Abstract
Viruses are the primary etiological agents responsible for numerous mosquito-borne diseases, which pose a global threat to humans and their carriers. In this study, we conducted a comprehensive analysis of the simultaneous presence of different viruses in a total of 237 transcriptomic datasets derived from Aedes aegypti (AA), obtained from the SRA and ENA databases to understand its impact on transmission of arboviruses such as Dengue, Zika, and Chikungunya to humans. We investigated the co-occurrence patterns of 21 distinct viruses, encompassing 52 isolates, which are known to be transmitted by mosquitoes. In addition, we used heatmaps, correlation coefficients, estimated count values, and differential gene expression to examine the viral abundance and the significant genes involved in viral co-transmission. We provide novel evidence of a robust antagonistic relationship between the Chikungunya virus and flaviviruses during their co-transmission in AA. Our study confirms that the genes involved in RNAi, ATP-binding, and venom allergen pathways are the pivotal vector interaction hubs in response to viral co-transmission in AA. For the first time, we highlight the critical role of the DDX5 gene in modulating responses to the relative loads of flaviviruses and togaviruses, which could determine the transmission dominance of one over the other.
Similar content being viewed by others
Introduction
Mosquito species such as Aedes aegypti (AA), Anopheles gambiae, and Culex pipiens have been widely acknowledged as carriers of numerous devastating disease-causing viruses. Several prominent examples include the Zika virus (ZIKV), the Chikungunya virus (CHIKV), the Dengue virus (DENV), the West Nile virus (WNV), and the Yellow fever virus (YFV)1. Various factors, including temperature, humidity, the presence of vertebrate hosts, the density of the vector population, the survival rate of mosquitoes, and the capacity of the virus to replicate, might collectively impact the vector’s ability to acquire a disease and efficiently transmit it to a vulnerable host2,3,4,5,6,7,8. Certain viruses, such as ZIKV, DENV and CHIKV, have the ability to replicate within Aedes mosquitoes subsequent to the acquisition from an infected host during an ensuing blood meal5. On the other hand, several viruses such as the Rift Valley fever virus (RVFV) do not replicate inside the vector, but rather utilize it solely for mechanical transmission. Yet some viruses are just transient in nature and neither interact with the vectors nor are transmitted by them3.
The vector-borne viruses must contend with innate immune responses of the vector and cross multiple tissue barriers linked to the midgut and salivary glands to systemically infect the vector9,10,11,12,13. The Toll, IMD (Immune Deficiency), RNA interference (RNAi), and JAK/STAT pathways are crucial processes associated in the immune response against viral infections14,15,16. Mosquitoes possess pattern recognition receptors that enable the detection of viral components, including double-stranded RNA. Additionally, they synthesize antimicrobial peptides, such as defensins, that disrupt the integrity of viral membranes and impede the process of viral entrance into host cells17. When multiple viruses coexist within the same vector, some may engage in resource competition or contend for vector competency, while others might cooperate in ways that enhance their transmissibility18,19,20,21. For example, many viruses such as WNV, Eastern Equine Encephalitis Virus (EEEV), DENV, or CHIKV co-occur inside mosquito vectors22,23,24. Emerging studies indicate that the coexistence of DENV and ZIKV within a shared mosquito vector might potentially leading to reciprocal effects on their respective replication abilities25. This overlap increases the risk of co-infections in humans and raises important questions about virus–virus and virus–vector interactions26,27,28. Instances of co-infection involving DENV and CHIKV have been documented in countries such as India and Colombia29,30. These epidemiological studies help identify patterns, causes, and risk factors of viral infections across populations, which aid in early detection and outbreak prevention, ultimately guiding effective control measures and health policies31. Furthermore, it was discovered that co-infection with these viruses had no effect on the mosquito’s ability to disseminate them32,33.
In this study, we hypothesized that viral co-infections may modulate gene expressions in the mosquito and potentially influence the efficiency of viral co-transmission. To test this, we conducted meta-analysis on RNA-seq datasets (totaling 237) of AA from the public repositories. Specifically, we aimed to detect the presence of 21 different viruses known to be transmitted by AA, quantify their abundance, and analyze patterns of co-occurrence. Our data showed synergistic co-transmission patterns for flaviviruses and antagonistic behavior for flavi- and togaviruses. We further examined the epidemiological data from India, Colombia and the Americas to correlate the transcriptomic insights with regional patterns of viral spread. Our results reveal how viral co-infections influence mosquito biology and virus transmission, which may aid targeted surveillance and control efforts.
Results
Viral prevalence and its correlation
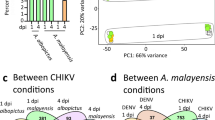
The analysis of 237 RNA-seq datasets of AA, Although 21 viral species were screened, only five were detected in a set of 149 libraries, while the rest were completely devoid of any virus. It is worth mentioning that the ZIKV was present in all the datasets that showed viral presence; nevertheless, there was a discernible variation in its abundance across the samples. The CHIKV was co-discovered alongside ZIKV in a limited number of libraries, though their ECs were significantly lower. The correlation matrix provided supporting evidence that there is negative association between CHIKV and the other members of flaviviruses, especially DENV and ZIKV (Fig. 2). The presence of Usutu virus (UV) and JEV, however, was observed in a limited number of libraries, often in conjunction with DENV and ZIKV (Fig. 1). The simultaneous presence of ZIKV and DENV was evident in multiple datasets, though their association was weak. However, there was synergism between DENV with UV and JEV (Fig. 2). YFV, as documented in the literature, was not detected in any of the libraries21,34,35.
Epidemiological data analysis
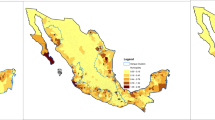
Considering the observed antagonism between DENV and CHIKV, we examined the epidemiological data on Dengue and Chikungunya cases in India from 2018 to 2023, given their high prevalence in the region. An investigation of the epidemiological data collected from the National Center for Vector Borne Diseases Control (NCVBDC), Ministry of Health & Family Welfare, Government of India (https://ncvbdc.mohfw.gov.in/index.php), reveals a clear pattern: Dengue cases outweigh Chikungunya in all the Indian states (Fig. 3). Curiously, in 2006, there was a massive Chikungunya epidemic in India, affecting thirteen states and 1.4 million people36,37, whereas dengue incidences were much lower 12,317.
shows the distribution of Dengue (in blue) and Chikungunya (in red) cases across Indian states from 2018 to 2023. The x-axis represents the different regions of India, while the y-axis indicates the respective number of cases. The graph shows that chikungunya cases were low whenever dengue cases were high.
The results had strong statistical support, as evidenced by the robust p-values obtained from the t-tests for DENV and CHIKV across all years (2018–2023), with the observed p-values ranging from 5.63 e-06 to 0.002 (Supplementary Table 3), which reflect a persistent and statistically significant difference in the average number of instances between the two diseases. This is consistent with DENV’s prevelance, as it is known for its higher transmission rates and wider geographical dispersion than CHIKV. Similarly, according to epidemiological bulletins from the National Health Institute (Instituto Nacional de Salud) of Colombia, in 2015, a total of 4,54,463 DENV and 3,59,728 CHIKV cases were reported, indicating a higher prevalence of DENV during the period of co-circulation. In 2016, both infections showed a substantial decline, with 103,822 DENV and 19,556 CHIKV cases; however, DENV remained predominant38. Between September 2014 and January 2015, Chikungunya incidence rates in Ovejas and Corozal of Columbia were found to be significantly higher (12,621 and 1,640 cases per 100,000 inhabitants, respectively). Conversely, during the corresponding time frame, the cumulative incidence of dengue in Colombia was 420 cases per 100,000 individuals39,40. From epidemiological weeks (EW) 1 to 53 of 2020, a total of 2,457,864 cases of arboviral diseases were reported across the Americas. Among these, DENV infections constituted the vast majority, accounting for 2,331,840 cases, or 94.9% of the total. In contrast, CHIKV infections were substantially lower, with 103,046 recorded cases, while ZIKV infections accounted for 22,978 ones, representing 4.2% and 0.9% of the overall burden, respectively (Source: Health Information Platform for The Americas (PLISA, PAHO/WHO) by the Ministries and Institutes of Health of the countries and territories of the Region, https://www.paho.org/plisa). A previous outbreak study of arboviruses in Thailand, which analyzed 1,806 serum samples from a chikungunya cohort, identified a very low incidence of co-infection (~ 0.7%), including DENV/CHIKV (8 cases; ~0.4%), ZIKV/CHIKV (3 cases; ~0.2%), and DENV/ZIKV (2 cases; ~0.1%)41. These observations underscore the consistent epidemiological dominance of Dengue over Chikungunya in both national and regional contexts.
DGE and GO enrichment analysis
In total, 77 gene expressions were quantified for 237 datasets, revealing that the gene expression profile of AA in response to the virus significantly differed from that of the control group. Four different situations were compared to the control: sole togaviral infection (TI), sole flaviviral infection (FI), flaviviral co-occurrence (FC), and flavi- and togaviral co-occurrence (FTC). Remarkably, only a tiny subset of genes was discovered to be significantly regulated when comparing each of these circumstances directly to the control Fig. 4.
The figure shows the up- and down-regulated genes in four different conditions. A total of nine and seven significant genes in FC (up-regulated) and FTC (down-regulated) conditions, respectively, could be observed among the set of 77 differentially expressed genes, which were analyzed. Genes were considered significant based on an adjusted p-value < 0.05 and |log2 fold change| > 1, using uninfected mosquitoes as the control group.
KEGG pathway analysis42,43,44 identified several genes, viz., 5572553, 5579076, 5574165, and 5575707 are associated with the spliceosome pathway (aag03040), and 5569076, 5571006, 5573872, and 5569796 belong to the nucleocytoplasmic transport (aag03013). Gene 5569076 (GO id), which is involved in nucleocytoplasmic transport and spliceosome function, showed increased expression in both FI and TI conditions but not in FTC. In a related study of AA, genes 5567877 was identified as a venom allergens and exhibited downregulation under FTC conditions45. Furthermore, genes 5569076 and 5571006 were associated with hydrolase activity and RNA helicase activity, found to be downregulated in the FTC condition46. STAT (5572256) was upregulated under FI conditions, highlighting its established role in mounting antiviral immune responses. A gene that is part of the Spliceosome pathway (5573872) was elevated in the FC condition, but a different gene that is also part of the same pathway (5571006) was downregulated in the FTC, suggesting that the genes related to spliceosomes are differentially regulated in response to distinct viral infection situations. Furthermore, FTC showed upregulation of the gene 5579924 (Dicer), which is an essential component of the siRNA pathway and an antiviral defence against viruses47. The zinc finger and BTB domain-containing protein 17-like (110675125) is upregulated in the FTC condition, indicating a possible role in host immune modulation and antiviral response48. Gene “eye shut” (5569703) was upregulated in all four conditions. An uncharacterized gene (5576948) was downregulated in both FC and FTC conditions. GO analysis of upregulated genes across all four conditions identified the following enriched biological process term: “GO:0006357 - regulation of transcription by RNA polymerase II,” and the molecular functions for both upregulated and downregulated genes were identical, including “GO:0003724 - RNA helicase activity,” “GO:0016787 - hydrolase activity,” “GO:0005524 - ATP binding”49, ”GO:0003723 - RNA binding,” “GO:0004386 - helicase activity,” and “GO:0003676 - nucleic acid binding.” Additionally, the upregulated genes were characterized by an extra term: “GO:0000978 - RNA polymerase II cis-regulatory region sequence-specific DNA binding.” The cellular components identified for upregulated genes include “GO:0005634 - nucleus” and “GO:0005737 - cytoplasm,” while for downregulated genes, the identified component is “GO:0010494 - cytoplasmic stress granule”.
The biological network (Fig. 5) of differentially expressed genes in different conditions revealed the small nuclear ribonucleoprotein U2 (AAEL003605), which is increased in the TI condition, as a key hub gene with a degree of 5. The U2 protein has been implicated in interactions with DDX5 in Saccharomyces cerevisiae and Drosophila melanogaster, as well50. The DDX5 isoform (AAEL010787) had a degree of 4, while DDX18 (AAEL005744) and the helicase (AAEL003968) showed network degrees of 5 each and a betweenness centrality of 0.5. Another helicase, UAP56/SUB2 (AAEL001216), which showed a degree of 5 and betweenness of 0.5, was upregulated in TI and downregulated in FTC. Interestingly, DDX5, along with DDX18 and the helicase (AAEL003968), which are upregulated in the FC condition, have a direct interaction with U2, suggesting a coordinated role in RNA processing. Curiously, another DDX5 isoform (AAEL008738), which is downregulated in the FTC condition, also interacts with the U2 protein, indicating that different isoforms of DDX5 might have context-specific regulatory roles in splicing or RNA metabolism across infection types. No interactions were found for the genes AAEL018055, AAEL024064, AAEL021082, AAEL019658, and AAEL009230 based on the STRING network analysis.
The interaction network of differentially expressed genes spanning the four conditions. Those genes that are differentially expressed across multiple conditions are colored in purple. Only genes with an adjusted p-value < 0.05 are colored; the rest of the nodes are shown in pastel green. All nodes are of the same size and each represents a distinct gene. The edges indicate known or predicted gene-gene interactions based on STRING database. The up-regulated genes of FTC are shown in red, while the down-regulated ones are colored in coral. The up-regulated genes of FC are depicted in green, TI in pink, and FI in blue.
Discussion
Humans have become co-infected with many viruses because of epidemiological synergy between viruses transmitted by AA, including the ZIKV and DENV22,51,52. Our results indicate that ZIKV (GenBank Acc. KY766069.1) can co-occur with either DENV (GenBank Acc. U87411.1) or CHIKV (GenBank Acc. AF369024.2). However, interestingly, all three viruses were hardly observed together in the datasets under study. The two are primarily spread by AA, co-circulate in the same geographic areas, and are increasingly causing co-infections in humans22. For example, DENV and CHIKV are known to co-circulate in Delhi region, leading to overlapping outbreaks. Although both viruses are present in the same environment, actual co-infection within a single mosquito is uncommon53. DENV-3 showed that co-infection depends on the relative viral loads; DENV-3 could suppress CHIKV when present at higher loads. Co-infection was only established when CHIKV had a higher titer than DENV-3 and both were introduced simultaneously, while super-infections failed to result in dual infection54. However, our findings indicate that such co-infections likely result from bites by different mosquitoes rather than simultaneous transmission from one. While co-transmission of ZIKV and CHIKV is possible, it remains relatively rare in global datasets55. Additionally, evidence from another study suggests that ZIKV, CHIKV, and DENV-2 can co-circulate within the same mosquito populations, although the prevalence varies regionally. Co-infection in mosquitoes has been reported but at relatively low rates (7.1–8.3%)56. These findings align with our study, which demonstrates that flaviviruses are strongly co-transmitted57, while their spread is negatively correlated with CHIKV.
Further, the statistical results of the epidemiological data from Indian territories between 2018 and 2023, where both dengue and chikungunya are quite prevalent, consistently support the hypothesis that DENV exhibits a greater capacity for survival and dissemination within the vector compared to CHIKV. Multiple virus transmission and susceptibility to viral co-infection in mosquitoes can be influenced by various factors, including the mosquito’s genetic makeup and immune response58. Prior infection with a particular virus may confer partial immunity to related viruses in certain circumstances59. We believe that one or more such factors might render CHIKV less potent than DENV and other flaviviruses in AA. The results of our investigation corroborate the hypothesis that DENV boasts a superior capacity for survival and dissemination within the vector in comparison to CHIKV. This is further supported by the epidemiological data from different countries. Additionally, there is an antagonistic interaction between the two viruses as supported by a controlled in vitro investigation of the concurrent infection of AA with DENV, CHIKV, and ZIKV60. The study revealed a significant decrease in the transmission rates of CHIKV when mosquitoes were co-infected with DENV, as opposed to mosquitoes that were only exposed to CHIKV. The prevalence of DENV can be attributed to several causes, including the extended presence of the virus in infected humans, which heightens the probability of transmission to mosquitoes and, consequently, to new hosts. Occasionally, viruses that are transmitted by the same mosquito vector may contend for scarce resources, including nutrients or host cell apparatus, which can have an impact on the success of their survival. Due to the limited availability of entomological and environmental data, their influence on epidemiological outcomes could not be directly evaluated. Further studies integrating these parameters with disease incidences may aid comprehensive understanding of the mechanisms driving the predominance of DENV in the epidemiological landscape.
An analysis of the differentially regulated AA genes reveals that they are associated with essential antiviral defense. The salivary proteins of AA, LOC5566624, and LOC5567877, have been determined to exhibit a strong binding affinity towards the envelope protein of the ZIKV46. The presence of the dicer gene also strengthens its involvement in the AA’s antiviral response. The differential regulation of genes 5573872 and 5571006 (both encoding DDX5/p68), which are involved in the spliceosome pathway, suggests that these genes are conditionally regulated in response to distinct viral infections. DDX5 is known to modulate viral replication through interactions with viral helicases and host transcriptional regulators, facilitating the processing of viral RNA and enhancing replication efficiency61,62. Additionally, it engages with immune-related proteins and spliceosomal components, thereby influencing the host’s antiviral response. Depending on the viral context, DDX5 serves either as a proviral factor supporting replication or as a host defense element that restricts it63. In humans, DDX5 plays a multifaceted role in viral infections by participating in viral replication and modulating several virus-related signaling pathways. Specifically, it is known to inhibit viruses like HBV and EBV while supporting the replication of viruses such as HIV and JEV64. DDX5 has been implicated in the replication cycles of flaviviruses like DENV65 and JEV66, but its role in CHIKV67 infection is less understood. Notably, our finding that DDX5 (gene 5571006) is downregulated when both togaviruses (CHIKV) and flaviviruses (DENV, ZIKV, JEV and UV) co-occur may indicate a complex interplay between viral infections, potentially involving a suppression mechanism to balance the host’s antiviral response or modulate the spliceosome activity during co-infection. This discovery could pave the way for further investigation into the interaction between DDX5 and CHIKV, as there have been no previous studies establishing a connection between them.
Conclusion
Our research bridges a critical gap in the existing understanding of how viruses interact with their vector and how this impacts the transmission of the disease, particularly involving multi-viral infections. The comprehensive approach of transcriptomic analysis, viral abundance assessment, differential gene expression analysis, epidemiological data analysis, and functional annotation enables us to learn more about the interactions between viral co-infections and vectors. Consequently, we discovered many instances of positive and negative correlations between viruses, which may influence the specificity, severity, and transmission rates of diseases. We show strong evidence for CHIKV incompatibility with other flaviviruses such as DENV as well as a high cooperative effect within flaviviruses. The findings imply a connection between the epidemiological characteristics of the virus and its relationship to the vector. In addition, it is worth noting that viruses that co-occur not only engage in communication with one other, but their prevalence can serve as an indicator of their interactions with their vector and, perhaps, their capacity to infect a novel host. This knowledge is crucial for the formulation of focused strategies aimed at mitigating vector-borne diseases, as well as for the identification of possible areas for intervention in disease control.
Materials and methods
Data retrieval from NCBI
The transcriptome datasets of AA were obtained from the National Centre for Biotechnology Information Sequence Read Archive (NCBI SRA, https://www.ncbi.nlm.nih.gov/sra) and European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena/browser/) (Supplementary data, as of Aug 24). A total of 237 publicly available RNA-Seq datasets of AA mosquitoes exposed to arboviruses such as DENV, ZIKV, and CHIKV were retrieved. Only those with clearly defined infection status, raw FASTQ files, and complete metadata were included in the analysis. Uninfected mosquito samples designated “control” or “wild” in the original study served as controls. The healthy/normal AA samples were also included and screened for viral presence prior to analysis. Both field-collected and experimentally reared laboratory mosquito populations were considered. Studies involving confounding factors such as antiviral exposure, bacterial co-infections, immune priming, or genetically modified strains were excluded. To ensure consistency, only Illumina-based RNA-Seq data were used for downstream analysis. FastQC (version 0.12.1)68 was used to assess the quality of the datasets and identify low-quality sequences and adapter contamination. Trimmomatic (v0.39)69 and Cutadapt (v4.0)70 were then used to remove adapters and low-quality reads, maintaining a Phred score of 30. The high-quality reads were aligned to the reference genome AaegL5 using Bowtie2 (v2.5.4)71. Viral genomes were collected from the NCBI Genome database (https://www.ncbi.nlm.nih.gov/genome) and the information regarding mosquito genes was acquired from the NCBI Gene database (https://www.ncbi.nlm.nih). A comprehensive review of the available literature pertaining to the transmissibility of mosquitoes led to the identification of a total of 21 viruses belonging to eight distinct families (Table 1).
The tobacco mosaic virus (TMV) and brome mosaic virus (BMV), both of which are mechanically transmitted, were included as negative controls in this study. The vector datasets exhibited substantial variations with respect to library preparation techniques, sequence lengths, collection locations, as well as physiological and developmental stages during the sampling process.
Quantifying abundance and statistical analysis
Kallisto (version 0.46.1), a k-mer-based tool for quantifying transcript abundances from bulk and single-cell RNA-Seq data or other target sequences using high-throughput sequencing reads, was used with default parameters for transcript alignment, characterization, and evaluation of the estimated count (EC)72,73. Transcriptomic datasets without evidence of viral presence were discarded from subsequent analysis. A heatmap and a Spearman correlation matrix were generated using the R programming (version 4.3.1) packages ggplot, Hmisc, and corrplot74. The ECs were log10 transformed prior to visualization. To account for erroneous discoveries, a Benjamini & Hochberg False Discovery Rate (FDR) correction was applied, where FDR = (m/n) * p, with “m” representing the total number of tests run and “n” indicating the rank of a given p-value in proportion to the total number of tests undertaken. For the epidemiological data, a t-test was carried out for dengue and chikungunya using R for different years with data from individual states and UTs, and the resulting significance estimates were tabulated and analyzed (Supplementary Table 3). DESeq2 version 1.38.375 was used to perform the differential expression analysis. Genes associated with AA in response to the virus were categorized as significantly regulated if their adjusted p-value was less than 0.05 and log2 fold change |log2FC| = 1 (|FC| = 2). Furthermore, the differentially expressed genes were subjected to functional enrichment analysis using DAVID76. The Benjamini & Hochberg method was used to apply FDR correction, and a significance criterion of < 0.05 was used to determine the statistical significance to discover enriched gene ontology (GO) terms. The STRING database77 was utilized to predict and analyze potential interactions among the genes of interest and Cytoscape version 3.10.378 was used for visualization.
Epidemiological study
The epidemiological data from the National Center for Vector Borne Diseases Control (NCVBDC), Ministry of Health & Family Welfare, Government of India (https://ncvbdc.mohfw.gov.in/index.php), were downloaded, which reported the total number of arboviral disease-positive cases in twenty-eight states and seven union territories, at different time periods (2018–2023, Supplementary Table 3). The data on potential confounding factors such as vector density or other environmental variables were not available. To evaluate the statistical significance of observed differences across years and regions, two-tailed t-tests were conducted.
Data availability
The data that support the findings of this study are freely available in the NCBI SRA and ENA databases. Additional Figures and data are provided in the supplementary file.
References
Beerntsen, B. T., James, A. A. & Christensen, B. M. Genetics of mosquito vector competence. Microbiol. Mol. Biol. Rev. 64, 115–137 (2000).
Kuno, G. & Chang, G. J. J. Biological transmission of arboviruses: reexamination of and new insights into components, mechanisms, and unique traits as well as their evolutionary trends. Clin. Microbiol. Rev. 18, 608–637 (2005).
Fischer, D. et al. Climate change effects on Chikungunya transmission in europe: Geospatial analysis of vector’s Climatic suitability and virus’ temperature requirements. Int J. Health Geogr 12, 51 (2013).
Bellone, R. & Failloux, A. B. The role of temperature in shaping Mosquito-Borne viruses transmission. Front. Microbiol. 11, 584846 (2020).
Mayer, S. V., Tesh, R. B. & Vasilakis, N. The emergence of arthropod-borne viral diseases: A global prospective on dengue, Chikungunya and Zika fevers. Acta Trop. 166, 155–163 (2017).
Lee, B. Y. et al. Virus-based piezoelectric energy generation. Nat. Nanotechnol. 7, 351–356 (2012).
Souza-Neto, J. A., Powell, J. R. & Bonizzoni, M. Aedes aegypti vector competence studies: A review. Infect. Genet. Evol. 67, 191–209 (2019).
Parry, R., James, M. E. & Asgari, S. Uncovering the worldwide diversity and evolution of the Virome of the mosquitoes aedes aegypti and aedes albopictus. Microorganisms 9, 1653 (2021).
Ledermann, J. P. et al. Aedes hensilli as a potential vector of Chikungunya and Zika viruses. PLoS Negl. Trop. Dis 8, 1–9 (2014).
Schulz, C. & Becker, S. C. Mosquitoes as arbovirus vectors: from species identification to vector competence. 163–212 (2018). https://doi.org/10.1007/978-3-319-94075-5_9
Franz, A. W. E., Kantor, A. M., Passarelli, A. L. & Clem, R. J. Tissue barriers to arbovirus infection in mosquitoes. Viruses 7, 3741–3767 (2015).
Simmonds, P. et al. ICTV virus taxonomy profile: flaviviridae. J. Gen. Virol. 98, 2 (2017).
Dahmana, H. & Mediannikov, O. Mosquito-Borne Diseases Emergence/Resurgence and How to Effectively Control It Biologically. Pathog. Vol. 9, Page 310 9, 310 (2020). (2020).
Wang, Y. H., Chang, M. M., Wang, X. L., Zheng, A. H. & Zou, Z. The immune strategies of mosquito Aedes aegypti against microbial infection. Dev. Comp. Immunol. 83, 12–21 (2018).
Clayton, A. M., Dong, Y. & Dimopoulos, G. The anopheles innate immune system in the defense against malaria infection. J. Innate Immun. 6, 169–181 (2014).
Olson, K. E. & Blair, C. D. Arbovirus-mosquito interactions: RNAi pathway. Curr. Opin. Virol. 15, 119–126 (2015).
Agiesh Kumar, B. & Paily, K. P. Actin protein up-regulated upon infection and development of the filarial parasite, Wuchereria bancrofti (Spirurida: Onchocercidae), in the vector mosquito, Culex quinquefasciatus (Diptera: Culicidae). Exp. Parasitol. 118, 297–302 (2008).
Smith, D. R. An update on mosquito cell expressed dengue virus receptor proteins. Insect Mol. Biol. 21, 1–7 (2012).
Terradas, G. & McGraw, E. A. Using genetic variation in Aedes aegypti to identify candidate anti-dengue virus genes. BMC Infect. Dis 19, 580 (2019).
Novelo, M. et al. Dengue and Chikungunya virus loads in the mosquito Aedes aegypti are determined by distinct genetic architectures. PLoS Pathog 19, e1011307 (2023).
Danet, L. et al. Midgut barriers prevent the replication and dissemination of the yellow fever vaccine in Aedes aegypti. PLoS Negl. Trop. Dis 13, e0007299 (2019).
Göertz, G. P., Vogels, C. B. F., Geertsema, C., Koenraadt, C. J. M. & Pijlman, G. P. Mosquito co-infection with Zika and Chikungunya virus allows simultaneous transmission without affecting vector competence of Aedes aegypti. PLoS Negl. Trop. Dis. 11, e0005654 (2017).
Blagrove, M. S. C. et al. Co-occurrence of viruses and mosquitoes at the vectors’ optimal climate range: an underestimated risk to temperate regions? PLoS Negl. Trop. Dis. 11, e0005604 (2017).
Sallam, M. F. et al. Co-occurrence probabilities between mosquito vectors of West nile and Eastern equine encephalitis viruses using Markov random fields (MRFcov). Parasites Vectors. 16, 1–12 (2023).
Lin, D. C. D., Weng, S. C., Tsao, P. N., Chu, J. J. H. & Shiao, S. H. Co-infection of dengue and Zika viruses mutually enhances viral replication in the mosquito Aedes aegypti. Parasites Vectors. 16, 1–14 (2023).
Kazazian, L., Lima Neto, A. S., Sousa, G. S., Nascimento, O. J. & Castro, M. C. do Spatiotemporal transmission dynamics of co-circulating dengue, Zika, and chikungunya viruses in Fortaleza, Brazil: 2011–2017. PLoS Negl. Trop. Dis. 14, e0008760 (2020).
Rückert, C. et al. Impact of simultaneous exposure to arboviruses on infection and transmission by Aedes aegypti mosquitoes. Nat. Commun. 8, 15412 (2017).
Peng, J. et al. Biased virus transmission following sequential coinfection of Aedes aegypti with dengue and Zika viruses. PLoS Negl. Trop. Dis. 18, e0012053 (2024).
Saswat, T. et al. High rates of co-infection of dengue and Chikungunya virus in Odisha and maharashtra, India during 2013. Infect. Genet. Evol. 35, 134–141 (2015).
Edwards, T. et al. Co-infections with Chikungunya and dengue viruses, guatemala, 2015. Emerg. Infect. Dis. 22, 2003–2005 (2016).
Burrell, C. J., Howard, C. R. & Murphy, F. A. Epidemiology of Viral Infections. in Fenner and White’s Medical Virology 185–203 (Elsevier, 2017). https://doi.org/10.1016/B978-0-12-375156-0.00013-8
Yu, X., Zhu, Y., Xiao, X., Wang, P. & Cheng, G. Progress towards Understanding the Mosquito-Borne virus life cycle. Trends Parasitol. 35, 1009–1017 (2019).
Altinli, M., Schnettler, E. & Sicard, M. Symbiotic interactions between mosquitoes and mosquito viruses. Front. Cell. Infect. Microbiol. 11, 694020 (2021).
Mutebi, J. P. et al. Mosquitoes of Western Uganda. J. Med. Entomol. 49, 1289–1306 (2012).
Gabiane, G., Yen, P. S. & Failloux, A. B. Aedes mosquitoes in the emerging threat of urban yellow fever transmission. Rev. Med. Virol. 32, e2333 (2022).
Jagadesh, A. et al. Current status of Chikungunya in India. Front Microbiol 12, 59–63 (2021).
Agarwal, A. et al. Molecular and phylogenetic analysis of Chikungunya virus in central India during 2016 and 2017 outbreaks reveal high similarity with recent new Delhi and Bangladesh strains. Infect Genet. Evol 75, 103940 (2019).
Warnes, C. M., Santacruz-Sanmartin, E., Carrillo, F. B. & Velez, I. D. Surveillance and epidemiology of dengue in medellín, Colombia from 2009 to 2017. Am. J. Trop. Med. Hyg. 104, 1719 (2021).
Oviedo-Pastrana, M., Méndez, N., Mattar, S., Arrieta, G. & Gomezcaceres, L. Epidemic outbreak of Chikungunya in two neighboring towns in the Colombian caribbean: A survival analysis. Arch. Public. Heal. 75, 1–8 (2017).
Gutierrez-Barbosa, H., Medina-Moreno, S., Zapata, J. C. & Chua, J. V. Dengue infections in colombia: epidemiological trends of a hyperendemic country. Trop Med. Infect. Dis 5, (2020).
Khongwichit, S., Chuchaona, W., Vongpunsawad, S. & Poovorawan, Y. Molecular surveillance of arboviruses circulation and co-infection during a large Chikungunya virus outbreak in thailand, October 2018 to February 2020. Sci. Rep. 12, 22323 (2022).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677 (2025).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M. & Goto, S. K. E. G. G. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Valenzuela-Leon, P. C. et al. Multiple Salivary Proteins from Aedes aegypti Mosquito Bind to the Zika Virus Envelope Protein. Viruses 14, (2022).
Shi, R. Z., Pan, Y. Q. & Xing, L. RNA helicase A regulates the replication of RNA viruses. Viruses 13, (2021).
Dong, Y. et al. The Aedes aegypti SiRNA pathway mediates broad-spectrum defense against human pathogenic viruses and modulates antibacterial and antifungal defenses. PLOS Biol. 20, e3001668 (2022).
Zhang, L., Zhu, Y. & Cheng, G. Comparison of innate immune responses against arboviruses in mammalian hosts and mosquito vectors. hLife https://doi.org/10.1016/j.hlife.2025.02.007 (2025).
Bonora, M. et al. ATP synthesis and storage. Purinergic Signal. 8, 343 (2012).
Xing, Z., Ma, W. K. & Tran, E. J. The DDX5/Dbp2 subfamily of DEAD-box RNA helicases. WIREs RNA 10, e1519 (2019).
Vogels, C. B. F. et al. Arbovirus coinfection and co-transmission: A neglected public health concern? PLOS Biol. 17, e3000130 (2019).
Furuya-Kanamori, L. et al. Co-distribution and co-infection of Chikungunya and dengue viruses. BMC Infect. Dis 16, 84 (2016).
Vikram, K. et al. Co-distribution of dengue and Chikungunya viruses in Aedes mosquitoes of delhi, India. J. Vector Borne Dis. 58, 386–390 (2021).
Jain, J., Dubey, S. K., Shrinet, J. & Sunil, S. Dengue Chikungunya co-infection: A live-in relationship?? Biochem. Biophys. Res. Commun. 492, 608–616 (2017).
Ahmed, S. et al. Global prevalence of Zika and Chikungunya coinfection: A systematic review and Meta-Analysis. Diseases 12, 31 (2024).
Mourya, D. T. et al. Experimental Zika virus infection in Aedes aegypti. Indian J. Med. Res. 147, 88–96 (2018).
Vaddadi, K., Gandikota, C., Jain, P. K., Prasad, V. S. V. & Venkataramana, M. Co-circulation and co-infections of all dengue virus serotypes in hyderabad, India 2014. Epidemiol. Infect. 145, 2563–2574 (2017).
Lewis, J. et al. Intrinsic factors driving mosquito vector competence and viral evolution: a review. Front Cell. Infect. Microbiol 13, 1330600 (2023).
Rodriguez-Andres, J., Axford, J., Hoffmann, A. & Fazakerley, J. Mosquito transgenerational antiviral immunity is mediated by vertical transfer of virus DNA sequences and RNAi. iScience 27, 108598 (2024).
Rückert, C. et al. ARTICLE impact of simultaneous exposure to arboviruses on infection and transmission by Aedes aegypti mosquitoes. Nat Commun 8, 15412 (2017).
Cheng, W., Chen, G., Jia, H., He, X. & Jing, Z. DDX5 RNA helicases: emerging roles in viral infection. Int. J. Mol. Sci. 2018. 19, 1122 (2018).
Squeglia, F., Romano, M., Ruggiero, A., Maga, G. & Berisio, R. Host DDX helicases as possible SARS-CoV-2 proviral factors: A structural overview of their hijacking through multiple viral proteins. Front. Chem. 8, 602162 (2020).
Zan, J. et al. RNA helicase DDX5 suppresses IFN-I antiviral innate immune response by interacting with PP2A-Cβ to deactivate IRF3. Exp. Cell. Res. 396, 112332 (2020).
Hu, M. et al. DDX5: an expectable treater for viral infection- a literature review. Ann. Transl Med. 10, 712 (2022).
Cervantes-Salazar, M. et al. The Nonstructural Proteins 3 and 5 from Flavivirus Modulate Nuclear-Cytoplasmic Transport and InnateCervantes-, The Nonstructural Proteins 3 and 5 from Flavivirus Modulate Nucle. (2018) (2018). https://doi.org/10.1101/375899
Li, C. et al. The DEAD-box RNA helicase DDX5 acts as a positive regulator of Japanese encephalitis virus replication by binding to viral 3′ UTR. Antiviral Res. 100, 487–499 (2013).
Kumar, R. et al. Chikungunya virus non-structural protein nsP3 interacts with Aedes aegypti DEAD-box helicase RM62F. https://doi.org/10.1007/s13337-021-00734-y
Brown, J., Pirrung, M. & McCue, L. A. FQC dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139 (2017).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with bowtie 2. Nat. Methods. 9, 357–359 (2012).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Balakrishnan, B., Arul, S. S., Ravindran, A. & Venkataraman, S. Brain Virome in neurodegenerative disorders: insights from transcriptomic data analysis. ACS Chem. Neurosci. https://doi.org/10.1021/ACSCHEMNEURO.3C00432 (2023).
Gentleman, R., Huber, W., Carey, V., Irizarry, R. & Dudoit, S. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. (2005).
Love, M. I., Huber, W. & Anders, S. Moderated Estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Dennis, G. et al. Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3 (2003).
Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
SV thanks University Grants Commission-Faculty Recharge Program (UGC-FRP), New Delhi, India, for salary and research support. The authors thank BIC at DoBT, AU (BT/PR40163/BTIS/137/31/2021), DBT, Govt. of India for computational facilities.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
B.BK. and SS.A. were involved in conceptualization, data analysis, and manuscript writing; A.R. participated in design, analysis, and reviewing the manuscript. S.V. was involved in conceptualization, design, analysis, and finalizing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Balakrishnan, B., Arul, S.S., Ravindran, A. et al. Competitive Viral Interference Controls Arbovirus Co-Transmission in Aedes aegypti. Sci Rep 15, 45679 (2025). https://doi.org/10.1038/s41598-025-08286-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-08286-7