell infiltration and inflammatory responses5. For instance, differential gene expression analysis of peripheral blood (e.g., weighted gene co-expression network analysis, WGCNA) can identify hub genes associated with disease staging or prognosis. In patients with myocardial infarction (MI), peripheral blood gene modules correlate with the extent of myocardial injury and repair processes, and certain genes (e.g., inflammatory cytokine-encoding genes) may serve as potential biomarkers for predicting the progression of local lesions48. Future studies should validate these findings using multi-tissue samples to further elucidate the molecular mechanisms underlying CAD and other related conditions.
By constructing the TF_mRNA and mRNA-miRNA-lncRNA networks, this study thoroughly explored the molecular regulatory mechanisms of SUMO1 and PPARG. These networks not only unveiled the complex regulatory relationships governing gene expression but also identified potential targets for future research, particularly in the context of CAD therapy. Understanding the molecular regulation of CAD biomarkers is crucial for elucidating its pathophysiological processes and developing novel therapeutic strategies. CAD biomarkers are primarily linked to inflammatory factors, lipid metabolism, cellular damage, and repair-related molecules. In patients with CAD, the expression levels of inflammatory markers such as C-reactive protein (CRP) and interleukin-6 (IL-6) are elevated, which contribute to the progression of CAD49. LDL, oxLDL, plays a significant role in CAD development50. Molecules released during myocardial injury and repair, such as cardiac troponins and brain natriuretic peptide (BNP), have been extensively studied as CAD biomarkers51. The functionality of high-density lipoprotein (HDL) in patients with CAD is influenced by the expression of atherosclerotic protective genes (S100A8) and atherogenic genes (SREBF1, LCAT, PRKACG, PRKACB, and PRKCQ) in relation to HDL cholesterol levels14,52.
Drug prediction and molecular docking are key strategies for exploring new treatment approaches. The present study highlights the potential of drug interventions targeting SUMO1 and PPARG, offering new avenues for CAD treatment. Cardiomyocyte death is a critical factor in myocardial infarction resulting from CAD. Molecular docking techniques enable the identification of drugs that can protect cardiomyocytes and mitigate myocardial injury, such as small molecules that activate the PI3K/Akt signaling pathway, which is essential for cell survival53. Preventing thrombosis is essential for CAD treatment and prevention. Molecular docking can identify compounds that target platelet ADP receptors or coagulation factors, potentially enhancing antiplatelet and anticoagulant effects while reducing the risk of bleeding54. However, while these techniques provide valuable tools for drug prediction and screening, the efficacy and safety of the final drug candidates must be validated through laboratory studies and clinical trials.
In CAD research, disease association analysis of biomarkers is crucial for identifying disease risk, monitoring progression, and evaluating treatment responses. This study utilized the biomarkers SUMO1 and PPARG to explore new avenues for investigating CAD and its complications. While SUMO1 plays a role in the development of various tissues, its direct association with oral and maxillofacial malformation 10 (OFC10), individual tooth loss, or extensive tooth loss has been scarcely studied. In contrast, PPARG directly influences adipogenesis and fat metabolism by promoting the differentiation and maturation of preadipocytes into adipocytes, as well as regulating lipid storage and release in adipose tissue55. Specific mutations in the PPARG gene impair normal adipocyte differentiation, disrupt lipid metabolism, compromise adipocyte function, and indirectly affect insulin sensitivity32. The disease association analysis of these biomarkers provides a significant molecular foundation for the diagnosis, risk assessment, disease monitoring, and treatment of CAD. However, it is important to recognize that the expression levels of these biomarkers may vary across individuals and disease stages, highlighting the need to consider various factors in clinical applications.
In the present study, the expression levels of biomarkers in single-cell sequencing data were analyzed following data filtering and dimensionality reduction annotation. Differences in SUMO1 expression were observed in cardiomyocytes. Cardiomyocytes (clusters 0 and 3) were characterized by indicator genes such as TTN, MYH7, MYH6, and TNNT2, which are myocardial structural protein genes, and their abnormalities may impair heart contractility. Endothelial cells (cluster 1) were marked by VWF, IFI27, and PECAM1, highlighting their role in vascular endothelial injury and repair. Fibroblasts (cluster 2), characterized by DCN, C7, LUM, FBLN1, and COL1A2, are involved in collagen synthesis and tissue repair, contributing to vascular wall fibrosis. Macrophages (cluster 5), marked by immune- and inflammation-related genes (PTPRC, CD163, CCL4, and CXCL8), play a significant role in plaque formation and progression in CAD. Smooth muscle cells (cluster 4) are characterized by ACTA2, CALD1, and MYH11, participating in vascular wall tension regulation and plaque stability52,56. The elevated expression of SUMO1 in cardiomyocytes in the control group may be linked to the normal function and stress response of these cells, as SUMOylation significantly impacts the stability and function of various proteins5. Consequently, the downregulation of SUMO1 expression in the disease group could lead to disturbances in cardiac protein modifications, affecting cardiomyocyte function and survival. In the disease group, PPARG is primarily expressed in endothelial cells. As a nuclear receptor, PPARG regulates lipid metabolism, inflammatory responses, and insulin sensitivity32. Variations in PPARG expression may indicate altered endothelial cell function and involvement in the inflammation and atherosclerosis processes that characterize CAD. These changes in the expression levels of SUMO1 and PPARG may reflect the stress response and dysfunction of cardiomyocytes and endothelial cells in CAD. As biomarkers, their expression levels could serve as valuable indicators for early diagnosis and monitoring disease progression in CAD. Furthermore, the targeted regulation of their expression or function through pharmacological interventions could offer a novel strategy for CAD treatment.
The findings of this study provide valuable insights into the molecular mechanisms of CAD and may aid in the development of new therapeutic strategies. However, while these biomarkers and pathways show statistical significance, their universality and relevance across different populations and disease stages require further investigation. Therefore, additional large-scale and in-depth mechanistic studies are essential to validate and expand upon these preliminary findings. Moreover, the potential clinical applications of these biomarkers, especially in disease monitoring, prognostic evaluation, and therapeutic interventions, warrant further exploration and discussion.
Conclusions
This study identified two genes, SUMO1 and PPARG, as SUMOylation-related biomarkers in CAD, offering a novel perspective for the treatment of patients with CAD.
Data availability
The datasets used and analysed during the current study available from the corresponding author on reasonable request.
References
Mortality, G. B. D. Causes of death C. global, regional, and National life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the global burden of disease study 2015. Lancet 388 (10053), 1459–1544 (2016).
The, W. Report on cardiovascular health and diseases in China 2022: an updated summary. Biomed. Environ. Sci. 36 (8), 669–701 (2023).
Cassar, A. et al. Chronic coronary artery disease: diagnosis and management. Mayo Clin. Proc. 84 (12), 1130–1146 (2009).
Knuuti, J. et al. 2019 ESC guidelines for the diagnosis and management of chronic coronary syndromes. Eur. Heart J. 41 (3), 407–477 (2020).
Feng, X. et al. Identification of diagnostic biomarkers and therapeutic targets in peripheral immune landscape from coronary artery disease. J. Transl Med. 20 (1), 399 (2022).
Chang, H. M., Yeh, E. T. H. & Sumo From bench to bedside. Physiol. Rev. 100 (4), 1599–1619 (2020).
Tilemann, L. et al. SUMO-1 gene transfer improves cardiac function in a large-animal model of heart failure. Sci. Transl Med. 5 (211), 211–159 (2013).
Wang, C. et al. MMP9, CXCR1, TLR6, and MPO participant in the progression of coronary artery disease. J. Cell. Physiol. 235 (11), 8283–8292 (2020).
Wang, L. et al. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat. Cell. Biol. 22 (1), 108–119 (2020).
Ritchie, M. E. & Phipson, B. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47 (2015).
Mullan, K. A. et al. GgVolcanoR: A Shiny app for customizable visualization of differential expression datasets. Comput. Struct. Biotechnol. J. 19, 5735–5740 (2021).
Gu, Z. et al. Circlize implements enhances circular visualization R. Bioinformatics 30 (19), 2811–2812 (2014).
Gao, C. H., Yu, G. & Cai, P. ggVennDiagram: An intuitive, easy-to-use, and highly customizable R package to generate Venn diagram. Front. Genet. 12, 706907 (2021).
Wu, T. et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. (Camb). 2 (3), 100141 (2021).
Ito, K. & Murphy, D. Application of ggplot2 to pharmacometric graphics. CPT Pharmacometrics Syst. Pharmacol. 2 (10), e79 (2013).
Beck, M. W. & NeuralNetTools Visualization and analysis tools for neural networks. J. Stat. Softw. 85 (11), 1–20 (2018).
Robin, X. et al. pROC: an open-source package for R and S + to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
Goodsell, D. S. et al. The AutoDock suite at 30. Protein Sci. 30 (1), 31–43 (2021).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42 (2), 293–304 (2024).
Sun, J. et al. LCox: a tool for selecting genes related to survival outcomes using longitudinal gene expression data. Stat. Appl. Genet. Mol. Biol. 18 (2) (2019).
Baatarkhuu, O. et al. Acute hepatitis A, B and C but not D is still prevalent in Mongolia: a time trend analysis. Clin. Mol. Hepatol. 23 (2), 147–153 (2017).
Marshall, J. B. & Berger, W. L. End-expiratory pressure best approximates intrinsic lower esophageal sphincter pressure. Report of a patient with Cheyne-Stokes respiration. Dig. Dis. Sci. 35 (2), 267–270 (1990).
Da Costa, I. C. & Schmidt, C. K. Ubiquitin-like proteins in the DNA damage response: the next generation. Essays Biochem. 64 (5), 737–752 (2020).
Liu, Y. Z. et al. SUMOylation in atherosclerosis. Clin. Chim. Acta. 508, 228–233 (2020).
Libby, P., Ridker, P. M. & Maseri, A. Inflammation and atherosclerosis. Circulation 105 (9), 1135–1143 (2002).
Geiss-Friedlander, R. & Melchior, F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell. Biol. 8 (12), 947–956 (2007).
Takamura, H. et al. SUMO1 modification of Tau in progressive supranuclear palsy. Mol. Neurobiol. 59 (7), 4419–4435 (2022).
Imbert, F. & Langford, D. Comprehensive SUMO proteomic analyses identify HIV latency-associated proteins in microglia. Cells 14 (3), 1 (2025).
Wilkinson, K. A. & Henley, J. M. Mechanisms, regulation and consequences of protein sumoylation. Biochem. J. 428 (2), 133–145 (2010).
Mandel, N. & Agarwal, N. Role of sumoylation in neurodegenerative diseases. Cells 11 (21), 1 (2022).
Chen, Y. A new immuno-oncology target—SUMOylation. Trends Cancer. 9 (8), 606–608 (2023).
Tontonoz, P. & Spiegelman, B. M. Fat and beyond: the diverse biology of PPARgamma. Annu. Rev. Biochem. 77, 289–312 (2008).
Kintscher, U. et al. Peroxisome proliferator-activated receptor and retinoid X receptor ligands inhibit monocyte chemotactic protein-1-directed migration of monocytes. Eur. J. Pharmacol. 401 (3), 259–270 (2000).
Lind, C. M. et al. The relationship between plasma steroid hormone concentrations and the reproductive cycle in the Northern Pacific rattlesnake, Crotalus oreganus. Gen. Comp. Endocrinol. 166 (3), 590–599 (2010).
Erman, B. A fast approximate method of identifying paths of allosteric communication in proteins. Proteins 81 (7), 1097–1101 (2013).
Alsaleh, A., Sanders, T. A. & O’Dell, S. D. Effect of interaction between PPARG, PPARA and ADIPOQ gene variants and dietary fatty acids on plasma lipid profile and adiponectin concentration in a large intervention study. Proc. Nutr. Soc. 71 (1), 141–153 (2012).
Hartge, M. M., Unger, T. & Kintscher, U. The endothelium and vascular inflammation in diabetes. Diab Vasc Dis. Res. 4 (2), 84–88 (2007).
Glass, C. K. & Witztum, J. L. Atherosclerosis, the road ahead. Cell 104 (4), 503–516 (2001).
Higashi, Y. et al. Endothelial function and oxidative stress in cardiovascular diseases. Circ. J. 73 (3), 411–418 (2009).
Newby, A. C. Metalloproteinase expression in monocytes and macrophages and its relationship to atherosclerotic plaque instability. Arterioscler. Thromb. Vasc Biol. 28 (12), 2108–2114 (2008).
Ridker, P. M. & Luscher, T. F. Anti-inflammatory therapies for cardiovascular disease. Eur. Heart J. 35 (27), 1782–1791 (2014).
Moore, K. J. & Tabas, I. Macrophages in the pathogenesis of atherosclerosis. Cell 145 (3), 341–355 (2011).
Hansson, G. K. & Hermansson, A. The immune system in atherosclerosis. Nat. Immunol. 12 (3), 204–212 (2011).
Kyaw, T. et al. Conventional B2 B cell depletion ameliorates whereas its adoptive transfer aggravates atherosclerosis. J. Immunol. 185 (7), 4410–4419 (2010).
Drechsler, M. et al. Hyperlipidemia-triggered neutrophilia promotes early atherosclerosis. Circulation 122 (18), 1837–1845 (2010).
Bentzon, J. F. et al. Mechanisms of plaque formation and rupture. Circ. Res. 114 (12), 1852–1866 (2014).
Gambarino, S. et al. Characteristics of RNA stabilizer RNApro for peripheral blood collection. Diagnostics (Basel). 14 (10), 1 (2024).
Yang, X. et al. Identification of PFKFB2 as a key gene for the transition from acute to old myocardial infarction in peripheral blood. Front. Cardiovasc. Med. 9, 993579 (2022).
Ridker, P. M. Clinical application of C-reactive protein for cardiovascular disease detection and prevention. Circulation 107 (3), 363–369 (2003).
Steinberg, D. The LDL modification hypothesis of atherogenesis: an update. J. Lipid Res. 50 (Suppl), S376–S381 (2009).
Thygesen, K. et al. Third universal definition of myocardial infarction. J. Am. Coll. Cardiol. 60 (16), 1581–1598 (2012).
Dergunov, A. D. et al. Differential expression of subsets of genes related to HDL metabolism and atherogenesis in the peripheral blood in coronary artery disease. Curr. Issues Mol. Biol. 45 (8), 6823–6841 (2023).
Manning, B. D. & Toker, A. AKT/PKB signaling: navigating the network. Cell 169 (3), 381–405 (2017).
Gresele, P., Momi, S. & Falcinelli, E. Anti-platelet therapy: phosphodiesterase inhibitors. Br. J. Clin. Pharmacol. 72 (4), 634–646 (2011).
He, W. et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. U S A. 100 (26), 15712–15717 (2003).
Mosquera, J. V. et al. Integrative single-cell meta-analysis reveals disease-relevant vascular cell States and markers in human atherosclerosis. Cell. Rep. 42 (11), 113380 (2023).
Author information
Authors and Affiliations
Contributions
XZ and FL analyzed and interpreted the datasets. XZ was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Animal care and experimental procedures were conducted in accordance with the ethical guidelines for experimental animal welfare and were approved by the Ethics Committee of the Experimental Animal Center at Central South University (Hunan, China) (202408136). All the methods are performed in accordance with ARRIVE guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, X., Luo, F., Xiang, B. et al. The working mechanism of biomarkers related to sumoylation modification in coronary artery disease. Sci Rep 15, 17055 (2025). https://doi.org/10.1038/s41598-025-02099-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-02099-4