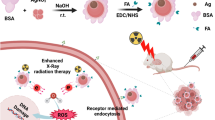
Abstract
Our goal in this study was to develop bismuth sulfide nanoparticles (NPs) that were functionalized with chitosan and incorporated with decoy oligodeoxynucleotides (ODNs) specifically targeting the NANOG transcription factor (designated as Bi@Chi-DEC NPs) in triple-negative breast cancer cells. FT-IR, UV–vis, FESEM, EDX, TEM, DLS, release kinetics, and hemolysis assays were done to validate the successful synthesis of Bi@Chi-DEC NPs. The synthesized spherical particles exhibited a size distribution averaging 213.8 nm, with a zeta potential measured at -3.27 mV. The anticancer properties of the synthesized nanoparticles, along with X-ray irradiation (2Gy), were assessed through a series of cellular assays, including MTT, cellular uptake, apoptosis, cell cycle analysis, scratch, and tumorsphere formation assays on MDA-MB-231 breast cancer cells. Treatment with the synthesized nanoparticles and X-irradiation resulted in a significant reduction in cell viability, tumorsphere formation, and cellular migration, while concurrently enhancing the rate of apoptotic cells and inducing cell cycle arrest at the G2/M phase. It can be inferred that Bi@Chi-DEC NPs possess the potential to serve as a therapeutic modality for cancer treatment, particularly when utilized along with radiation therapy. Further, in vivo studies are warranted to substantiate the efficacy of this therapeutic approach.
Similar content being viewed by others
Introduction
Breast cancer persists as one of the most widespread and formidable malignancies globally, necessitating the development of innovative therapeutic modalities to enhance patient prognoses.
Recent studies have indicated mutation incidence and overexpression of the NANOG transcription factor in breast cancer. NANOG, in addition to being a cancer stem cell (CSC) marker, can promote CSC-like characteristics in various cancers, and its binding to the promoter region of cyclin D1 can regulate cell cycle and proliferation1,2,3.
Recent progressions in the realms of nanotechnology and gene therapy have unveiled novel pathways for oncological treatment, particularly in augmenting the efficacy of radiotherapeutic interventions4. Two promising strategies that have emerged are the application of bismuth (Bi2S3) nanoparticles as radiosensitizing agents and the utilization of decoy oligodeoxynucleotides (ODNs) aimed at stemness-associated gene regulation5,6,7.
The application of the decoy ODNs strategy in the clinic is accompanied by challenges such as low cellular uptake of oligos (if used naked) and degradation by cellular nucleases. In recent studies, various nanocarriers have been used to overcome the drawback of low cellular uptake, which has been largely successful. In addition, to overcome the problem of degradation of decoy ODNs, various structural modifications (i.e., phosphorothioated modification, locked nucleic acids substitutions, ribbon type structure) have been made to the structure of oligos7,8,9. The use of various nanostructures containing decoy ODNs as a drug delivery tool has been demonstrated to be a successful remedy for several types of cancer, and it has a synergistic effect when combined with other therapies9,10.
Radiotherapy, while an integral component of oncological treatment regimens, frequently encounters constraints due to radioresistance and the potential for collateral damage to non-cancerous tissues11. The enhancement of radiosensitivity in neoplastic cells, while concurrently minimizing adverse effects, is imperative for the advancement of therapeutic outcomes. Furthermore, the targeting of cancer stem cells, which are postulated to contribute to tumor recurrence and metastasis, offers a promising strategy to surmount treatment resistance and hinder disease advancement12. Notwithstanding the promise exhibited by Bi2S3 nanoparticles (BiNPs) and decoy ODNs as individual therapeutic modalities, their integrated application in the treatment of breast cancer remains predominantly underexplored. Prior investigations have illuminated the radiosensitizing properties of BiNPs across various cancers, while decoy ODNs have demonstrated potential in downregulating genes associated with stemness. Nevertheless, the synergistic efficacy of these two strategies in amplifying X-ray-induced cytotoxicity in breast cancer cells, particularly through the targeting of the NANOG transcription factor, signifies a substantial lacuna in the extant scholarly literature. Our previous studies demonstrated other potential applications of the current combinational therapy strategy using various NPs containing decoy ODNs along with X-irradiation on different cancer cell lines such as colon (HT29)13, cancer stem-like cells (NTERA-2)14, glioblastoma multiforme (U87)15, Prostate (LNCaP)10.
This study endeavors to bridge this knowledge deficit by exploring a novel therapeutic strategy that amalgamates BiNPs and NANOG-decoy ODNs to enhance X-ray-induced cytotoxicity in breast cancer cells. The innovation resides in the convergence of these two disparate approaches; leveraging BiNPs to bolster radiosensitivity and incorporating NANOG-decoy ODNs to specifically target cancer stem-like cells. By concurrently addressing the challenges of radioresistance and stemness, this integrative strategy possesses the potential to markedly augment the efficacy of radiotherapeutic interventions in breast cancer.
The structure of the article is delineated as follows: Initially, we elucidate the synthesis and characterization of BiNPs and NANOG-decoy ODNs. This encompasses a detailed exposition of the methodologies employed to fabricate and validate the quality and functionality of these therapeutic agents. Subsequently, we assess their individual and synergistic impacts on MDA-MB-231 cells, with a particular emphasis on radiosensitization and combined therapeutic effects (Fig. 1).
Materials and methods
Materials
Phosphate buffer saline (PBS), 3-(4,5-dimethylthialzol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and Dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (MO, USA). Fetal bovine serum (FBS), Dulbecco’s modified Eagle’s medium (DMEM) high glucose, and penicillin/streptomycin were obtained from Gibco, Life Technologies (NY, USA). Bi (NO3)3, Bovine Serum Albumin (BSA), Sodium hydroxide (NaOH), Chitosan, and Acetic acid were purchased from Merck (Darmstadt, Germany). MDA-MB-231 cell line (IBRC C11325) was provided by the Cell Bank of Pasteur Institute of Iran (Tehran, Iran).
Design and synthesis of ODNs
Phosphorothioate (PS) modified 18-mer NANOG decoy (DEC) and as a negative control, scrambled (SCR) ODNs were designed according to the promoter region of the human Sox2 gene16 and synthesized by Bioneer Inc. Lyophilized ODNs were prepared as described previously17. Forward and reverse strands of DEC and SCR oligos were dissolved in sterile TE buffer solution, annealed at 95 °C for 10 min, and then cooled at room temperature to obtain double-stranded ODNs. A NanoDrop spectrophotometer was used to quantify each ODN. Cy3 fluorescent dye was added to the 3′ end of the ODNs in order to track them within the cells. The core-binding site (boldface), PS alterations at 3′ and 5′ (asterisk), and three mutations (italics/underlined) are displayed in the ODN sequences:
-
NANOG decoy ODN sequence,
-
F 5′-G*GCCTTTGTCATGCTGA*C-3′
-
R 5′-C*CGGAAACTGTACGACT*G-3′
-
NANOG scramble ODN sequence,
-
F 5′-G*GCATGGGTCATGCTGA*C-3′
-
R 5′- C*CGTACCCAGTACGACT*G-3′
Preparation of nanostructures
The environmentally friendly synthesis of Bismuth nanoparticles (BiNPs) was executed utilizing a modified variant of a previously documented methodology, which is fundamentally based on a redox reaction involving Bismuth (III) nitrate and bovine serum albumin (BSA), with the latter acting as the reducing agent. Initially, BSA was prepared at a concentration of 10% w/v in 10 mL of PBS at pH 7.4, to which 0.4% w/v of Bismuth (III) nitrate was incorporated. This resultant mixture was subsequently subjected to vigorous agitation at ambient temperature for 10 min to ensure the attainment of a homogenous solution. Following this, the pH was meticulously adjusted to 8.5 through the incremental addition of 1 N NaOH. After that, the reaction mixture was kept at 90 °C for an hour while being constantly stirred. The successful synthesis of BiNPs was shown by the noticeable color change of the colloidal solution from colorless to black at the end of this procedural process. The colloidal solution was centrifuged at 1000 rpm for five minutes in order to separate any undissolved BSA and purify the produced nanoparticles. The BiNP-containing supernatant was subsequently subjected to dialysis using a 12-kDa molecular weight cut-off membrane against ultrapure water for a whole night18. The removal of any unreacted silver nitrate and related byproducts required this specific process. A pure colloidal solution of BiNPs produced using an environmentally friendly process consists of the final product.
To prepare chitosan-coated bismuth nanoparticles (Bi@Chi NPs) containing ODNs (Bi@Chi-DEC NPs and Bi@Chi-SCR NPs), a 1% w/v chitosan solution was made by dissolving chitosan in distilled water containing 1% acetic acid, with or without 100 µM ODNs (DEC or SCR), and the temperature was adjusted to 70 °C. Subsequently, the chitosan solution was added dropwise to colloidal bismuth nanoparticles and the mixture was stirred continuously at 60 °C for 24 h to obtain final formulation19.
Characterization of synthesized nanostructure
Fourier transform infrared (FT-IR) & ultraviolet–visible (UV–vis) spectroscopy
To provide insight into the chemical structure of the produced materials and investigate potential interactions between the drug molecules and the nanoparticle carriers, Fourier transform infrared (FT-IR) spectroscopy studies were performed. A Bruker Tensor 27 spectrometer, based in Germany, was used for the analysis. It can scan in the mid-infrared spectrum between 4000 and 400 cm−1. When it comes to recognizing chemical interactions and functional groups, this spectral range is particularly valuable. The materials were carefully prepared for the analysis by dispersing them in a matrix of potassium bromide (KBr). Because of its inertness and transparency in the mid-infrared spectrum, which guarantees that it will not hinder the identification of the sample of distinctive absorption bands, KBr is the preferred matrix for these kinds of applications. In order to create a consistent and transparent surface for infrared analysis, the sample-KBr mixture was further compacted into a disc20. In order to ascertain the existence of particular functional groups, bonding patterns, and possible interactions between the BiNPs, ODNs, and the chitosan coating, the resulting spectra were captured and analyzed. Additionally, a Shimadzu UV-160 Spectrophotometer (Japan) was used to evaluate the optical characteristics of the nanostructures using UV–vis spectroscopy21.
Morphology analysis by FESEM & TEM
The morphological characteristics and particle dimensions of the synthesized bismuth nanoparticles were meticulously examined utilizing field emission scanning electron microscopy (FESEM, MIRA3, TESCAN, Brno, Czech Republic). The specimen was subjected to a gold coating before analysis through the FE-SEM methodology. Furthermore, a transmission electron microscope (TEM) was utilized to ascertain the dimensions and morphology of the synthesized nanoparticles, employing a Philips EM208S 100 kV electron microscope22.
Determination of hydrodynamic average size and Zeta potential
The hydrodynamic average size, polydispersity index (PDI), and zeta potential of the nanoparticles integrated into the drug delivery system (DDS) were assessed using zeta dynamic light scattering (DLS) and hydrodynamic average size determination techniques. These factors are essential for evaluating the stability and uniformity of suspensions of nanoparticles, which in turn affects their biological interactions and drug-carrying efficacy. The DLS analyses were carried out using a Malvern Nano ZS device, a state-of-the-art device produced by Malvern Instruments, located in Worcestershire, UK, and renowned for its precision in nanoparticle characterization. 0.5 mL of the nanoparticle suspension was extracted and then diluted with 2 mL of deionized water to prepare the samples that were about to be measured10. To reduce the possibility of contamination that would skew the results, this dilution process was carried out inside a sterile Malvern sample vial. The DLS analysis was started after the dilution process was finished, providing thorough insights into the behavior of the suspended nanoparticles. In order to calculate the average hydrodynamic diameter—which includes the size of the nanoparticles as well as the solvent layer that is dynamically connected to their surface—the device measured the Brownian motion of the particles. Additionally, the PDI—a dimensionless indicator of the size distribution’s breadth within the nanoparticle sample—was determined. A more uniform size distribution is indicated by a low PDI, which is usually beneficial for therapeutic applications to guarantee reliable drug delivery and biodistribution. Ultimately, the zeta potential—a measure of the nanoparticles’ surface charge—was measured. Because it influences particle stability in suspension, this parameter is especially important. Higher zeta potentials (positive or negative) tend to reject one another, which lowers the possibility of aggregation.
In vitro drug release
The in vitro release profile of the drugs DEC and SCR from the formulations of Bi@Chi-DEC NPs and Bi@Chi-SCR NPs was executed utilizing a dialysis diffusion methodology within a PBS solution supplemented with 5% (v/v) ethanol. In addition, to replicate the drug release dynamics under both acidic and normal physiological conditions, the in vitro drug release experiments were conducted at acidic and neutral pH levels (specifically 4.8 and 7.4). In summary, a predetermined quantity of Bi@Chi-DEC NPs and Bi@Chi-SCR NPs was introduced into the PBS solution (30 ml containing 5% v/v ethanol) and subjected to incubation at 37 °C with gentle agitation at 100 rpm. Subsequently, at various designated time intervals, a specified volume (2 ml) of the external medium was extracted for spectrophotometric analysis before being returned to the medium to maintain a constant volume throughout the drug release assessment. The concentrations of DEC and SCR within the external medium were ascertained using a UV–visible spectrophotometer at a wavelength of 260 nm14.
Hemolysis study
Hemolysis test is used to check the blood biocompatibility of nanostructures. Informed consent was obtained from all individual participants included in the study. This test is performed by checking the amount of heme released in the plasma due to damage to the red blood cell membrane by nanoparticles. The higher the toxicity of the nanoparticle, the higher the amount of heme released from RBCs to the environment. The group of nanoparticles can be mixed with red blood cells and then the compatibility of the nanoparticles can be checked based on the amount of heme released in the solution and measured using spectrophotometry at a wavelength of 540 nm. Three repetitions of the experiment were conducted and the percentage of hemolysis was calculated as:
In this equation, A treated sample, A negative control, and A positive control are representative of the mean absorbance of the sample, negative control, and positive control, respectively.
Cellular uptake assay
The entry rate of nanoparticles into cancer cells (MDA-MB-231) was analyzed using nanostructures that contain oligodeoxynucleotides labeled with Cy3 fluorescent dye (Bi@Chi-labeled ODNs). In brief, the cells were seeded in a 24-well plate at a density of 3 × 104 cells and incubated at 37◦C in a 5% CO2 incubator. Once the cells reached 70–80% confluency, they were treated with Bi@Chi-labeled ODNs at different concentrations (25, 50, 100, 200, and 300 nM). Following 5 h of incubation, the treatment medium was removed and replaced with a fresh medium9. After 24 h of incubation, the cells were rinsed with PBS and followed by trypsinization, collection, and analysis by using flow cytometry (BD Biosciences, San Jose, CA), and FlowJo v7 software (Tree Star, Ashland, OR).
Cytotoxicity assay
The cytotoxicity effects of nanostructures on MDA-MB-231 cells, both with and without irradiation, were evaluated using the MTT test based on our previous study10. The 8 × 103 cells were seeded in 96-well plates and incubated at 37 °C for 24 h under a 5% CO2 atmosphere. Afterward, the cells were treated with different concentrations (100, 200, 300, and 400 nM) of BiNPs, Bi@Chi NPs, Bi@Chi-SCR NPs, Bi@Chi-DEC NPs, and cells with no treatment were regarded as a control. The treatment medium was eliminated after 6 h of treatment, and 500 μL of fresh medium was added to every well. The cells were then exposed to a 2 Gy fractionated X-ray and incubated for 24 h. After the cell culture medium was removed, the prepared MTT solution (5 mg/mL) was added and incubated for 4 h. Subsequently, 100 μL of DMSO was added to each well to dissolve the formed formazan crystal, and their absorbance was measured at 570 nm wavelength using a microplate (ELISA) reader. All previously mentioned steps were also performed without an X-ray exposure condition.
Cell cycle assay
Based on our previous investigations, flow cytometry was used to assess the effect of nanostructures on the cell cycle of MDA-MB-231 cells, both with and without X-irradiation17. Briefly, 5 × 105 cells were cultivated under conventional conditions after being seeded in a 12-well plate. Cells were treated with 300 nM of different nanostructures (BiNPs, Bi@Chi NPs, Bi@Chi-SCR NPs, and Bi@Chi-DEC NPs) once they had reached 70% confluence, and cells with no treatment were regarded as control. All cell groups had their medium removed after 6 h of treatment, and each well received 500 μL of fresh complete medium. Then, a 2 Gy fractionated X-ray was applied to the cell plate. The cells were collected after 24 h and centrifuged for three minutes at 1200 rpm to pellet them. The cell pellets were washed with 50 μL of PBS and fixed with cold 70% ethanol and subsequently, extra ethanol was removed by centrifugation. The cells were treated with 1 mL of PI Master MIX solution (950 μL of PBS, 40 μL of PI, and 10 μL of RNase) and incubated for 30 min at room temperature. Finally, the cells were analyzed using flow cytometry (BD Biosciences, San Jose, CA) and FlowJo v7 software (Tree Star, Ashland, OR) to determine the percentage of cell population in each phase of the cell cycle. All previously mentioned steps were also performed without an X-ray exposure condition.
Apoptosis assay
Based on our recent investigation, we used flow cytometry to assess the effect of nanostructures on the apoptotic rate of MDA-MB-231 cells, both with and without X-irradiation9. Briefly, a 6-well plate was seeded with cells at a density of 5 × 105, and the cells were then incubated until they achieved the proper confluency. The cells were then treated with 300 nM concentrations of BiNPs, Bi@Chi NPs, Bi@Chi-SCR NPs, and Bi@Chi-DEC NPs, whereas cells that received no treatment served as a control. All cell groups had their media withdrawn after 6 h of treatment, and 500 μL of fresh complete medium was added to each well. Each well was then subjected to a 2 Gy fractionated X-ray, and the cells were incubated for 24 h. Following the harvesting and washing process, the treated cells were first suspended in Annexin V binding buffer (100 μL) and then stained with Annexin V-FITC and propidium iodide before flow cytometry analysis. Finally, the cells were analyzed using flow cytometry (BD Biosciences, San Jose, CA) and FlowJo v7 software (Tree Star, Ashland, OR) to determine the apoptosis rate. All previously mentioned steps were also performed without an X-ray exposure condition.
Wound-healing (scratch) assay
A scratch test based on our previous studies was used to assess the effect of nanostructures on the migration inhibition rate of MDA-MB-231 cells, both with and without X-irradiation23. In brief, cells were seeded at a density of 5 × 105 cells/well in a 12-well plate and then incubated until they attained the proper confluency. Then, using a 200 µL sterile pipette tip, wounds were made on the cell layer. The cells were treated with 150 nM concentrations of BiNPs, Bi@Chi NPs, Bi@Chi-SCR NPs, and Bi@Chi-DEC NPs, and cells that received no treatment were considered a control. After 6 h of treatment, the medium was removed from all cell groups, and 500 μL of fresh complete medium was added to each well and subsequently, exposed to a 2 Gy fractionated X-ray. The scratched area was photographed at 0 h and 72 h for wound gap closure assessment. Finally, the migration inhibition rate percent of the cells was determined by ImageJ software (1.52a)17. All previously mentioned steps were also performed without an X-ray exposure condition.
Tumorsphere assay
A tumorsphere test based on our previous investigations was used to assess the effect of nanostructures on the capacity of MDA-MB-231 cells to form tumorspheres, both with and without X-irradiation13. Agar-coated 24-well (nonadherent) plates (1.8% agarose) and 2% serum-free stem cell harboring (SCM) DMEM containing fibroblast growth factor (Sigma, St. Louis, MO) and epidermal growth factor (EGF) (Sigma, Gaithersburg, MD) were used to seed the cells (100/well) for two weeks. After being treated with 0.05% trypsin/EDTA, the cells were centrifuged at 1000 rpm to create a single-cell suspension. Under standard conditions, cells were grown in serum-free agar-coated 12-well plates under treatment with 150 nM of BiNPs, Bi@Chi, Bi@Chi-SCR, and Bi@Chi-DEC nanostructures or DMEM medium as controls in nonadherent agar. Immediately after this step, the cells were exposed to fractionated X-rays with a 2 Gy dose. Tumorspheres were examined with a light microscope and followed for 8 days. All previously mentioned steps were also performed without an X-ray exposure condition. Finally, light microscopy and ImageJ software (1.52a) were employed to evaluate the size of the formed tumorsphere.
Statistical analysis
The data was analyzed and expressed as mean ± standard deviation using GraphPad Prism 6. Analysis of variance (ANOVA) was used both one-way and two-way to assess the significance of differences between experimental groups. At least three repetitions of each experiment were conducted. Whenever the p-value is *p < 0.05, **p < 0.01, ***p < 0.001, or ****p < 0.0001, the significance can be taken into account.
Results
The systemic interactions and molecular biological behavior of nanoparticles are significantly influenced by their physicochemical characteristics. In order to elucidate the physical and chemical attributes of nanoparticles—including surface charge, dimensionality, polydispersity index (PDI), morphology, the release kinetics of oligonucleotides (ODNs), and enhanced radiation absorption.
FT-IR & UV–vis spectroscopy reveals successful chitosan coating and drug loading in BiNPs
The FT-IR spectroscopy technique was employed to validate the successful synthesis of the nanoparticles. The outcomes of the FTIR spectroscopy are illustrated in Fig. 2, which displays the individual surfactants alongside their corresponding nanoparticles. The binding of chitosan (Chi) to the BiNPs was substantiated by a comparative analysis of the FT-IR spectra for BiNPs, the chitosan substrate, and the Bi@Chi NPs (Fig. 2A). The emergence of peaks at 1651 cm-1 and 1576 cm-1, associated with amide and primary amine linkages in the FT-IR spectrum of BiNPs, signifies the presence of bovine serum albumin (BSA) on the nanoparticles. The peak observed at 3400 cm-1 across all three nanostructures and substrates is attributable to the hydroxyl functional group (O–H). Peaks detected at 2908 cm-1 and 2865 cm-1 in the FT-IR spectrum correspond to chitosan, indicating the presence of C−H and C−C bonds, respectively. Upon the incorporation of chitosan into BiNPs, alterations in the 1651 cm-1 and 1576 cm-1 peaks were noted. Furthermore, the peaks at 2908 cm-1 and 2865 cm-1 were absent in the Bi@Chi nanostructure, attributed to interactions between the amine and hydroxyl groups, which supports the successful formation of Bi@Chi nanoparticles. Additionally, the absence of a peak within the range of 1100 cm−1 suggests that the chitosan coating has been executed proficiently. In Fig. 2B, the FT-IR spectra of free SCR, Bi@Chi NPs, and Bi@Chi-SCR NPs were analyzed to authenticate the successful encapsulation of SCR within chitosan. In Fig. 2C, the FT-IR spectra of free DEC, Bi@Chi NPs, and Bi@Chi-DEC were compared to validate the successful encapsulation of DEC within chitosan. The SCR and DEC were characterized by FT-IR spectra, exhibiting peaks at 1400 cm−1 associated with hydroxyl groups, 1600 cm−1 related to the carbonyl moiety, and 3440 cm−1 corresponding to the N–H bond; all of these spectral features were detected in Bi@Chi-SCR and Bi@Chi-DEC, albeit with slight shifts and modifications.
The assessment of the progress of reactions and the investigation of compound interactions are key applications of UV–vis spectroscopy. Figure 2D presents the UV–vis absorption spectrum for BiNPs, Bi@Chi NPs, Bi@Chi-DEC NPs, Bi@Chi-SCR NPs, Free Chi, Free DEC, and Free SCR. The surface plasmon resonance (SPR) peak for BiNPs was identified at a wavelength of 280 nm, signifying the formation of BiNPs. For Bi@Chi NPs, the shift in wavelength peak, contingent upon the presence of chitosan, reached approximately 300 nm. The peak observed in the range of 420 nm is associated with decoy and scramble oligonucleotides, thereby indicating their interaction with chitosan.
FESEM and TEM confirm the influence of chitosan on BiNP dimensions and morphology
The FESEM was employed to elucidate the morphological characteristics and average dimensions of Bi@Chi-DEC NPs. As illustrated in Fig. 3A, Bi@Chi-DEC NPs exhibit nearly uniform and spherical morphologies. This compression is mitigated in the case of Bi@Chi-DEC NPs, a phenomenon likely resulting from incorporating chitosan and establishing electrostatic repulsion between the negatively charged bismuth and the positively charged chitosan. The weight percentage of the bismuth element was ascertained through energy-dispersive X-ray spectroscopy (Fig. 3B). The analytical outcomes indicated that the Bi@Chi-DEC NPs contain 42.53% bismuth. The observed reduction in the bismuth content of Bi@Chi-DEC NPs is attributable to the presence of chitosan.
The dimensions and morphology of the Bi@Chi-DEC NPs were further examined utilizing transmission electron microscopy (TEM) analysis. The TEM imagery demonstrates that the Bi@Chi-DEC NPs (Fig. 3C) possess relatively uniform and spherical configurations. The findings indicate that the average size of Bi@Chi-DEC NPs is 171.10 ± 39.19 nm (Fig. 3D). Notably, the findings derived from the TEM and FESEM analyses corroborate one another.
DLS analysis shows the impact of surface modifications on nanoparticle size and charge
The mean hydrodynamic diameter, zeta potential, and polydispersity index (PDI) were ascertained utilizing dynamic light scattering (DLS) methodology. The measurements indicated that the dimensions of the BiNPs, Bi@Chi NPs, Bi@Chi-SCR NPs, and Bi@Chi-DEC NPs were 82.28, 178.80, 218.8, and 213.8 nm, respectively, with corresponding PDI values of 0.114, 0.267, 0.292, and 0.328 (Fig. 4A, B).
The average zeta potentials for the BiNPs, Bi@Chi NPs, Bi@Chi-SCR NPs, and Bi@Chi-DEC NPs were recorded at -18.46, 3.94, -3.81, and -3.27 mV, respectively. The observed negative charge of the BiNPs can be attributed to the incorporation of albumin, whereas the positive charge of the Bi@Chi NPs arises from the application of chitosan, which has effectively neutralized the negative charge of the BiNPs and facilitated the absorption of a certain degree of positive charge. The Bi@Chi-SCR NPs and Bi@Chi-DEC NPs have acquired a negative charge due to the inherent negative charge of the decoy and scramble oligonucleotides, resulting in the loss of the positive charge previously exhibited (Fig. 4C, D).
Demonstrating biocompatibility and controlled release of ODNs from BiNPs
The findings of the drug loading efficacy assessment indicated that the ultimate loading percentage of ODNs within the nanostructures was recorded at 27.45%. The release kinetics of ODNs were evaluated utilizing spectrophotometric methods across a temporal scale from 0 to 48 h under two distinct pH conditions of 7.4 and 5.8 (Fig. 5). The release rate at pH 7.4 (representative of physiological conditions) after 48 h for the Bi@Chi-DEC and Bi@Chi-SCR nanostructures was quantified at 69% and 65%, respectively (see Fig. 5A). Conversely, at pH 5.8 (simulating the acidic environment typical of cancer cells), the release rate of ODNs from Bi@Chi-DEC and Bi@Chi-SCR was approximately 82% and 80%, respectively, over the same 48-h period (see Fig. 5B). Consequently, the release percentage of oligonucleotides from the nanostructures in acidic pH conditions exceeds that observed at physiological pH, which posits a favorable characteristic for nanoparticles given the acidic milieu prevalent in cancerous cellular environments.
In the present investigation, the hemocompatibility of BiNPs, Bi@Chi NPs, Bi@Chi-SCR NPs, and Bi@Chi-DEC NPs was assessed utilizing human blood, a critical consideration for potential in vivo applications. The hemolytic characteristics of these synthesized nanoparticles were examined at varying concentrations (65, 125, 250, 500, 1000, and 2000 nM). For this experimental assay, PBS served as the negative control, while Triton X100 was employed as the positive control. As illustrated in Fig. 5C, the incubation of blood with BiNPs, Bi@Chi NPs, Bi@Chi-SCR NPs, and Bi@Chi-DEC NPs for 4 h yielded hemolytic ratios that ranged from 2.53 to 13.29, 2.96 to 11.83, 2.81 to 12.10, and 3.62 to 10.82%, respectively.
In vitro cellular uptake of Bi@Chi-labeled ODNs nanostructure
As shown in Fig. 6, the cellular uptake rate of Bi@Chi NPs as a negative control (lack of Cy3-labeled oligos) with a concentration of 300 nm is 2.78%, which is not significant compared to the control group (untreated cells). The obtained results showed that the cellular uptake rate of the investigated concentrations of Bi@Chi-labeled ODNs nanostructure such as 25, 50, 100, 200, and 300 nM, is 16.3, 41.9, 58.6, 88.0, and 99.4%, respectively. The rate of cellular uptake in all five different concentrations of Bi@Chi-labeled ODNs has a significant difference compared to the untreated control group. Furthermore, the uptake rate at a concentration of 300 nM was higher than other concentrations.
(A) The cellular uptake rate of Bi@Chi NPs nanostructures (negative control) and Bi@Chi-labeled ODNs (ODNs labeled with Cy3) in the MDA-MB-231 cell line at concentrations of 25, 50, 100, 200, and 300 nM (B) Statistical graph showing uptake rate of nanostructured groups in different concentrations. **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Cellular toxicity was increased with and without X-ray exposure
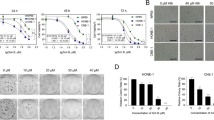
In the without X-ray exposure conditions (Fig. 7A), by increasing all concentrations of BiNPs, Bi@Chi NPs (except 100 and 200 nM), Bi@Chi-SCR NPs, and Bi@Chi-DEC NPs the cell growth inhibition was enhanced in a dose-dependent manner compared to the cell control group (untreated group). In Bi@Chi NPs nanoparticles, the cellular toxicity level has decreased due to the coating of bismuth nanoparticles with chitosan. The level of cytotoxicity in the group treated with Bi@Chi-DEC nanoparticles was high compared to the groups treated with Bi@Chi-SCR, and this could indicate the specific function of Decoy compared to scramble.
Cytotoxicity evaluation of different groups of nanostructured BiNPs, Bi@Chi, Bi@Chi-SCR, and Bi@Chi-DEC NPs at concentrations of 50, 100, 200, 300, and 400 nM on MDA-MB-231 cells in the condition of (A) without X-ray exposure and (B) with X-ray exposure. *p < 0.05, **p < 0. 01, *** p < 0.001, and ****, p < 0.0001.
In the X-ray exposure condition (Fig. 7B), the effect of treatment with various concentrations of BiNPs, Bi@Chi, Bi@Chi-SCR, and Bi@Chi-DEC nanostructures along with applying 2 Gy of X-rays (divided into 0.25Gy doses) has been investigated on cytotoxicity of MDA-MB-231 cells. The obtained results showed that all concentrations of different groups of nanostructures (except 50 nM concentration of Bi@Chi nanoparticles and Bi@Chi-SCR nanoparticles) led to a significant increase in cytotoxicity in a dose-dependent manner compared to the control group. As mentioned, this issue can indicate the specific performance of the decoy compared to the scramble.
Cell cycle was arrested with and without X-ray exposure
In the without X-ray exposure condition (Fig. 8A, C), the number of cells arrested in the G1 phase has increased compared to the control group. The percent of cells arrested in this phase after treatment with 300 nM of BiNPs, Bi@Chi, Bi@Chi-SCR, and Bi@Chi-DEC nanostructures were 38.16, 44.54, 40.92, 41.86, and 49.71%, respectively. Furthermore, treatment with Bi@Chi-DEC NPs caused significant G1 arrest compared to the Bi@Chi-SCR NPs and Bi@Chi NPs (p < 0.001) and BiNPs (p < 0.05). The rate of cell arrest in the group treated with Bi@Chi-DEC NPs is higher than in other groups, which indicates the greater effect of decoy in arresting the MDA-MB-231 cells in this phase.
In the X-ray exposure condition (Fig. 8B, D), cell groups treatment with 300 nM of BiNPs, Bi@Chi, Bi@Chi-SCR, and Bi@Chi-DEC nanostructures along with applying 2 Gy of X-rays (divided into 0.25Gy doses) led to the cell cycle arrest at G2/M phase equal to 19.30, 30.20, 23.16, 24.51 and 42.12% respectively. The results revealed that treated with different nanostructures which were also subjected to X-ray, resulted in significant G2/M arrest compared to the control group. Treatment with Bi@Chi-DEC NPs compared to other treated groups significantly arrested the MDA-MB-231 cells in the G2/M phase (p < 0.0001). Also, in the group treated with Bi@Chi-DEC NPs, the highest amount of Sub G1 was observed, which indicates a higher rate of apoptosis in this group.
Apoptosis rate was increased with and without X-ray exposure
In the without X-ray exposure, as shown in parts A and C of Fig. 9, the total apoptosis rate (early and late) after treatment with 300 nM of BiNPs, Bi@Chi, Bi@Chi-SCR, Bi@Chi-DEC nanostructures and control group (Ctrl) were 4.59, 20.82, 15.34, 19.5, 25.75 and 5.59%, respectively. The obtained results showed that the apoptosis rate in all treated groups with different nanostructures increased compared to the untreated control group. The amount of apoptosis induced in the cell group treated with Bi@Chi-DEC NPs is significantly higher than in other treated groups.
In the X-ray exposure condition, as shown in parts B and D of Fig. 9, cell groups treated with 300 nM of BiNPs, Bi@Chi, Bi@Chi-SCR, and Bi@Chi-DEC nanostructures and control group (Ctrl) along with applying 2 Gy of X-rays (divided into 0.25Gy doses) led to induce total apoptosis rate equal to 10.52, 39.6, 20.8, 28.9, 50.4 and 10.52%, respectively. All groups treated with different nanostructures have shown a significant apoptosis rate compared to the control group. The cells treated with Bi@Chi-DEC NPs compared to the BiNPs, Bi@Chi-SCR NPs and Bi@Chi NPs groups (p < 0.0001) showed significantly more apoptosis. These results show the specific function and the role of Nanog’s specific decoy in increasing the apoptosis rate of the MDA-MB-231 cells. Bi@Chi NPs group showed more apoptosis than Bi@Chi NPs (p < 0.001), which could be due to the reduction of toxicity of Bi@Chi NPs due to coating with chitosan.
Cell migration was inhibited with and without X-irradiation exposure
In the without X-ray exposure (Fig. 10B), the percent of cell migration inhibition 72 h after treatment with 150 nM of BiNPs, Bi@Chi, Bi@Chi-SCR, and Bi@Chi-DEC nanostructures was investigated compared to time zero (Fig. 10A). The obtained results showed that in the cell group treated with Bi@Chi nanoparticles compared to the control group, there was no significant change in gap filling (cell migration) in 72 h, which could be caused by the chitosan (Chi) coating effect. At the same time (72 h), the treatment of cell groups with BiNPs, Bi@Chi-SCR, and Bi@Chi-DEC nanostructures resulted in the inhibition of cell migration significantly compared to the control group. The percentage of inhibition of cell migration in the group treated with Bi@Chi-DEC is significantly different (p < 0.0001) and gap filling did not occur to a large extent in these groups (Fig. 10D).
Cell migration inhibition evaluation after treatment with 150 nM of BiNPs, Bi@Chi, Bi@Chi-SCR, and Bi@Chi-DEC NPs nanostructures on MDA-MB-231 cells at (A) 0h after treatment, (B) 72h later in without X-ray exposure conditions and (C) 72h later in with X-ray exposure conditions. (D) Statistical analysis of the data from the percentage of cell migration inhibition. **p < 0.01 and ****p < 0.0001.
In the X-ray exposure condition (Fig. 10C), the cell migration speed was significantly delayed in the treated groups compared to the without X-ray condition. In the BiNPs, Bi@Chi-SCR, and Bi@Chi-DEC treatment group inhibition of cell migration was significant compared to the control group. It is worth mentioning that, cell migration inhibition in the Bi@Chi-DEC-treated group was significantly higher than other groups (Fig. 10D).
Tumorsphere formation was reduced with and without X-ray exposure
In the without X-ray exposure, as shown in Fig. 11A, the ability to form tumorspheres after treatment with 150 nM BiNPs, Bi@Chi, Bi@Chi-SCR, and Bi@Chi-DEC nanostructures was investigated for 8 days. In all treated groups, the inhibition of the formation of the tumorspheres was significant compared to the control group. The group treated with Bi@Chi-DEC NPs showed a significant decrease in tumorsphere formation (size and number) compared to the control (p < 0.01) and Bi@Chi-SCR NPs (p < 0.05) groups (Fig. 11C).
Tumorsphere formation evaluation after treatment with 150 nM of BiNPs, Bi@Chi, Bi@Chi-SCR, and Bi@Chi-DEC NPs nanostructures on MDA-MB-231 cells in conditions (A) without X-ray exposure and (B) with X-ray exposure. (C) Statistical analysis of the data obtained from the examination of tumorsphere forming ability. * p < 01 and **p < 0.01.
In the X-ray exposure condition, as shown in Fig. 11B, the size of formed tumorspheres decreased in all groups compared to the control group and without the X-ray exposure condition. A significant decrease was observed in the groups treated with Bi@Chi-DEC NPs in comparison with Bi@Chi-SCR NPs, which indicates the specific function of Decoy in reducing tumor sphere formation. In both with and without X-ray exposure conditions, the results indicated that treatment with Bi@Chi-DEC NPs significantly inhibited tumorsphere formation compared with the control group (Fig. 11C).
Discussion
Triple-negative breast cancer (TNBC) exhibits clinical attributes such as heightened aggressiveness and metastatic capability, rendering it susceptible to recurrence and associated with a dismal prognosis. Conventional therapeutic interventions, including surgical resection, chemotherapy, and radiotherapy, demonstrate limited efficacy in managing breast cancer24,25,26. Investigators are actively exploring novel therapeutic modalities as well as combinatorial approaches for a spectrum of diseases, with a particular emphasis on malignancies. The utilization of transcription factor decoy (TFD) strategies represents an efficacious methodology within oligodeoxynucleotide-based small molecule therapies aimed at mitigating the overexpression of various oncogenes implicated in neoplastic conditions7,17,21. Nonetheless, the precise delivery of the specific decoy targeting oncogenes into neoplastic cells and effectively obstructing the targeted oncogene presents a formidable challenge. Concurrently, the research acknowledges the essential contribution of cancer stem cells to treatment resistance and disease advancement. NANOG, a pivotal transcription factor, is crucial in preserving stem cell characteristics and is often re-expressed in a variety of malignancies, including breast cancer. The researchers aim to obstruct the molecular pathways that facilitate the survival and proliferation of cancer stem cells through the development of decoy ODNs specifically targeting NANOG.
Recently, nanoparticles have been employed to enhance radiosensitivity and to treat a myriad of tumor types27. In particular, nanoparticles composed of bismuth (Bi) are particularly advantageous for the detection of neoplastic diseases and exhibit heightened sensitivity to X-ray radiation attributable to their elevated atomic number (Z = 83) and significant X-ray attenuation coefficient28,29. This class of nanoparticles may serve as effective radiosensitizers, thereby augmenting radiation absorption in both combinatorial and synergistic cancer treatment modalities30. BiNPs are characterized by their excellent biocompatibility and minimal toxicity. Furthermore, this nanoparticle has been developed as innovative nanoplatforms for targeted drug delivery31. The integration of BiNPs and NANOG decoy oligonucleotides presents a multifaceted therapeutic paradigm. This methodology addresses two critical obstacles in oncological treatment: the inherent resistance of cancer cells to radiation and the molecular frameworks that sustain the viability of cancer stem cells. The synergistic potential of these two components may yield a more holistic and efficacious treatment approach. Furthermore, the strategy aspires to mitigate collateral damage to healthy tissues, a chronic concern associated with conventional radiotherapy. The initiative aims to enhance treatment precision and diminish potential adverse effects by selectively targeting cancer cells and stem cells. This targeted strategy signifies a considerable progression in personalized cancer therapy, transcending the traditional one-size-fits-all treatment model. The prospective ramifications of this research extend beyond immediate therapeutic efficacy. By disrupting the mechanisms associated with cancer stem cells and amplifying radiation-induced cytotoxicity, the study could elucidate more effective, less invasive strategies for cancer treatment. This investigation not only tackles the pressing challenge of breast cancer therapy but also enriches the broader comprehension of cancer stem cell biology and therapeutic interventions. Altogether, in the present study, the combinational therapy effects of synthesized bismuth nanoparticles loaded with NANOG transcription factor-specific decoy sequences along with radiotherapy were investigated on TNBC (MDA-MB-231) cell lines.
FT-IR spectroscopic analyses have been conducted to validate the successful fabrication of nanostructures. In accordance with prior research, the emergence of the bending vibration peak associated with the O–H bond (3400 cm−1) in the BiNP spectrum signifies the appropriate bonding between bismuth and bovine serum albumin (BSA)6,32. Furthermore, the bending vibration peak corresponding to the N–H bond (2800–2900 cm−1) is observable for chitosan; however, this peak is absent in the Bi@Chi NPs due to the effective interaction between the amine and hydroxyl moieties, thereby affirming the synthesis of this particular nanostructure. Ultimately, the identification of novel peaks at wave numbers 1700 cm-1 and 1600 cm-1 in the FT-IR spectrum of Bi@Chi-DEC substantiates the successful physical linkage of the decoy oligonucleotide (ODN) and the establishment of the final formulation. The UV–vis spectrophotometer was employed to examine the intermolecular interactions of the nanoparticles. In this assessment, alterations in the nanoparticle surface and their successful synthesis are discerned through variations in the λmax of the absorption spectrum. The absorption peak for BiNP is approximately located at 280 nm, which shifts to 300 nm following the formation of the Bi@Chi NPs. Nosrati et al. demonstrated that the peak of the UV–vis spectrum for Bi2S3@BSA nanoparticles is 264 nm, which aligns with the findings of this study33. Bismuth nanoparticles exhibit a distinct absorption spectrum that may vary depending on the synthesis method employed or the substrate or surfactant used for functionalization.
FESEM and TEM microscopy techniques were employed to investigate the geometric characteristics and dimensions of nanoparticles. The morphological analysis of Bi@Chi-DEC NPs revealed that they possess a spherical and uniform structure, with their average diameters, as inferred from TEM imagery, measuring approximately 171.10 ± 39.19 nm. The dimensions acquired through these methodologies are smaller than those determined by DLS. In dynamic light scattering, the hydrodynamic dimensions of nanoparticles are assessed in wet and aqueous environments, whereas in TEM and FESEM, the dimensions of nanoparticles in dry form are examined. Consequently, the dimensions derived from DLS are expected to exceed those measured by TEM and FESEM. The findings from the study conducted by Lee et al. indicated that the morphology of Bi2S3@BSA-Bio-MTX nanoparticles is also spherical and homogeneous, with an average dimension of 10.4 ± 0.71 nm34. The sizes of bismuth nanoparticles may vary depending on their synthesis methods and the specific materials with which they have been functionalized.
The outcomes of size characterization via Dynamic Light Scattering (DLS) reveal that the mean dimension of bismuth nanoparticles is 82.31 nm, which escalates to 215.7 nm after stabilization with chitosan and the incorporation of decoy oligodeoxynucleotides. In the research conducted by Nosrati et al. the DLS measurements for Bi2S3@BSA and Bi2S3@BSA-FA-CUR (which incorporates curcumin and is conjugated with folic acid) were recorded at 78.9 nm and 170.9 nm, respectively32. Additionally, in an alternative investigation, DLS findings indicated that Bi2S3@BSA@CUR Hybrid Nanoparticles (HNPs) possess a diameter of approximately 103 nm, as demonstrated by the facile green synthesis of bismuth sulfide radiosensitizers through the biomineralization of albumin, a natural biomolecule employed in chemoradiation therapy. The polydispersity index (PDI) serves as a crucial metric for assessing the size distribution of nanoparticles. The results indicated that the dispersion index for the Bi@Chi-DEC nanostructure is calculated to be 0.311. A PDI value below 0.5 is deemed acceptable35, and the results derived from the present study suggest that the nanoparticle size distribution is indeed appropriate. Zeta potential, which quantifies the electrostatic charge surrounding nanoparticles, represents one of the significant physicochemical characteristics of nanoparticles. The mean zeta potential of the Bi@Chi-DEC nanostructure is recorded at -2.98 mV. A zeta potential range from -30 mV to + 30 mV has been reported as acceptable, as this interval facilitates adequate repulsive forces necessary for achieving colloidal stability of nanoparticles. A cumulative size below 500 nm has been validated for nanoparticles. Sizes exceeding 500 nm may lead to the entrapment of nanoparticles within blood vessels. The electrostatic charge of various nanoparticles will vary contingent upon the specific substrate or surfactant with which they are loaded or conjugated (functionalized).
One notable characteristic associated with the application of nanostructures in the pharmaceutical domain is the regulated release of therapeutic agents. To evaluate the drug release kinetics from the nanostructure containing decoy ODNs, pH levels of 5.8 (which corresponds to the pH present in cellular endosomes) and 7.4 (the pH level of human blood plasma) were employed. The oligonucleotide release rate after 48 h reached approximately 69% at pH 7.4 and around 82% at pH 5.8. These findings indicate that the release rate in an acidic milieu surpasses that in a physiological context, aligning with the environmental conditions prevalent in neoplastic tissues and acidic endosomes. In the hemolysis assay, the cytotoxicity of the nanoparticles is directly correlated with the liberation of heme. The maximal concentration of nanoparticles utilized in this investigation was 2000 nM, which resulted in a hemolysis rate of 13% within the Bi@Chi-DEC cohort. The exposure of erythrocytes to nanoparticles for 4 h yielded a hemolytic ratio of less than 5% for the tested concentrations of 65, 250, and 500 nM. According to the guidelines established by ISO/TR 7406, samples exhibiting hemolytic rates below 5% (the critical threshold for safe hemolytic ratios in biomaterials) are classified as nonhemolytic, thereby suggesting that Bi@Chi-DEC NPs demonstrate favorable compatibility with blood.
Due to their small size, nanoparticles are actively or passively absorbed by cells when they are in contact with tumor cells, and the efficiency of cellular absorption of nanoparticles and the resulting intracellular concentration determine the cytotoxic potential. In addition, the cellular uptake efficiency is mainly dependent on the concentration of nanoparticles. In this study to investigate the cellular uptake of nanostructures containing ODNs, Cy3 fluorescent dye-labeled oligos and flow cytometry were used. The cellular uptake rate of 25, 50, 100, 200, and 300 nM concentrations of the Bi@Chi-labeled ODNs nanosystem was 16.3, 41.9, 58.6, 88.0, and 99.4 percent, respectively. The uptake rate increased in a dose-dependent manner and the concentration of 300 nM was chosen as the optimal concentration.
The toxicity of different nanoparticles was observed based on the results of MTT in all treated groups. Cytotoxicity was dose-dependent in all groups treated with different nanoparticles. The level of cytotoxicity in the group treated with Bi@Chi NPs was lower compared to other groups. The reason for this could be the coating of bismuth nanoparticles with chitosan, which has reduced the toxicity of this nanoparticle. Based on the results of this test, the level of toxicity in the group treated with Bi@Chi-DEC is much higher than the Bi@Chi- group, which can indicate the specific function of DEC. The results showed that Bi@Chi-DEC in the presence of 2 Gy radiation therapy causes a greater decrease in the survival of MDA-MB-231 cells compared to the absence of radiation therapy. This issue can indicate the synergistic effect of bismuth nanoparticles, Nanog TFs-specific decoy, and radiotherapy. Therefore, these nanoparticles can reduce the dose of the drug and also the radiation used in radiotherapy. Nasrati et al. evaluated Bi2S3@BSA HNPs under 2 Gy and 6 Gy radiation. Their results showed that these nanoparticles rarely affect cell viability without radiation, which can be due to the coating of bismuth nanoparticles with BSA. The result revealed that Bi2S3@BSA@CUR HNPs can be used as both radiosensitizers and vehicle agents for the delivery of CUR in the HT-29 cell line33. The results of Azizi et al.'s study showed that Bi2S3@BSA-Bio-MTX nanoparticles together with radiotherapy have excellent anti-cancer activity36. Moreover, the study of Lu et al. showed that bismuth nanoparticles decrease the survival rate of HeLa cancer cells and this toxicity increases with the increasing concentration of bismuth nanoparticles37. Faghfoori et al. demonstrated that treatment of cells with Bi2S3@BSA-MTX NPs along with X-irradiation led to a lower degree of viability in comparison with the condition of the absence of irradiation, which confirmed the ability of this nanoparticle as a radio-sensitizer agent6. Colak et al. revealed that implanted 3D-printed Alg-Bi2S3@BSA (containing bismuth nanoparticles) under tumor tissue in a murine model by reducing the tumor volume growth rate under single-dose X-irradiation shows high therapeutic efficacy38. In this study, the concentration of 400 nM, especially in the treatment group (Bi@Chi-DEC), showed a toxic effect so the concentration of 300 nM was used as the appropriate concentration to perform the rest of the tests. Johari et al. showed that the Nanog decoy oligodeoxynucleotide-encapsulated niosomes nanocarriers have anti-cancer effects on human glioblastoma multiforme (U87) cells15.
In examining the cell cycle, the number of cells in the G1, S, and G2/M phases of the cell cycle is determined according to the amount of cellular DNA in the examination of the cell cycle in the conditions without the use of X-rays, the number of cells arrested in the G1 phase was high in all treated groups. The amount of this cell arrest in the group treated with Bi@Chi-DEC NPs was higher than in other groups. There is a significant difference in the number of cells arrested in the G1 phase in the Bi@Chi-DEC NPs group compared to the Bi@Chi-SCR NPs group. This result can indicate the specific performance of DEC compared to SCR. In the cell cycle test under the condition of applying X-rays with a dose of 2 Gy, cells treated with different nanostructures arrest in the G2/M phase. A significant difference was observed in the amount of cell arrest in the group treated with Bi@Chi-DEC NPs compared to the groups of BiNPs, Bi@Chi NPs, and Bi@Chi-SCR NPs. In the BiNPs and Bi@Chi NPs groups, more Sub-G1 (indicating the induction of apoptosis in the cell) was found compared to the other treated groups, which was also increased by the use of radiotherapy. Increased cell arrest in the Sub-G1 phase indicates more apoptosis in the MDA-MB-231 cell line. Studies have shown that Nanog knockdown reduces cell proliferation and induces G0/G1 cell cycle arrest in stem cells39,40. Han et al. study showed that RNA interference-mediated silencing of Nanog reduces cell proliferation and induces G0/G1 cell cycle arrest in breast cancer cells2.
In the apoptosis test without applying X-rays, in the group transfected with Bi@Chi-DEC with a concentration of 300 nm, 25.75% apoptosis, and 9.85% necrosis were observed. These percentages are significantly different from the control group, which indicates the induction of apoptosis in MDA-MB-231 cells by inhibition of Nanog by its specific decoy. Compared to the group treated with Bi@Chi-SCR, the apoptosis rate was high in the group treated with Bi@Chi-DEC, which can indicate the specific function of DEC compared to SCR. Due to the presence of chitosan in the Bi@Chi NPs group, the toxicity of nanoparticles is reduced and as a result, less apoptosis is observed. According to the obtained results, the amount of apoptosis has more increased with the application of X-rays with 2 Gy in all groups. Also, the amount of necrosis has increased in all groups, which is due to the effect of radiotherapy. In 2022, researchers concluded that conjugating Bi2S3@BSA NPs with metronidazole and using it simultaneously with radiotherapy significantly increased apoptosis in 4T1 cells41. It has been shown that BiNPs along with XRT (X-irradiation) led to increased efficacy of radiotherapy (RT) sensitization by decreasing the expression of thioredoxin reductases (TrxR). TrxR has an important role in the regulation of apoptosis5. Another study showed that Nanog inhibits apoptosis42, therefore, inhibiting this transcription factor can induce apoptosis. Previous studies have shown that the presence of bismuth (Bi) in the final nanosystem composition can lead to the induction of apoptosis and increased cytotoxicity in the cancer cell line under treatment compared to the normal cell line, which results are consistent with the present study in terms of anticancer effects43,44.
To evaluate the effect of nanostructure synthesized on cell migration and metastasis, the scratch or wound repair method was used. In this assay, all groups of nanoparticles with a concentration of 150 nM were used. It was observed that in the condition without applying X-rays, inhibition of cell migration in the group treated with Bi@Chi-DEC was well done after 72 h. In the Bi@Chi group, due to the chitosan coating effect, the amount of cell migration was lower compared to other groups. According to the results obtained after radiotherapy, the inhibition rate of gap-filling increased significantly. In vitro studies have shown that inhibiting Nanog expression results in decreased cellular migration, invasiveness, and proliferation45,46,47,48. Siu et al. demonstrated that when Nanog is overexpressed, it leads to an increase in cell growth and migration, as well as a decrease in E-cadherin mRNA levels. According to these findings, Nanog is connected to the abnormal migration and metastasis of cancer cells49. In the tumorsphere experiment, we concluded that Bi@Chi-DEC nanoparticles have the greatest effect on the MDA-MB-231 cell line and prevent the formation of tumorspheres. After applying radiotherapy, tumorsphere reduction is evident in all groups of nanoparticles.
In summary, BiNPs function by increasing the efficiency of radiation therapy treatment on account of their highly dense atomic numbers, thus permitting the effective absorption of X-ray energy over living tissues, releasing secondary electrons, which produce reactive oxygen species (ROS) and localized DNA damage in cancer cells; here in the study, BiNPs were coated with chitosan and loaded with NANOG decoy oligodeoxynucleotides (ODNs), which enhanced radiosensitivity in MDA-MB-231 breast cancer cells as evident from reduced cell viability, tumorsphere formation, and migration, together with enhanced apoptosis and G2/M cell cycle arrest with BiNPs being powerful radiosensitizers, amplifying X-ray-induced cytotoxic effects with a relatively high confidence level50,51.
Conclusion
The obtained results from nanostructure characterization confirmed that the bismuth sulfide nanoparticle coated with chitosan and containing decoy oligodeoxynucleotide for the Nanog transcription factor (Bi@Chi-DEC NPs) has been properly synthesized. Cell treatment with Bi@Chi-DEC NPs along with X-ray exposure showed reduced cell viability, arrested cell cycle, induced apoptosis, inhibited cell migration, and decreased tumorsphere formation ability on the MDA-MB-231 cells. Taken together, Bi@Chi-DEC NPs have the potential to be used along with radiotherapy as a combined treatment in breast cancer therapy, and in vivo studies are warranted to substantiate the efficacy of this therapeutic approach.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Lu, X., Mazur, S. J., Lin, T., Appella, E. & Xu, Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene 33(20), 2655–2664 (2014).
Han, J. et al. RNA interference-mediated silencing of NANOG reduces cell proliferation and induces G0/G1 cell cycle arrest in breast cancer cells. Cancer Lett. 321(1), 80–88 (2012).
Choi, S. C. et al. Nanog regulates molecules involved in stemness and cell cycle-signaling pathway for maintenance of pluripotency of P19 embryonal carcinoma stem cells. J. Cell. Physiol. 227(11), 3678–3692 (2012).
Sun, Y.-S. et al. Risk factors and preventions of breast cancer. Int. J. Biol. Sci. 13(11), 1387 (2017).
Badrigilan, S. et al. Bismuth-based nanomaterials: Recent advances in tumor targeting and synergistic cancer therapy techniques. Adv. Healthc. Mater. 9(7), 1901695 (2020).
Faghfoori, M. H. et al. Anticancer effect of X-Ray triggered methotrexate conjugated albumin coated bismuth sulfide nanoparticles on SW480 colon cancer cell line. Int. J. Pharm. 582, 119320 (2020).
Johari, B. et al. Myc decoy oligodeoxynucleotide inhibits growth and modulates differentiation of mouse embryonic stem cells as a model of cancer stem cells. Anti Cancer Agents Med. Chem. 17(13), 1786–1795 (2017).
Osako, M. K., Nakagami, H. & Morishita, R. Development and modification of decoy oligodeoxynucleotides for clinical application. In Nucleic Acid Drugs 49–59 (Springer, 2011).
Johari, B. et al. Combinational therapy with Myc decoy oligodeoxynucleotides encapsulated in nanocarrier and X-irradiation on breast cancer cells. Oncol. Res. 32(2), 309 (2023).
Ghorbani, R. et al. Targeted anti-tumor synergistic effects of Myc decoy oligodeoxynucleotides-loaded selenium nanostructure combined with chemoradiotherapy on LNCaP prostate cancer cells. Oncol. Res. 32(1), 101 (2023).
Babaei, M. & Ganjalikhani, M. The potential effectiveness of nanoparticles as radio sensitizers for radiotherapy. Bioimpacts 4(1), 15 (2014).
Nagpal, K., Singh, S. K. & Mishra, D. N. Chitosan nanoparticles: A promising system in novel drug delivery. Chem. Pharm. Bull. 58(11), 1423–1430 (2010).
Johari, B. et al. Increasing the colon cancer cells sensitivity toward radiation therapy via application of Oct4–Sox2 complex decoy oligodeoxynucleotides. Mol. Biol. Rep. 47, 6793–6805 (2020).
Johari, B. et al. Suppress the cell growth of cancer stem-like cells (NTERA-2) using Sox2-Oct4 decoy oligodeoxynucleotide—encapsulated niosomes-zinc hybrid nanocarriers under X-irradiation. Heliyon 10(13), e34096 (2024).
Gharbavi, M. et al. NANOG decoy oligodeoxynucleotide–encapsulated niosomes nanocarriers: A promising approach to suppress the metastatic properties of U87 human glioblastoma multiforme cells. ACS Chem. Neurosci. 11(24), 4499–4515 (2020).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133(6), 1106–1117 (2008).
Rahmati, M., Johari, B., Kadivar, M., Rismani, E. & Mortazavi, Y. Suppressing the metastatic properties of the breast cancer cells using STAT3 decoy oligodeoxynucleotides: A promising approach for eradication of cancer cells by differentiation therapy. J. Cell. Physiol. 235(6), 5429–5444 (2020).
Gharbavi, M., Johari, B., Tabar, R. M. & Sharafi, A. Selenium-doped albumin nanoparticles enhance tamoxifen-induced anticancer effects in 4T–1 mouse breast cancer cells. Appl. Organomet. Chem. 38(2), e7327 (2024).
Asadi, N., Gharbavi, M., Rezaeejam, H., Farajollahi, A. & Johari, B. Zinc nanoparticles coated with doxorubicin-conjugated alginate as a radiation sensitizer in triple-negative breast cancer cells. Int. J. Pharm. 659, 124285 (2024).
Jafari, B. et al. Mitigated oxidative stress and cognitive impairments in transient global ischemia using niosomal selegiline-NBP delivery. Behav. Neurol. 2022(1), 4825472 (2022).
Ghorbani, R., Gharbavi, M., Keshavarz, B., Madanchi, H. & Johari, B. Targeting c-Myc with decoy oligodeoxynucleotide-loaded polycationic nanoparticles inhibits cell growth and induces apoptosis in cancer stem-like cells (NTERA-2). Mol. Biol. Rep. 51(1), 623 (2024).
Felenji, H., Johari, B., Moradi, M., Gharbavi, M. & Danafar, H. Folic acid-conjugated iron oxide magnetic nanoparticles based on bovine serum albumin (BSA) for targeted delivery of curcumin to suppress liver cancer cells. Chem. Afr. 5(5), 1627–1639 (2022).
Johari, B. et al. Evaluation of STAT3 decoy oligodeoxynucleotides’ synergistic effects on radiation and/or chemotherapy in metastatic breast cancer cell line. Cell Biol. Int. 44(12), 2499–2511 (2020).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69(1), 7–34 (2019).
Yazdi Rouholamini, S. E. et al. Effect of silibinin-loaded nano-niosomal coated with trimethyl chitosan on miRNAs expression in 2D and 3D models of T47D breast cancer cell line. Artif. Cells Nanomed. Biotechnol. 46(3), 524–535 (2018).
Hajighasemlou, S. et al. Preparation of immunotoxin herceptin-botulinum and killing effects on two breast cancer cell lines. Asian Pac. J. Cancer Prev. 16(14), 5977–5981 (2015).
Rasouli, N. et al. Assessment of oxaliplatin-loaded iodine nanoparticles for chemoradiotherapy of human colorectal cancer (HT-29) cells. Polymers 14(19), 4131 (2022).
Ai, K. et al. Large-scale synthesis of Bi2S3 nanodots as a contrast agent for in vivo X-ray computed tomography imaging. Adv. Mater. 42(23), 4886–4891 (2011).
Kinsella, J. M. et al. X-ray computed tomography imaging of breast cancer by using targeted peptide-labeled bismuth sulfide nanoparticles. Angew. Chem. Int. Ed. 50(51), 12308–12311 (2011).
Glatz, M. et al. Emollient use alters skin barrier and microbes in infants at risk for developing atopic dermatitis. PLoS ONE 13(2), e0192443 (2018).
Bulmahn, J. C. et al. Laser-ablative synthesis of stable aqueous solutions of elemental bismuth nanoparticles for multimodal theranostic applications. Nanomaterials 10(8), 1463 (2020).
Nosrati, H., Charmi, J., Salehiabar, M., Abhari, F. & Danafar, H. Tumor targeted albumin coated bismuth sulfide nanoparticles (Bi2S3) as radiosensitizers and carriers of curcumin for enhanced chemoradiation therapy. ACS Biomater. Sci. Eng. 5(9), 4416–4424 (2019).
Nosrati, H. et al. Facile green synthesis of bismuth sulfide radiosensitizer via biomineralization of albumin natural molecule for chemoradiation therapy aim. Artif Cells Nanomed. Biotechnol. 47(1), 3832–3838 (2019).
Lee, C. High-Z metal encapsulated carbon dots for enhancing radiation therapy. University of Georgia (2020).
Byun, Y. et al. Formulation and characterization of α-tocopherol loaded poly ɛ-caprolactone (PCL) nanoparticles. LWT Food Sci. Technol. 44(1), 24–28 (2011).
Azizi, S., Nosrati, H., Sharafi, A. & Danafar, H. Preparation of bismuth sulfide nanoparticles as targeted biocompatible nano-radiosensitizer and carrier of methotrexate. Appl. Organomet. Chem. 34(1), e5251 (2020).
Luo, Y. et al. In vitro cytotoxicity of surface modified bismuth nanoparticles. J. Mater. Sci. Mater. Med. 23, 2563–2573 (2012).
Colak, B. & Ertas, Y. N. Implantable, 3D-printed alginate scaffolds with bismuth sulfide nanoparticles for the treatment of local breast cancer via enhanced radiotherapy. ACS Appl. Mater. Interfaces 16(13), 15718–15729 (2024).
Chen, T., Du, J. & Lu, G. Cell growth arrest and apoptosis induced by Oct4 or Nanog knockdown in mouse embryonic stem cells: a possible role of Trp53. Mol. Biol. Rep. 39, 1855–1861 (2012).
Pitrone, M. et al. Knockdown of NANOG reduces cell proliferation and induces G0/G1 cell cycle arrest in human adipose stem cells. Int. J. Mol. Sci. 20(10), 2580 (2019).
Javani, S. et al. Metronidazole conjugated bismuth sulfide nanoparticles for enhanced X-ray radiation therapy. J. Drug Deliv. Sci. Technol. 71, 103336 (2022).
Gawlik-Rzemieniewska, N. & Bednarek, I. The role of NANOG transcriptional factor in the development of malignant phenotype of cancer cells. Cancer Biol. Ther. 17(1), 1–10 (2016).
Golrokh, F. J. et al. Apoptosis induction in colon cancer cells (SW480) by BiFe2O4@ Ag nanocomposite synthesized from Chlorella vulgaris extract and evaluation the expression of CASP8, BAX and BCL2 genes. J. Trace Elem. Med. Biol. 83, 127369 (2024).
Moradi, A. et al. Synthesize of Bi2O3/Gln-TSC nanoparticles and evaluation of their toxicity on prostate cancer cells and expression of CASP8, BAX, and Bcl-2 genes. Sci. Rep. 12(1), 21245 (2022).
Siu, M. K. et al. Overexpression of NANOG in gestational trophoblastic diseases: Effect on apoptosis, cell invasion, and clinical outcome. Am. J. Pathol. 173(4), 1165–1172 (2008).
Xu, F. et al. Nanog: A potential biomarker for liver metastasis of colorectal cancer. Dig. Dis. Sci. 57, 2340–2346 (2012).
Borrull, A. et al. Nanog and Oct4 overexpression increases motility and transmigration of melanoma cells. J. Cancer Res. Clin. Oncol. 138, 1145–1154 (2012).
Bourillot, P.-Y. et al. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells 27(8), 1760–1771 (2009).
Siu, M. K. et al. Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene 32(30), 3500–3509 (2013).
Sisin, N. N. T. et al. Radiosensitization effects by bismuth oxide nanoparticles in combination with cisplatin for high dose rate brachytherapy. Int. J. Nanomed. 14, 9941–9954 (2019).
Talik Sisin, N. N. et al. Synergetic influence of bismuth oxide nanoparticles, cisplatin and baicalein-rich fraction on reactive oxygen species generation and radiosensitization effects for clinical radiotherapy beams. Int. J. Nanomed. 15, 7805–7823 (2020).
Acknowledgements
The authors would like to thank the staff of the Medical Biotechnology Department of Zanjan University of Medical Sciences
Funding
The present study was supported by Zanjan University of Medical Sciences, Zanjan, Iran (Grant Number: A-12-1244-22).
Author information
Authors and Affiliations
Contributions
S.H.: methodology, writing – original draft. M.G.: conceptualization, methodology, investigation, software, writing – review and editing. B.J.: conceptualization, supervision, software, writing – review and editing, formal analysis, approval of final manuscript. R.G.: Methodology, H.R.: Methodology in radiotherapy.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Informed consent was obtained from all individual participants included in the study. This study was approved by the Research Ethics Committees of Zanjan University of Medical Sciences, Zanjan, Iran (Ethical Code: IR.ZUMS.REC.1400.518). Moreover, all methods were performed in accordance with the relevant guidelines and regulations.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Heidari, S., Gharbavi, M., Ghorbani, R. et al. Combined treatment using bismuth sulfide nanoparticles loaded with NANOG decoy oligodeoxynucleotides under X-ray radiation for breast cancer cells. Sci Rep 15, 23131 (2025). https://doi.org/10.1038/s41598-025-05074-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-05074-1