Abstract
Cholangiocarcinoma (CCA) is a prevalent bile duct cancer with limited treatment options. Cisplatin-based chemotherapy is a common approach, but response rates vary. Recently, chromosome aberrations have emerged as predictors of chemotherapy response in various malignancies. This study aimed to identify chromosomal aberrations associated with cisplatin response in CCA. A histoculture drug response assay was conducted on 20 fresh CCA tissues to determine cisplatin sensitivity. In parallel, chromosome aberration analysis was performed using chromosome microarray. Using chromosome microarray analysis on 20 CCA samples, we found distinct chromosomal aberration patterns between cisplatin responders and non-responders. Gains in 18q and 20q, and losses in 1p and 8p were more common in non-responders. Conversely, the responders exhibited gains in 8q and losses in 16q. Our heatmap analysis of log-2 ratio demonstrated difference in various chromosomes including 2, 3p, 5q, 6q, 7, 9p, 11q, 14q, 15q, 18q, 20q, and 22q between the response and the non-response to cisplatin. Further analysis using machine learning with random forest model identified genes like NCOA3, TPK1, CEP57L1, DEPDC5, and PLXNA4 as potential predictors of cisplatin response. Validation in a separate cohort of 33 CCA samples confirmed the association of NCOA3 and DEPDC5 with cisplatin response. Our findings suggest that chromosomal aberrations and specific genes may predict cisplatin response in CCA, potentially guiding personalized treatment strategies.
Similar content being viewed by others
Introduction
Cholangiocarcinoma (CCA) is higher incidence in Asia than in other continents, especially in Southeast Asia, Laos and Thailand, where the incidence is more than 6 per 100,000 people1. In addition, CCA is a major public health problem and has a highest incidence in the Northeast of Thailand, notably in Khon Kaen province2. It is mainly caused by eating raw freshwater fish with liver flukes from Opisthorchis viverrini and high mortality rate, which affects DNA damage and cause of cell proliferation, cell differentiation and genetic alterations, leading to promote the development of cholangiocarcinogenesis. Furthermore, the early stage of the disease usually does not have symptoms, and this cancer is mostly diagnosed at a late stage. Therefore, chemotherapy is a common treatment in CCA patient with unable to undergo surgery3. Cisplatin is one of the most common chemotherapies used for cancer treatment including CCA4. Generally, cisplatin is one of the first-line chemotherapeutic drug combination with gemcitabine in advanced stages CCA5. Although most of patients are response to cisplatin, but some still resistant to cisplatin and affects to survival time. In essence, while gemcitabine and cisplatin are a common and effective treatment, it is important to tailor the treatment plan to the specific needs of each patient. This personalized approach can help maximize the benefits of chemotherapy and improve patient outcomes. Recent data describes the impact of chromosomal abnormalities to predicting immunotherapy and chemotherapy response in numerous cancers. In addition to influencing drug response through various pathways, chromosomal abnormalities can also contribute to chromosomal instability and altered gene expression6. These are also associated with progression and aggressive disease. Previous research indicated that aneuploidy could induce complex genetic alterations affecting individual genes. These alterations have been correlated with enhanced cancer aggressiveness, increased recurrence rates, and varied drug responses7. For example, chromosomal abnormalities, such as trisomy of chromosomes 7 and 17, have been observed in CCA patients and are associated with poor prognosis and metastasis8. Furthermore, chromosomal aneuploidy and structural chromosomal abnormalities have been shown to predict chemotherapy response in various cancers, including hepatocellular carcinoma (HCC), breast invasive carcinoma, and colon and rectum adenocarcinoma6. An amplification of the 3p25.3 region has been associated with cisplatin resistance in germ cell tumors9. Marchio et al. compared the detection of chromosome 17 abnormalities associated with HER2 gene amplification using FISH and array comparative genomic hybridization (aCGH) in breast cancer. They found that aCGH microarray could detect HER2 amplification in some cases where FISH failed10. This suggests that chromosomal microarray (CMA), a newer technology, may be more sensitive in detecting chromosomal abnormalities. CMA analysis revealed large deletions within chromosomes 6q and 13q in both hepatocellular-like and cholangiocarcinoma-like areas of the combined hepatocellular-cholangiocarcinoma tumor11. Copy number variation (CNV) analysis using CMA in breast cancer biopsy specimens identified deletions in multiple ABC transporter genes associated with favorable responses to neoadjuvant chemotherapy. Patients with deletions in ABCB1, ABCB3, and ABCC5, as well as those with deletions in ABCG1, ABCG2, ABCG5, and ABCG8, were more likely to respond positively to treatment12. Previous studies have not reported the use of CMA to investigate chromosomal abnormalities in CCA, particularly in relation to predicting chemotherapy response. Therefore, the identification of specific chromosomal abnormal using CMA analysis may be a valuable tool for predicting cisplatin response in CCA patients.
This study aims to investigate the signature pattern of chromosomal abnormalities in CCA patients using CMA analysis to predict cisplatin response. A deeper understanding of these chromosomal alterations could potentially improve chemotherapeutic drug response prediction and treatment selection, leading to enhanced overall survival in CCA patients.
Results
Characteristics of CCA patients and HDRA results
This study investigated the association between chromosomal abnormalities and cisplatin response in CCA patients. The study included two cohorts of patients: 20 cases for discovery using CMA and 33 cases for validation using real-time PCR. The Discovery cohort consisted of 14 extrahepatic CCA and 6 intrahepatic CCA samples. The patients were 62.5 years old on average, with 13 males and 7 females. The validation study included 33 CCA tissue samples, with 20 male and 13 female patients. The average age was 62.5 years. All cases were diagnosed by a pathologist as either intrahepatic (9 cases) or extrahepatic (24 cases). HDRA analysis was performed on the discovery cohort to classify samples into cisplatin responders and non-responders. A cisplatin concentration of 20 µl/mg and an IR% cutoff of 39.05% were used. The results identified 8 responders and 12 non-responders. The validation cohort underwent real-time PCR analysis to validate the cisplatin response classification. These cases had been evaluated for cisplatin response, with 16 responders and 17 non-responders. Tables 1 and 2 summarize the patient characteristics and cisplatin response classifications for the discovery and validation cohorts, respectively.
Chromosome abnormalities between response and resistance to cisplatin groups
CMA analysis was performed on frozen CCA tissue samples to investigate the association between chromosomal abnormalities and cisplatin response. Both responder and non-responder groups exhibited a higher frequency of chromosome gains than losses, particularly in the cisplatin responder group, as illustrated in the karyoview (Fig. 1). In response to the cisplatin group, most chromosome abnormalities were a gain of chromosomes. Overall, the frequency of chromosomal abnormalities was lower in the cisplatin responder group compared to the non-responder group. Whilst the non-response group displayed both gain and loss abnormalities in almost the whole chromosome. Chromosomal abnormalities commonly observed in the non-responder group included gains in chromosomes 1q, 5p, 6p, 7q, 8, 9q, 14q, 17q, 18q, and 20. While some chromosomal abnormalities were shared between the two groups, including gains in 5p, 8, 18q, and 20, the non-responder group displayed distinct patterns of abnormalities. A gain in the 14q32.33 region was observed in 10 out of 20 cases (50%) in both responder and non-responder groups. Karyoview analysis further highlighted the differences in chromosomal abnormalities between the two groups. The non-responder group exhibited a higher frequency of gains in the 18q11.2q12.1 and 20q13.32q13.33 regions (5 out of 12 cases, 41.66%) and a higher frequency of losses in the 1p35.3 and 8p23.1 regions (4 out of 12 cases, 33.33%). In contrast, the responder group primarily showed gains in the 8q12.1q24.21 region and losses in the 16q22.1 region (2 out of 8 cases, 25%). While a clear pattern of chromosomal abnormalities differentiating responders and non-responders was not identified, the presence of specific chromosomal disturbances in the non-responder group suggests that chromosomal aberrations may contribute to chemoresistance in CCA. These findings, in addition to the common chromosomal abnormalities observed in both groups, highlight the complex interplay between chromosomal alterations and cisplatin response in CCA. (Fig. 1, Table S1).
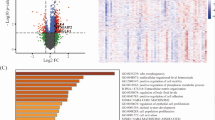
Comparative analysis of Log2 ratio levels in cisplatin responders and non-responders
Heatmap analysis was performed using Pearson, t-test, ANOVA to select the top 50 chromosome probes with the most significant differences between the cisplatin responder and non-responder groups in each chromosome. These top 50 chromosome probes of each chromosome were then analyzed using PCA, OPLS-DA, and heatmap to investigate group differentiation and clustering (Fig. 2, Table S2). The 50 selected markers, ranked by their log2 ratio differences, were further analyzed using multivariate techniques. PCA and OPLS-DA were employed to explore the potential separation of the two groups based on these probe markers. The heatmap visualization provided a visual representation of the log2 ratio differences across the selected chromosome regions (Fig. 3; Table 3). Analysis of the top 50 markers revealed significant differences in log2 ratio within multiple chromosomal regions, including 2, 3p, 5q, 6q, 7, 9p, 11q, 14q, 15q, 18q, 20q, and 22q. Chromosome 7 exhibited the highest number of differences in log2 ratio between the cisplatin responder and non-responder groups. Log2 ratios for chromosome 7 were generally higher in the non-responder group compared to the responder group, particularly in the 7q region. Both responder and non-responder groups displayed low log2 ratios for chromosomes 22q and 12q of the top 50 markers, 37 were coding genes, representing 29 genes. Notable genes with multiple chromosome probe markers included DEPDC5 (22q12.2), GLT8D2 (12q23.3), CEP57L1 (6q21), TPK1 (7q35), LINC01331 (5q13.3), and TRB (7q34).
Schematic representation of the research methodology for identifying chromosomal abnormalities associated with cisplatin response in cholangiocarcinoma patients. The flowchart outlines the steps involved, including sample collection histoculture drug response assay, chromosomal microarray analysis, data analysis, machine learning, and biomarker validation.
Random forest model to extract cisplatin response in CCA patient
Machine learning technique using random forest model was employed to develop models capable of predicting cisplatin response based on the log2 ratio levels of top 50 chromosome probe markers from previous data of heatmap analysis. A random forest model was trained using the top 50 markers with the most significant differences in log2 ratio between the response and non-response groups. The data was split into training (80%) and testing (20%) sets to evaluate model performance. The feature importance of each marker was assessed to identify those with the greatest impact on cisplatin response prediction. The top 10 markers included 5 non-coding gene regions and 5 regions involving genes: NCOA3 (20q13.12), TPK1 (7q35), CEP57L1 (6q21), DEPDC5 (22q12.3), and PLXNA4 (7q32.3). These markers were found to be statistically significantly different between the two groups (p-value < 0.001). (Fig. 4; Table 4). Analysis of log2 ratio levels in these gene panels revealed distinct patterns between responders and non-responders. NCOA3, TPK1, and PLXNA4 tended to have higher log2 ratios in the response group, while DEPDC5 and CEP57L1 exhibited higher levels in the non-response group (Fig. 5). Testing the model on the held-out test set using these gene panels resulted in 100% accuracy in predicting cisplatin response. This suggests that the differences in log2 ratio within these gene panels may serve as potential biomarkers for predicting cisplatin response in CCA patients.
Feature importance analysis of the top 50 chromosome probe markers. A random forest model was used to determine the relative importance of each marker in predicting cisplatin response. The graph represents the value of feature important of each chromosome probe marker. *The top 10 markers with the highest feature importance.
Identification of differentially expressed genes associated with cisplatin response
A gene expression panel consisting of NCOA3, TPK1, DEPDC5, and PLXNA4 was investigated to assess their association with cisplatin response in CCA patients. Real-time PCR analysis was performed on frozen tissue samples from 33 CCA patients, including 16 responders and 17 non-responders. NCOA3 expression was significantly higher in the responder group (p = 0.029), consistent with the log2 ratio data. However, DEPDC5 expression was inconsistent with the log2 ratio, as it was significantly higher in the responder group (p = 0.047) (Fig. 6). Although TPK1 and PLXNA4 were not statistically significant, their expression levels between the two groups exhibited a similar trend to the log2 ratio data. Both genes tended to have higher expression levels in the non-responder group. Overall, these findings suggest that NCOA3 and DEPDC5 may be potential biomarkers for predicting cisplatin response in CCA patients.
Gene expression analysis of NCOA3, DEPDC5, TPK1, and PLXNA4 using real-time PCR. The graph represents the mRNA expression of (A) NCOA3, (B) DEPDC5, (C) TPK1, and (D) PLXNA4 in CCA frozen tissues. Significant differences were observed in NCOA3 and DEPDC5 between responders and non-responders. TPK1 and PLXNA4 showed a trend towards differential expression between the two groups.
Discussion
Cholangiocarcinoma has a high incidence, especially in the Northeast of Thailand13 and usually has no symptoms in the early stage3. Therefore, although most of the patients can be resectable, but there are some patients diagnosed in advanced stages and unresectable14. Consequently, chemotherapy is a common treatment for CCA patients who are unresectable, and one of the most common chemotherapy treatments usually used is gemcitabine combined with cisplatin15. Unfortunately, most patients eventually become resistant to the treatment, and their tumors continue to grow within twelve months16. Recently, several chemotherapy drugs and targeted therapies have been reported to accumulate17 such as pembrolizumab (FGFR2 inhibitor)18 immunotherapy, nabpaclitaxel19 and oxaliplatin20. Therefore, predicting drug response to cisplatin might be necessary to consider alternative drug combinations with gemcitabine instead of cisplatin in cases of cisplatin non-response.
This study is the first to identify distinct chromosome abnormality patterns associated with response and non-response to cisplatin in CCA patients. Our study found that 60% (20 of 33 cases) of patients received adjuvant chemotherapy, with common regimens including cisplatin plus gemcitabine, Carboplatin, Capecitabine, 5-FU, and Gemcitabine, though 39.4% (13 of 33 cases) did not. Due to a lack of patients receiving cisplatin monotherapy, we couldn’t definitively assess the efficacy of individual chemotherapy treatments from our clinical data alone. Therefore, we grouped patients into response and non-response categories using HDRA, a method previously demonstrated to be promising in predicting clinical response in cancers such as ovarian cancer, where it exhibited an impressive 88% sensitivity and 86% specificity21. Our findings of CMA analysis revealed that the non-response group frequently exhibited chromosome amplification on chromosomes 1q, 7q, and 20, as well as loss of chromosome 9p, while these abnormalities were not observed in the response group. Previously, Andersen and Snorri indicated the genetic profiling of intrahepatic CCA using the CGH technique, they found the amplification of chromosomes 1q, 20q, and deletion of 9p. Apart from our result, they also found the amplification of chromosomes 5p, 7p, 17, and deletion of 1p, 4q, 8p, 17p, and 19q22. Interestingly, gain of 1q which contains oncogene such as MCL1 (1q21.3) and MDM4 (1q32.1)23. Gain of 7q in non-response group, consistent with previous studies showing a gain of this chromosome region and gene mutation consisting of proto-oncogene MET (7q31.2) is also associated with various cancers, including HCC24. According to loss of 9p21, it was likewise found in numerous cancers, including adenosquamous carcinoma of the pancreas, pancreatic ductal adenocarcinoma, primary sclerosing cholangitis-associated CCA, and intrahepatic CCA25,26,27. Previous studies have identified chromosome abnormalities and gene mutations associated with drug response mechanisms in various cancers. Biomarkers are being investigated to aid in selecting appropriate treatments for specific cancers, such as germ cell tumors. For example, a study on nonseminomatous germ cell tumors found that amplification of the 3p25.3 region was associated with cisplatin resistance, leading to reduced overall survival and progression-free survival compared to patients without this amplification28. In addition, ARID1A genetic alterations detected using targeted next-generation sequencing can predict whether advanced biliary tract cancer will initially resist treatment with gemcitabine and cisplatin29.
In this study, we compared the log2 ratios of individual chromosome probe markers between the cisplatin-responsive and non-responsive groups. To develop a prediction model for cisplatin response, we used a machine learning approach with a random forest model, incorporating the top 50 chromosome probe markers identified through heatmap analysis.
Our investigation demonstrated that generative predictive models are unable to achieve optimal performance using a single biomarker alone. We therefore employed features selection technique using the Recursive Feature Elimination (REF) to identify the minimal marker panel required for high accuracy predictions. We prioritized markers based on feature importance by ranking the top 10 markers with high feature importance. Then, the panel 5 markers were selected from the top 10 markers, focusing on genes that most significantly influence cisplatin response prediction. This approach would enhance clinical utility while reducing patient costs. This analysis revealed several key regions associated with cisplatin response, including NCOA3 (20q13.12), TPK1 (7q35), CEP57L1 (6q21), DEPDC5 (22q12.3), and PLXNA4 (7q32.3). Since the limitation of our study is the small sample size, which potentially increases the risk of overfitting our models. Especially when employing machine learning models such as random forest that are susceptible to sample limitations. The specific requirement for tumors to be ≥ 10 cm in diameter to allow for multi-site evaluation of cisplatin response via the HDRA technique and treatment with hepatectomy inherently led to a small cohort. While we explored and implemented strategies to address overfitting by customizing hyperparameters within our prediction model, such as the number of estimators and maximum depth, the main constraint imposed by the limited sample size could not be entirely overcome through these adjustments alone30. Therefore, the mRNA level of these gene set from predictive model was validated. The validation of mRNA expression using real-time PCR not only confirms differential expressions and abnormalities in our gene panel derived from genomic analysis but also holds substantial potential for transforming clinical practice by overcoming current turnaround time constraints in gene expression analysis. This approach can enhance efficiency and guide doctors toward more effective treatment strategies. Furthermore, the mRNA expression may lead to information for understanding the mechanisms of cancer development and developing future treatments. Hence, it can serve as a predictive biomarker, where validating gene expression associated with drug activity mechanisms or resistance genes can aid in selecting appropriate chemotherapy for personalized medicine and might be used as a targeted molecule.
In addition, we observed higher differential gene expression and log2 ratios for NCOA3, TPK1, and PLXNA4 in the non-responsive group compared to the responsive group. Previous studies have identified NCOA3 as an oncogene in thyroid cancer, and it frequently exhibits copy number gains in various cancers, including thyroid31 breast, colorectal, HCC8,31,32,33 consistent with our finding, especially in the non-response to cisplatin.
Previous reports revealed that NCOA3, TPK1, and PLXNA4 were correlated with increased tumor progression, cell migration, and cell proliferation through various mechanisms. Our results demonstrated a consistent gain in the NCOA3 log2 ratio by CMA and the elevated high NCOA3 mRNA expression detected by real-time PCR. Prior studies have established a correlation between high NCOA3 expression and tumor progression across multiple cancers, including colorectal cancer and HCC8,32,33,34. Furthermore, NCOA3 has been showed to promote and activate STAT3, influence cell proliferation, metastasis, and tumorigenesis30 and to upregulate multiple drug resistance 1 (MDR1)35 a key factor in chemotherapy resistance. The previously reported association of high NCOA3 mRNA with taxol resistance in breast cancer36 further supports the possibility that NCOA3 overexpression may contribute to cisplatin resistance, potentially through the STAT3-MDR1 pathway, consistent with our results. NCOA3 activates TERT signaling, leading to induced angiogenesis, anti-cell apoptosis, and metastasis8. Additionally, NCOA3 gains are associated with AIB1 overexpression, promoting cell proliferation and tumor progression in HCC37. Moreover, NCOA3 amplification in breast cancer has been linked to chemotherapy resistance through the upregulation of NF-κB, an inflammatory cytokine32. In this study, we found that TPK1 expression and log2 ratios were trending higher in the non-responsive group compared to the responsive group, consistent with previous findings. While TPK1 plays a role in the phosphorylation of thiamine into thiamine pyrophosphate (TPP), a coenzyme in cellular metabolism, and acts as an antioxidant, its overexpression has been observed in breast and gastric cancer, increasing cellular resistance to oxidative stress38,39,40,41. PLXNA4 encodes the plexin-A4 protein, a receptor that binds to semaphorins in neural tissue and dendritic cells. In cancer cells, plexin-A4 binding to semaphorin 6 A (Sema6A) promotes cell proliferation through VEGF and bFGF signaling, potentially influencing angiogenesis and tumorigenesis. Disruptions in plexin-A4 expression have also been implicated in cell proliferation and cell migration42,43. Whilst Balakishnan A. indicated that copy number gain of PLXNA4 in melanoma, pancreatic ductal adenocarcinoma44 and lung cancer43 and its mutation also involved in cancer progression.
Our findings revealed that although DEPDC5 has a low of log2 ratio level in both groups, especially in the non-response groups, DEPDC5 expression levels are also higher in the non-response groups. This indicated that the gene expression level was not correlated with genetic abnormalities, the studies about the mechanisms may improve better understanding. DEPDC5 is the component of the GATOR complex and is related to controlling the mTORC1 pathway through GAP activity. However, DEPDC5 displayed deletion and mutation in various cancers, including HCC, glioblastoma, and renal cell carcinoma45,46. Moreover, DEPDC5 is a member of the DEPTOR protein family, which typically inhibits the mTOR1 pathway, a key regulator of cell division. In contrast, mTOR1 also suppresses the PI3K/AKT pathway. Overexpression of DEPTOR inhibits the mTOR1 pathway and affects the increase of activity the PI3K/AKT, leading to increase cell proliferation. Therefore, DETOR overexpression may be one of the mechanisms selected that contribute to cancer cell survival by activating the PI3K/AKT47. In previous studies, Miki D. suggested that the differential single nucleotide polymorphisms (SNP) in DEPDC5 correlated to hepatocellular carcinoma risk in chronic hepatitis C virus. Still, the expression level of DEPDC5 increased in tumor tissue compared to the adjacent tissue, which may be influenced by gender48. In contrast, the cell proliferation rate was higher in knockout DEPDC5 than in wild type in the HCC cell line, and DEPDC5 expression was inversely related to p62 expression, which involved autophagy and reactive oxygen scavengers. Further research into the underlying mechanisms could provide valuable insights.
We constructed a molecular association network using the STRING database and observed pathway enrichment analysis using KEGG database49,50,51. There is pathway enrichment analysis within our genes resulting from multivariate analysis demonstrated correlations with pathway implicated in tumor progression and potential disruption of cisplatin-induced cytotoxicity. DEPDC5 and ITFG2 were associated with the mTOR1 signaling pathway, PEG10 with the TGF-β pathway, and PLXNA4 with the MAPK signaling pathway. These pathways correlation suggest potential mechanisms by which genes may contribute to tumor progression and disrupt with cisplatin-induced cell death. Moreover, our analysis identified multiple co-expression networks that may implicate promoting resistance. Through co-expression with CCND1 (Cyclin D1) and c-Myc to promote cell proliferation and co-expression with the amplification-dependent oncogene HEART6, we found that NCOA3 was associated with the Wnt and RANKL pathways. Important oncogenes c-Myc and CCND1 cooperate to encourage uncontrolled cell proliferation, which essentially enables cancer cells to outrun the effects of the drug by proliferating despite cisplatin-induced DNA damage. These substances also aid damaged cells in avoiding apoptosis, which keeps them from dying. Because they are strong transcription factors, they can enhance DNA repair pathways and upregulate other genes involved in drug efflux or other survival mechanisms, reducing the effectiveness of cisplatin’s DNA-damaging effects. Additionally, being potent transcription factors, they have the ability to upregulate other genes involved in drug efflux or other survival mechanisms, and their activity can improve DNA repair pathways, decreasing the effectiveness of cisplatin’s DNA-damaging impact. While PLXNA4 and SEMA6B exhibited associations with the MAPK signaling pathway. Moreover, TPK1 showed co-expression with THTPA and HEBP2, which function as antioxidants in thiamine metabolism, which is a potential mechanism where these antioxidant activities could reduce intracellular oxidative stress. These identified pathways and gene interactions provide critical insights into the complex cellular adaptations that lead to cisplatin resistance.
In summary, our analysis identified distinct patterns of chromosome abnormalities and gene expression levels (e.g., NCOA3, DEPDC5) between cisplatin-non-responders and responders in CCA patients. These findings suggest that these genetic markers could potentially serve as predictors of cisplatin response. Our findings demonstrate promising diagnostic potential for both CMA and gene expression testing, several practical considerations must be addressed before clinical integration. These include cost-effectiveness analysis, laboratory infrastructure requirements, standardization of protocols, turnaround time optimization, and healthcare provider training. Detecting these chromosomal region alterations and gene expression changes in individual CCA patients before treatment could help predict their response to cisplatin, leading to more effective treatment and improved survival rates. Targeting these genes or chromosomal regions may offer promising avenues for improving treatment outcomes in CCA patients who are resistant to cisplatin.
Methods
Ethical considerations, sample, and clinical data collection
Fresh and frozen CCA tissues obtained from patients who had undergone surgery and preserved in specimen bank of the cholangiocarcinoma Research Institute after were diagnosed with CCA at Srinagarind Hospital, Khon Kaen University. Frozen tissue samples and clinicopathological data of CCA patients were collected and assessed between January 1, 2017, and September 30, 2023. Data collection was completed on December 31, 2023. To maintain participant anonymity, the Cholangiocarcinoma Research Institute blinded identifiable information by replacing hospital numbers (HN) and other personal details with unique index numbers. This ensured that the authors only had access to de-identified data, preventing the identification of individual participants. The informed consent was obtained from all subjects or their legal guardians. Frozen tissue samples from 20 cases were analyzed using CMA to investigate chromosomal abnormalities. In addition, 33 cases were evaluated using real-time polymerase chain reaction (Real-time-PCR) to investigate gene expression. Furthermore, all cases of our study assessed the response to cisplatin treatment using histoculture drug response assay (HDRA) technique. All methods in this study were performed in accordance with relevant guidelines and regulations with the principles of the Declaration of Helsinki and approved by Ethics Committee for Human Research, Khon Kaen University (HE641237).
Histoculture drug response assay results
The protocol of HDRA has previously reported52. In brief, the tumor tissues were separated aseptically chopped from main tumor 10 mg pieces. The tumor specimen was placed on sponge gel and cultured in a 24-well plate with RPMI-1640 medium (Thermo Fisher Scientific, Waltham, USA.) with present and absent of 20 µg/ml cisplatin (Fresenius SE & Co. KGaA, Bad Homburg, Germany) for 4 days at 37˚C, 5% CO2. Then, the cell viability was determined by MTT assay (Sigma Aldrich, St. Louis, MO, USA.). The activity of MTT assay was measured at an absorbance of 540 nm. The chemotherapy efficacy for each well was evaluated with the percentage inhibition (% inhibition rate; % IR). Patients were categorized into cisplatin response and non-response groups based on a %IR cutoff of 39.05%.
Chromosome microarray technique
A total of 20 cases of frozen CCA tumor tissue were extracted DNA using QIAamp Fast DNA tissue kit: 51,404 according to manufacturer’s recommendations. DNA concentrations were measured by Nanodrop spectrophotometer, which has a concentration of more than 50 µg/ml to be sufficient for analysis with CMA technique. Processing of DNA samples in Affymetrix CytoScan® 750 K (Affymetrix CytoScan® 750 K Array, ATLAS Biolabs, Berlin, Germany) was performed according to the manufacturer’s protocols (www.affymetrix.com).
Analysis of chromosome microarray
The copy number variation and intensities of each marker probe hybridization were analyzed using Affymetrix Chromosome Analysis Suite (ChAS) with the default setting. The difference of intensity level in each chromosome marker probe representing as log2 ratio data between CCA with response and non-response was analyzed using Principal component analysis (PCA) and Orthogonal partial least squares discriminant analysis (OPLS-DA) by MetaboAnalyst 5.0, which is web-based tool multivariate analysis. In addition, heatmap analysis was used to clustering the group of response and non-response in each chromosome probe using MetaboAnalyst 5.0. The analysis workflow of this study demonstrated as Fig. 2.
Machine learning analysis by random forest model
The top 50 chromosome probes identified through heatmap analysis were further investigated for their predictive efficiency using a random forest model on GitHub website. The log2 ratios of these probes were analyzed to establish a cisplatin response prediction model. Feature importance was determined using tree-based feature importance within the random forest model to identify the chromosome probes with the greatest impact on cisplatin response prediction. Probes with high feature importance values were selected to construct the prediction model. CCA samples were randomly split into training (80%) and testing (20%) sets. The training set was used to train the model using the log2 ratios of the 50 selected chromosome probes (Table S3, S4).
Real-time PCR
The difference of gene expression level was measured mRNA by real-time RT PCR using. The 20 mg of all 33 Frozen CCA tissues were extracted RNA using PureLink RNA Mini Kit (12183018 A, Invitrogen, CA, USA) with standard protocol. In brief, the CCA frozen tissues were added lysis buffer and pestle by plastic pestle until the tissue is the same consistency as the solution. Centrifuge and discard supernatant, the specimen was added 70% ethanol to binding RNA. Then, DNA and protein were eliminated by wash buffer and RNase-free water was elute RNA follow by measure concentration and quality RNA with Nanodrop. Next, RNA was diluted to 50 ng/µl before reverse mRNA to cDNA with reverse transcription kit (4368814, Themo fisher scientific, Vilnius, Lithuania) with Bio-Rad C1000 PCR Thermal Cycler 96-Well Fast Block and incubate in PCR 2 h. Then, the cDNA plate adds TaqMan Probe with 4 genes including NCOA3, DEPDC5, PLXNA4, and TPK1. The mRNA probes for 4 targets gene expression using TaqMan assay (NCOA3 #cat No. 4351372, DEPDC5 #cat No. 4351372, PLXNA4 #cat No. 4351372, and TPK1 #cat No. 4351372; Applied Biosystem, USA.) (Table S5).
Statistical analysis of real-time PCR
The relative mRNA expression data were analyzed using SPSS 18.0 statistical software (SPSS Inc., Chicago, IL). The Mann-Whitney U test was used to determine if there was a statistically significant difference between two groups of samples. A p-value of less than 0.05 was considered statistically significant.
Data availability
All data analysed during this study are included in this published article and its supplementary information files are available in the additional files.
Abbreviations
- CCA:
-
Cholangiocarcinoma
- CIS:
-
Cisplatin
- CMA:
-
Chromosome microarray
- HDRA:
-
Histoculture drug response assay
- CNV:
-
Copy number variation
- HCC:
-
Hepatocellular carcinoma
- iCCA:
-
Intrahepatic Cholangiocarcinoma
- LOH:
-
Loss of heterozygosity
- OPLS-DA:
-
Orthogonal partial least squares-discriminant analysis
- PCA:
-
Principal Component Analysis
References
Virani, S. et al. National and subnational Population-Based incidence of Cancer in thailand: assessing cancers with the highest burdens. Cancers (Basel). 9 https://doi.org/10.3390/cancers9080108 (2017).
Sripa, B. et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 4, e201. https://doi.org/10.1371/journal.pmed.0040201 (2007).
Banales, J. M. et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 17, 557–588. https://doi.org/10.1038/s41575-020-0310-z (2020).
Chen, S. H. & Chang, J. Y. New insights into mechanisms of cisplatin resistance: from tumor cell to microenvironment. Int. J. Mol. Sci. 20 https://doi.org/10.3390/ijms20174136 (2019).
Rogers, J. E. et al. Second-line systemic treatment for advanced cholangiocarcinoma. J. Gastrointest. Oncol. 5, 408–413. https://doi.org/10.3978/j.issn.2078-6891.2014.072 (2014).
Kou, F., Wu, L., Ren, X. & Yang, L. Chromosome abnormalities: new insights into their clinical significance in Cancer. Mol. Ther. Oncolytics. 17, 562–570. https://doi.org/10.1016/j.omto.2020.05.010 (2020).
Shukla, A. et al. Chromosome arm aneuploidies shape tumour evolution and drug response. Nat. Commun. 11, 449. https://doi.org/10.1038/s41467-020-14286-0 (2020).
Li, W. et al. Targeting the NCOA3-SP1-TERT axis for tumor growth in hepatocellular carcinoma. Cell. Death Dis. 11, 1011. https://doi.org/10.1038/s41419-020-03218-x (2020).
Timmerman, D. M. et al. Chromosome 3p25.3 gain is associated with cisplatin resistance and is an independent predictor of poor outcome in male malignant germ cell tumors. 40, 3077–3087, (2022). https://doi.org/10.1200/jco.21.02809
Marchio, C. et al. Does chromosome 17 centromere copy number predict polysomy in breast cancer? A fluorescence in situ hybridization and microarray-based CGH analysis. J. Pathol. 219, 16–24. https://doi.org/10.1002/path.2574 (2009).
Wilhelm, A. B. et al. Pediatric combined hepatocellular-cholangiocarcinoma (cHCC-CC) with neuroendocrine features: distinguishing genetic alterations detected by chromosomal microarray. Diagn. Pathol. 18 https://doi.org/10.1186/s13000-023-01305-z (2023).
Litviakov, N. V. et al. Deletions of multidrug resistance gene loci in breast cancer leads to the down-regulation of its expression and predict tumor response to neoadjuvant chemotherapy. Oncotarget 7, 7829–7841. https://doi.org/10.18632/oncotarget.6953 (2016).
Alsaleh, M. et al. Cholangiocarcinoma: a guide for the nonspecialist. Int. J. Gen. Med. 12, 13–23. https://doi.org/10.2147/IJGM.S186854 (2019).
Tawarungruang, C. et al. Survival after surgery among patients with cholangiocarcinoma in Northeast Thailand according to anatomical and morphological classification. BMC Cancer. 21, 497. https://doi.org/10.1186/s12885-021-08247-z (2021).
Ramírez-Merino, N. Chemotherapy for cholangiocarcinoma: an update. World J. Gastrointest. Oncol. 5 https://doi.org/10.4251/wjgo.v5.i7.171 (2013).
Yoo, C., Hyung, J. & Chan, S. L. Recent advances in systemic therapy for advanced intrahepatic cholangiocarcinoma. Liver Cancer. 13, 119–135. https://doi.org/10.1159/000531458 (2024).
Harding, J. J., Khalil, D. N. & Fabris, L. Abou-Alfa, G. K. Rational development of combination therapies for biliary tract cancers. J. Hepatol. 78, 217–228. https://doi.org/10.1016/j.jhep.2022.09.004 (2023).
Kelley, R. K. et al. Pembrolizumab in combination with gemcitabine and cisplatin compared with gemcitabine and cisplatin alone for patients with advanced biliary tract cancer (KEYNOTE-966): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 401, 1853–1865. https://doi.org/10.1016/S0140-6736(23)00727-4 (2023).
Niger, M. et al. A phase II/III randomized clinical trial of cisplatin plus gemcitabine and Nabpaclitaxel (GAP) as pReoperative chemotherapy versus immediate resection in patients with resectable biliary tract cancers (BTC) at high risk for recurrence: PURITY study. BMC Cancer. 24, 436. https://doi.org/10.1186/s12885-024-12225-6 (2024).
Moris, D. et al. Advances in the treatment of intrahepatic cholangiocarcinoma: an overview of the current and future therapeutic landscape for clinicians. CA Cancer J. Clin. 73, 198–222. https://doi.org/10.3322/caac.21759 (2023).
Hoffman, R. M., Jung, P. S., Kim, M. B. & Nam, J. H. in 3D Sponge-Matrix Histoculture: Methods and Protocols (ed Robert M. Hoffman) 73–81Springer New York, (2018).
Andersen, J. B. & Thorgeirsson, S. S. Genetic profiling of intrahepatic cholangiocarcinoma. Curr. Opin. Gastroenterol. 28, 266–272. https://doi.org/10.1097/MOG.0b013e3283523c7e (2012).
Liao, H. et al. Whole-Genome DNA methylation profiling of intrahepatic cholangiocarcinoma reveals prognostic subtypes with distinct biological drivers. Cancer Res. 84, 1747–1763. https://doi.org/10.1158/0008-5472.CAN-23-3298 (2024).
Zhang, Y. et al. The genomic landscape of cholangiocarcinoma reveals the disruption of post-transcriptional modifiers. Nat. Commun. 13, 3061. https://doi.org/10.1038/s41467-022-30708-7 (2022).
Ahrendt, S. A. et al. Chromosome 9p21 loss and p16 inactivation in primary sclerosing cholangitis-associated cholangiocarcinoma. J. Surg. Res. 1, 88–93. https://doi.org/10.1006/jsre.1999.5615 (1999).
Jiang, Y. et al. Loss of chromosome 9p21 is associated with a poor prognosis in adenosquamous carcinoma of the pancreas. Precis Clin. Med. 6, pbad030. https://doi.org/10.1093/pcmedi/pbad030 (2023).
Sia, D. et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology 144, 829–840. https://doi.org/10.1053/j.gastro.2013.01.001 (2013).
Timmerman, D. M. et al. Chromosome 3p25.3 gain is associated with cisplatin resistance and is an independent predictor of poor outcome in male malignant germ cell tumors. J. Clin. Oncol. 40, 3077–3087. https://doi.org/10.1200/JCO.21.02809 (2022).
Lee, S. H. et al. ARID1A mutation from targeted Next-Generation sequencing predicts primary resistance to gemcitabine and cisplatin chemotherapy in advanced biliary tract Cancer. Cancer Res. Treat. 55, 1291–1302. https://doi.org/10.4143/crt.2022.1450 (2023).
Han, S., Williamson, B. D. & Fong, Y. Improving random forest predictions in small datasets from two-phase sampling designs. BMC Med. Inf. Decis. Mak. 21, 322. https://doi.org/10.1186/s12911-021-01688-3 (2021).
Spiliopoulou, P. et al. All is not lost: learning from 9p21 loss in cancer. Trends Immunol. 43, 379–390. https://doi.org/10.1016/j.it.2022.03.003 (2022).
Lira, M. C. et al. NCoA3 upregulation in breast cancer–associated adipocytes elicits an inflammatory profile. Oncol. Rep. 49 https://doi.org/10.3892/or.2023.8542 (2023).
Habermann, J. K. et al. Genomic instability and oncogene amplifications in colorectal adenomas predict recurrence and synchronous carcinoma. Mod. Pathol. 24, 542–555. https://doi.org/10.1038/modpathol.2010.217 (2011).
Li, Y. et al. NCOA3 is a critical oncogene in thyroid cancer via the modulation of major signaling pathways. Endocrine 75, 149–158. https://doi.org/10.1007/s12020-021-02819-6 (2022).
Ku, J. M. et al. SH003 overcomes drug resistance and immune checkpoints by inhibiting JAK-STAT3 signaling in MCF7/ADR cells. Phytomedicine Plus. 1, 100111. https://doi.org/10.1016/j.phyplu.2021.100111 (2021).
Ao, X. et al. Decreased expression of microRNA-17 and microRNA-20b promotes breast cancer resistance to taxol therapy by upregulation of NCOA3. Cell. Death Dis. 7, e2463. https://doi.org/10.1038/cddis.2016.367 (2016).
Xu, Y. et al. Overexpression of transcriptional coactivator AIB1 promotes hepatocellular carcinoma progression by enhancing cell proliferation and invasiveness. Oncogene 29, 3386–3397. https://doi.org/10.1038/onc.2010.90 (2010).
Hong, J. Y. et al. TPK1 as a predictive marker for the anti-tumour effects of Simvastatin in gastric cancer. Pathol. Res. Pract. 216, 152820. https://doi.org/10.1016/j.prp.2020.152820 (2020).
Zastre, J. A., Sweet, R. L., Hanberry, B. S. & Ye, S. Linking vitamin B1 with cancer cell metabolism. Cancer Metabolism. 1, 16. https://doi.org/10.1186/2049-3002-1-16 (2013).
Jonus, H. C. et al. The adaptive regulation of thiamine pyrophosphokinase-1 facilitates malignant growth during supplemental thiamine conditions. Oncotarget 9, 35422–35438. https://doi.org/10.18632/oncotarget.26259 (2018).
Kim, S. et al. Bioinformatic and metabolomic analysis reveals miR-155 regulates thiamine level in breast cancer. Cancer Lett. 357, 488–497. https://doi.org/10.1016/j.canlet.2014.11.058 (2015).
Kigel, B., Rabinowicz, N., Varshavsky, A., Kessler, O. & Neufeld, G. Plexin-A4 promotes tumor progression and tumor angiogenesis by enhancement of VEGF and bFGF signaling. Blood 118, 4285–4296. https://doi.org/10.1182/blood-2011-03-341388 (2011).
Toledano, S. & Neufeld, G. Plexins as regulators of Cancer cell proliferation, migration, and invasivity. Cancers (Basel). 15 https://doi.org/10.3390/cancers15164046 (2023).
Balakrishnan, A. et al. Molecular profiling of the plexinome in melanoma and pancreatic cancer. Hum. Mutat. 30, 1167–1174. https://doi.org/10.1002/humu.21017 (2009).
Caruso, S., O’Brien, D. R., Cleary, S. P., Roberts, L. R. & Zucman-Rossi, J. Genetics of hepatocellular carcinoma: approaches to explore molecular diversity. Hepatology 73 (Suppl 1), 14–26. https://doi.org/10.1002/hep.31394 (2021).
Liu, S. V. et al. Genomic analysis and selected molecular pathways in rare cancers. Phys. Biol. 9, 065004. https://doi.org/10.1088/1478-3975/9/6/065004 (2012).
Caron, A., Briscoe, D. M., Richard, D. & Laplante, M. DEPTOR at the Nexus of cancer, metabolism, and immunity. Physiol. Rev. 98, 1765–1803. https://doi.org/10.1152/physrev.00064.2017 (2018).
Miki, D. et al. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat. Genet. 43, 797–800. https://doi.org/10.1038/ng.876 (2011).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677. https://doi.org/10.1093/nar/gkae909 (2025).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Suksawat, M. et al. In vitro and molecular chemosensitivity in human cholangiocarcinoma tissues. PLoS One. 14, e0222140. https://doi.org/10.1371/journal.pone.0222140 (2019).
Acknowledgements
We would like to acknowledge Department of Pathology, Faculty of Medicine, and Cholangiocarcinoma Research Institute, Khon Kaen University, Khon Kaen, Thailand to support the patient information and tissue samples. This research was supported by NSRF under the Basic Research Fund of Khon Kaen University through Cholangiocarcinoma Research Institute.
Author information
Authors and Affiliations
Contributions
Conceptualization was performed by M.T., S.T., and S.W. Formal analysis by M.T., S.T. and Investigation by S.T., S.W., M.S., K.C., S.S., K.W., M.T. Project administration by M.T. , while S.T. and M.T. wrote the original manuscript text and prepared all figures. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
The methods used in this study follow the guidelines of the Declaration of Helsinki and ICH Good Clinical Practice (ICH GCP) guidelines. This study was approved by The Ethics Committee for Human Research, Khon Kaen University, Thailand and signed informed consent was obtained from all patients.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Techa-ay, S., Watcharadetwittaya, S., Deenonpoe, R. et al. Predicting cisplatin response in cholangiocarcinoma patients using chromosome pattern and related gene expression. Sci Rep 15, 20420 (2025). https://doi.org/10.1038/s41598-025-06851-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-06851-8