Abstract
Rhizomorphs are specialized structures formed by the intertwining of mycelia in Armillaria and certain other basidiomycete fungi. The growth and branching of rhizomorphs are essential for nutrient acquisition, host colonization, and environmental adaptation in fungi. Strigolactones (SLs), a class of plant-derived signaling molecules, are well known for their role in promoting hyphal germination and branching in arbuscular mycorrhizal (AM) fungi. Studies suggest that GR24, an SL analogue, may also influence the behavior of other fungi, including Armillaria, promoting the formation of highly specialized rhizomorphs for nutrient acquisition and host colonization. However, the underlying mechanism remains unclear. In this study, we found that rhizomorph branching in GR24-treated A. gallica 541 was significantly greater than in the blank control group (CK). To investigate the response mechanism, we conducted RNA sequencing to examine transcriptomic changes in A. gallica 541 following exogenous GR24 treatment. Differentially expressed gene (DEG) analysis comparing the treated and CK groups revealed 983 differentially expressed genes, including 670 upregulated and 313 downregulated genes. These DEGs were associated with cytochrome P450-related processes, GA synthesis, diterpenoid biosynthesis, NAD(P)-binding proteins, and pentose and glucuronate interconversions. The findings suggest that increased rhizomorph branching is linked to the upregulation of growth-related genes and enhanced carbon metabolism. This study offers new insights into the molecular basis of rhizomorph branching in response to GR24 in A. gallica 541 and highlights the potential role of GR24 in fungal development.
Similar content being viewed by others
Introduction
Armillaria belongs to Basidiomycotina, Physalacriaceae, and is widely distributed across North America, Europe, and Asia. To date, more than 70 species of Armillaria have been reported1. It is typically recognized as a destructive forest pathogen capable of infecting over 500 tree species, leading to forest root rot and causing significant losses in economic forestry2. However, Armillaria is not exclusively pathogenic; in some cases, its presence facilitates symbiotic relationships that support host growth. For instance, Gastrodia elata (“Tianma” in Chinese) and Polyporus umbellatus (“Zhuling” in Chinese), both widely used in traditional Asian medicine, depend on Armillaria rhizomorph invasion for nutrient acquisition, which is essential for their growth and reproduction3,4. In these cases, Armillaria functions as a symbiotic fungus.
The rhizomorph is a specialized structure of Armillaria, composed of numerous densely packed hyphae and resembling plant roots in form5. Compared to hyphae, rhizomorphs are highly differentiated, with a keratinized, dark brown surface. Most rhizomorphs develop within root tissues or soil, where they facilitate the transport of nutrients, water, and oxygen6. Young rhizomorphs are also capable of emitting fluorescence in dark environments7. The apical region of the rhizomorph consists of meristematic tissue, enabling continuous cell differentiation and branch formation. This feature allows Armillaria to grow to massive sizes, making it the largest known terrestrial organism8. Rhizomorphs exhibit either an apically bifurcated or uniaxial growth pattern, which influences their role in plant interactions. Rhizomorphs with bifurcated branches tend to be more invasive to tree seedlings, while those with uniaxial branching exhibit more saprophytic behavior9. Additionally, the extent of rhizomorph branching in Armillaria directly impacts the yield of its symbiotic partner, G. elata10. Consequently, rhizomorph branching not only reflects the growth and development status of Armillaria but also affects its host or symbiont.
Strigolactones (SLs) belong to the terpene class of small-molecule compounds and encompass both natural strigol analogs found in higher plants and synthetic derivatives11. The first identified SL compound, strigol, was isolated from the root exudates of cotton12. As a novel class of phytohormones, SLs have been reported to perform various biological functions, including the stimulation of hyphal branching in arbuscular mycorrhizal (AM) fungi. SLs secreted by plant roots can trigger spore germination and induce mycelial branching in AM fungi, increasing the likelihood of fungal contact with plant roots and facilitating the formation of infection structures, which initiates the symbiotic process13. Notably, this effect occurs even in the absence of direct contact between the host root and the fungus. The hyphal branching signal derived from host roots is also referred to as the branching factor14. Additionally, SLs have been identified as key signals in establishing the symbiotic relationship between G. elata and Armillaria, functioning in a manner similar to their role in promoting symbiosis between plants and AM fungi. Key genes involved in SL biosynthesis and transport, including carotenoid cleavage dioxygenases and ABC transporters, were found to be upregulated in G. elata. Controlled experiments confirmed that SLs promote rhizomorph branching in A. gallica, contributing to the formation of its symbiotic relationship with G. elata15.
In this study, the synthetic SL analog GR24 was applied to A. gallica 541 at different concentrations. By monitoring the growth and branching patterns of A. gallica 541 and analyzing differentially expressed genes at the transcriptome level, we examined the effects of GR24 on biological processes related to growth and development following rhizomorph branching. This work provides transcriptional profiling to explore the responses of A. gallica 541 to GR24 and offers insights into the interaction mechanisms between Armillaria and its host or symbiont.
Methods
Fungal materials and growth conditions
The A. gallica 541 strain used in this study was previously isolated from the sclerotium of P. umbellatus and identified in our earlier work16. A 1-cm-long segment of A. gallica 541 rhizomorph was placed in Potato Dextrose Agar (PDA) medium (1 L containing 200 g of potato, 20 g of glucose, and 12 g of agar) and incubated in the dark at 25℃ for 10 days. Once A. gallica 541 growth stabilized, a sterile 1 cm2 filter paper was placed about 2 mm to the central portion of the rhizomorph. A 20 µL aliquot of filtered GR24 solution, dissolved in 70% ethanol, was applied to the center of the filter paper at concentrations of 0.001, 0.01, 0.1, 1, and 10 µg·ml− 1. A control group (CK) without GR24 treatment was also included. After 4 days of continued incubation, the number of newly formed rhizomorph branches was recorded for all treatment and control groups. Samples were then collected separately and stored at -80 °C. Each group included at least three replicates.
RNA extraction, library construction and sequencing
The cDNA libraries were sequenced on the Illumina sequencing platform by Metware Biotechnology Co., Ltd. (Wuhan, China). Briefly, total RNA was extracted, and its concentration and quality were assessed using the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). mRNA was enriched using poly-T oligo-attached magnetic beads, and the fragmented mRNA was used as a template to synthesize the first-strand cDNA using a random hexamer primer and reverse transcriptase. The resulting cDNA underwent end repair, poly(A) tail addition, and sequencing adapter ligation. The ligation products were size-selected for PCR amplification. After passing library quality control, different libraries were pooled based on effective concentration and the required target sequencing depth for the Illumina HiSeq platform.
Bioinformatics analysis
Raw sequencing data were processed using fastp (version 0.19.3) with parameters set to -n_base_limit 15 and -qualified_quality_phred 20 to filter out low-quality reads. Reads containing adapter sequences, those with more than 10% unknown nucleotides (N), or those with over 50% low-quality bases (Q-value ≤ 20) were removed. Following data filtration, the average sequencing error rate was calculated at each position, and sample GC content distribution was examined. Clean reads were then aligned to the Armillaria gallica reference genome (accession NKEW00000000)17 using HISAT2 (version 2.1.0) to obtain genomic positional information and sequence-specific features for further analysis.
Differentially expressed gene analysis
Gene expression levels were quantified using fragments per kilobase of transcript per million mapped reads (FPKM). Differential expression analysis between the GR24-treated and CK groups was conducted using DESeq2 (version 1.22.1), with differentially expressed genes (DEGs) identified based on |Log2 Fold Change| ≥ 1 and FDR < 0.05. Gene Ontology (GO, http://geneontology.org/) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) pathway enrichment analyses were performed to classify the functional roles of the identified DEGs18,19.
Quantitative real-time PCR analysis (qRT-PCR )
To validate the transcriptome results, qRT-PCR was performed on 6 selected DEGs encoding cytochrome P450 and 6 randomly selected DEGs. Primer sequences used for amplification are listed in Table S1. cDNA was synthesized using the Reverse Transcription PrimeScript II cDNA Synthesis Kit (Takara, Kusatsu, Japan). The 20 µL qRT-PCR reaction mixture contained 10 µL ChamQ Universal SYBR qPCR Master Mix Kit (Vazyme, Nanjing, China), 0.5 µL each of forward and reverse gene-specific primers, 1 µL of cDNA, and 8 µL of sterile ddH2O. Amplification was performed using the LightCycler® 480 II system (Roche, Basel, Switzerland) under the following conditions: an initial denaturation step at 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. EF-1γ was used as the endogenous control20. Each sample was analyzed in triplicate, and relative gene expression was calculated using the 2−ΔΔCt method21. Statistical analysis was conducted using GraphPad Prism 9.5.
Results
GR24 induces rhizomorph branching in A. gallica 541
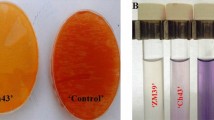
The effect of GR24 treatment at concentrations of 0.001, 0.01, 0.1, 1, and 10 µg·ml− 1 on rhizomorph branching in A. gallica 541 was assessed. The results showed that after A. gallica 541 growth stabilized, rhizomorph branching was significantly higher in the GR24-treated groups compared to the CK group (Fig. 1). The highest number of branches was observed in the 0.1 µg·ml− 1 GR24-treated group. To determine whether the solvent used for dissolving GR24 influenced branching, we tested the effect of 70% ethanol (solvent-only group) on A. gallica 541. The number of branches in the solvent-only group was comparable to that in the 10 µg·ml− 1 GR24-treated group, with no significant difference from the CK group. As GR24 concentration increased, the number of rhizomorph branches initially rose and then declined (Fig. 2). Based on these findings, 0.1 µg·ml− 1 GR24 was selected for further study.
RNA sequencing and mapping
A total of 53.98 GB of clean reads were generated through transcriptome sequencing, with each sample yielding between 51 million and 70.5 million reads, ensuring adequate sequencing depth. All samples exhibited high-quality sequencing data, with clean read rates exceeding 85% and error rates consistently maintained at 0.03%. The Q20 (percentage of bases with quality > 20 in clean reads) and Q30 values were above 92%, while GC content remained stable across samples, ranging from 51.42 to 51.76%. These metrics confirmed the high quality of the sequencing data, making them suitable for further transcriptome analysis (Table S2). The clean reads demonstrated a high mapping efficiency to the reference genome, with unique mapping rates ranging from 74.81 to 76.81% (Table S3). Principal component analysis (PCA) was conducted to compare the transcriptional profiles of GR24-treated and CK samples (Figure S1). The results revealed a clear separation between the two groups, indicating significant differences in gene expression patterns following GR24 treatment. The relatively tight clustering of biological replicates within each group (GR24 and CK) demonstrated high reproducibility and consistency, supporting further analysis of DEGs and their functional implications.
Differential gene expression analysis
Differential gene expression analysis revealed substantial transcriptional changes in A. gallica 541 in response to GR24 treatment. A total of 983 DEGs were identified between the GR24-treated and CK groups, with 670 genes upregulated and 313 downregulated in GR24-treated samples compared to CK. To visualize the overall distribution of DEGs, volcano plots were generated (Fig. 3A), where significantly upregulated genes (red dots) and downregulated genes (green dots) were clearly distinguished from non-differentially expressed genes (blue dots). A heatmap was constructed to illustrate gene expression patterns and relationships between samples (Fig. 3B). The hierarchical clustering dendrograms on the top and left indicated similarities in expression profiles among samples and genes, grouping those with comparable expression trends.
GO and KEGG pathway analysis of the DEGs
To further analyze the functions of DEGs, GO enrichment analysis was conducted. The results showed that DEGs were classified into three main GO ontologies: biological process, molecular function, and cellular component. The top 50 GO-enriched terms with the lowest q-values were selected, and a bar chart of the enriched categories was generated (Fig. 4). Among these, 30 GO-enriched terms were associated with biological processes, with DEGs primarily involved in polysaccharide metabolism, carbohydrate catabolism, polysaccharide breakdown, and cell wall organization. These genes played a crucial role in fungal growth and development while also influencing environmental adaptation and bioactive compound production. The enrichment of complex metabolic pathways and enzyme systems suggested a significant role in carbon source utilization. In the cellular component category, DEGs were mainly clustered in the cell wall, external encapsulating structures, fungal-type cell walls, and cell surfaces, highlighting structural modifications in these areas. In terms of molecular functions, DEGs were predominantly linked to hydrolase activity, monooxygenase activity, and oxidoreductase activity, which are key enzymatic processes in catalytic oxidation reactions. These genes contributed to redox reactions by incorporating oxygen atoms into substrate molecules or using molecular oxygen as an electron acceptor, catalyzed by NADH or NADPH. The GO pathway analysis indicated that GR24 treatment significantly influenced genes related to redox signaling pathways and carbon metabolism, particularly those involved in polysaccharide and carbohydrate catabolic processes in A. gallica 541.
The biological functions of DEGs were further examined through KEGG pathway enrichment analysis18,19. A total of 95 KEGG pathways were identified, with the top 20 significantly enriched pathways shown in Fig. 5. The majority of DEGs were associated with metabolic pathways (195 genes), as represented by the largest and darkest bubble, indicating a widespread influence on various metabolic processes. Additional pathways with significant DEG enrichment included diterpenoid biosynthesis (12 genes), pentose and glucuronate interconversions (19 genes), glycosphingolipid biosynthesis, globo and isoglobo series (6 genes), starch and sucrose metabolism (18 genes), biosynthesis of secondary metabolites (82 genes), sphingolipid metabolism (8 genes), butanoate metabolism (11 genes), taurine and hypotaurine metabolism (4 genes), and glycerolipid metabolism (13 genes), among others. Transcriptome analysis revealed that GR24 predominantly influenced metabolic pathways in A. gallica 541, affecting the transcript levels of metabolism-related genes.
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment classification of differentially expressed genes (top 20). The vertical axis represents the KEGG pathway, while the horizontal axis denotes the rich factor. A higher rich factor indicates a greater degree of enrichment. Smaller q-values reflect more statistically significant enrichment. Larger dots indicate a greater number of DEGs associated with the pathway.
Gene pathway enrichment analysis of A. gallica 541 in response to GR24 treatment
The KEGG analysis results indicated that GR24 treatment primarily affected metabolic pathways in A. gallica 541 (Fig. 5). Among these, two pathways were significantly enriched: diterpenoid biosynthesis (Figure S2) and pentose and glucuronate interconversions (Figure S3)22. In the diterpenoid biosynthesis pathway, 12 genes were upregulated in response to GR24 treatment: ARMGADRAFT_1005092, ARMGADRAFT_1008527, ARMGADRAFT_1079576, ARMGADRAFT_1100908, ARMGADRAFT_1164881, ARMGADRAFT_1014338, ARMGADRAFT_1014343, ARMGADRAFT_1166674, ARMGADRAFT_995035, ARMGADRAFT_1017443, ARMGADRAFT_1017449, and ARMGADRAFT_1018092. These genes encode cytochrome P450 enzymes, which function as ent-kaurene oxidases involved in redox reactions. These enzymes act on paired donors by incorporating or reducing molecular oxygen, with reduced flavin or flavoprotein serving as one donor and one oxygen atom being incorporated into the other donor. In the pentose and glucuronate interconversions pathway, 12 genes were upregulated following GR24 treatment: ARMGADRAFT_944276 (encoding pectin lyase-like protein), ARMGADRAFT_1007320 (glycoside hydrolase family 28 protein), ARMGADRAFT_1080574 (pectin lyase-like protein), ARMGADRAFT_1006889 (D-galacturonic acid reductase), ARMGADRAFT_951795 (pectin lyase-like protein), ARMGADRAFT_974108 (glycoside hydrolase family 28 protein), ARMGADRAFT_975938 (polygalacturonase), ARMGADRAFT_937074 (aldo/keto reductase), ARMGADRAFT_1007973 (NAD(P)-binding protein), ARMGADRAFT_1007976 (GroES-like protein), ARMGADRAFT_1011564 (GroES-like protein), and ARMGADRAFT_1006485 (oxidoreductase). Conversely, seven genes in this pathway were downregulated: ARMGADRAFT_1056077 (pectin lyase-like protein), ARMGADRAFT_1144626 (pectinesterase-like protein), ARMGADRAFT_944066 (carbohydrate esterase family 8 protein), ARMGADRAFT_1043944 (polygalacturonase), ARMGADRAFT_1074829 (endopolygalacturonase 2 precursor), ARMGADRAFT_527182 (endopolygalacturonase 2 precursor), and ARMGADRAFT_724685 (glycoside hydrolase family 28 protein).
Two other pathways were also significantly enriched. In the glycosphingolipid biosynthesis, globo and isoglobo series pathway, six genes were upregulated: ARMGADRAFT_1042677 (glycoside hydrolase family 27 protein), ARMGADRAFT_1060618 (glycoside hydrolase family 27 protein), ARMGADRAFT_636330 (hypothetical protein), ARMGADRAFT_636355 (glycoside hydrolase), ARMGADRAFT_69886 (glycoside hydrolase), and ARMGADRAFT_1013853 (glycoside hydrolase). These genes were associated with alpha-galactosidase function. In the starch and sucrose metabolism pathway, 11 genes were upregulated following GR24 treatment: ARMGADRAFT_72542 (glycoside hydrolase), ARMGADRAFT_667770 (exo-beta-1,3-glucanase), ARMGADRAFT_66798 (cytochrome P450), ARMGADRAFT_1019124 (glycogen synthase), ARMGADRAFT_971531 (putative alpha-amylase), ARMGADRAFT_1169808 (glycoside hydrolase), ARMGADRAFT_1044235 (glycoside hydrolase family 15 protein), ARMGADRAFT_1075864 (glucoamylase), ARMGADRAFT_1006169 (glycoside hydrolase), ARMGADRAFT_1015818 (glycogen phosphorylase), and ARMGADRAFT_100786 (glycoside hydrolase family 13 protein). Conversely, seven genes in this pathway were downregulated: ARMGADRAFT_1018067 (glycoside hydrolase family 1 protein), ARMGADRAFT_1049413 (glycoside hydrolase), ARMGADRAFT_985488 (endoglucanase V-like protein), ARMGADRAFT_996808 (endoglucanase V-like protein), ARMGADRAFT_104385 (cellobiohydrolase I), ARMGADRAFT_1165090 (cellobiohydrolase I), and ARMGADRAFT_929772 (cellobiohydrolase II-I).
RT-qPCR validation
Based on the significance of the KEGG enrichment analysis, 6 DEGs encoding cytochrome P450 that were associated with metabolic pathways in the transcriptome data combined with 6 random DGEs were selected for RT-qPCR validation (Fig. 6). The Ct values for each gene across technical replicates were calculated, and relative expression levels of DEGs were determined using the 2−ΔΔCt method. Expression levels for each transcript were normalized to FPKM. GraphPad Prism version 9.5 (GraphPad Software, CA, USA) was used for data visualization. The RT-qPCR results were highly consistent with the RNA-seq FPKM data, confirming the differential expression patterns of the selected genes.
Discussion
A defining and ecologically significant characteristic of Armillaria species is their ability to form rhizomorphs. These structures are crucial for survival, dispersal, and host infection. It is widely believed that a greater number of rhizomorph branches increases the likelihood of host infection by Armillaria9. Studies by Yuan et al. and Hua et al. demonstrated that GR24 promotes rhizomorph branching in A. gallica 54115,23. However, the underlying mechanisms governing the effects of GR24 on Armillaria growth remain largely unexplored. In this study, we examined the response of A. gallica 541 to GR24 treatment. The results indicated that GR24 significantly increased rhizomorph branching, following a pattern of initial improvement followed by a decline at higher concentrations. This trend aligns with previous findings on GR24’s effects on AM fungi and other organisms. Typically, SLs and their analogs function as phytohormones, playing roles in regulating root growth, leaf senescence, and interactions with symbiotic fungi. Previous studies have shown that SLs can induce extensive hyphal branching in AM fungi at very low concentrations (1 pg to 1 ng per disc)24. The synthetic SL analog GR24 has been found to stimulate mitosis and growth in AM fungi such as Gigaspora rosea by increasing energy metabolism25. AM fungi detect SL signals through specific receptors, such as D14 homologs, which activate downstream genes like RAM1 and RAM2, promoting hyphal branching and the formation of hyphopodia26. Additionally, studies using CRISPR/Cas9 to knock out the CCD7 gene in rice demonstrated that reduced SL secretion in mutant roots led to decreased mycelial branching in AM fungi, further confirming the essential role of SLs in fungal development27. These findings suggest that GR24 may influence rhizomorph production and branching in A. gallica 541 by acting as a signaling molecule that regulates metabolism and gene expression.
Gene Ontology (GO) annotation and KEGG pathway enrichment analysis of DEGs revealed that cytochrome P450-encoding genes were upregulated in A. gallica 541 following GR24 treatment. Cytochrome P450 is a ubiquitous monooxygenase superfamily and plays a key role in various fungal processes. For instance, in Nectria haematococca, cytochrome P450 enzymes can demethylate pisatin, a phytoalexin produced by the host, thereby reducing its antifungal toxicity and increasing fungal survival25. Based on their involvement in electron transfer, P450 monooxygenase systems are classified into ten groups, three of which have been identified in fungi28,29,30. In this study, genes from the CYP701 family within the cytochrome P450 superfamily were upregulated in response to GR24 treatment, coinciding with increased rhizomorph branching in A. gallica 541. The CYP701 family encodes enzymes that play a crucial role in gibberellin (GA) biosynthesis by catalyzing the cyclization of the diterpene precursor (E, E,E-geranylgeranyl diphosphate) into ent-labdadienyl/copalyl diphosphate, a key step in GA biosynthesis31. GAs are a class of phytohormones that regulate multiple developmental processes in higher plants, including seed germination, stem elongation, flowering, and fruit ripening32. In fungi, studies by Cao et al. demonstrated that the exogenous application of GA3 significantly improved the growth of A. gallica 012 m in both solid and liquid media33. In our study, GA-related genes were upregulated during the process of rhizomorph branching in A. gallica 541, suggesting that GAs may also play a critical role in its growth and development. Several diterpenoids, including ent-kaurene in Gibberella fujikuroi, are involved in GA biosynthesis, where the gene encoding ent-kaurene oxidase functions as a rate-limiting step34. Diterpenoids, as a significant class of secondary metabolites, are widely distributed among organisms and exhibit various biological functions, including anti-inflammatory and antioxidant activities35. In this study, KEGG pathway enrichment analysis showed that the diterpene biosynthesis pathway was significantly activated and regulated in response to GR24-induced rhizomorph branching in A. gallica 541. Cytochrome P450 monooxygenases have been reported to participate in complex diterpene biosynthesis in Aspergillus species36. Furthermore, diterpene biosynthesis genes are widely expressed in plant-associated fungi, suggesting a potential role in fungal-host interactions. Given that Armillaria can function as both a plant pathogen and a symbiont with G. elata and P. umbellatus, it is possible that diterpenoid biosynthesis during rhizomorph branching contributes to its saprophytic lifestyle and interaction with hosts.
Pentose and glucuronate interconversions play a crucial role in carbon utilization and energy metabolism in Armillaria, as observed in other fungi. For instance, transcriptome analysis of Morchella importuna revealed significant changes in gene expression related to this pathway, including transketolase, G6PD, and rpiA. These genes exhibited differential expression at various stages of mycelial growth, suggesting their involvement in fungal growth and development37. In our study, genes encoding NAD(P)-binding proteins within the pentose and glucuronate interconversion pathway were significantly upregulated during rhizomorph branching in A. gallica 541. NAD(P) is well recognized as a key energy-carrying molecule in cells. In Physcomitrium patens, NAD(P)-binding proteins have been implicated in chitosan-induced activation of peroxidase activity and LOX gene expression, highlighting their role in plant responses to environmental changes38. Similarly, in vascular plants, NAD(P)-binding enzymes, such as those belonging to the short-chain dehydrogenase/reductase superfamily, have been shown to increase pathogen resistance39. These findings suggest that A. gallica 541 may also undergo environmental stress responses alongside GR24-induced rhizomorph branching, potentially influencing its interactions with host plants. One upregulated gene, ARMGADRAFT_1007973, was annotated as D-xylose 1-dehydrogenase, an enzyme primarily involved in D-xylose metabolism with the assistance of NAD(P), converting D-xylose to D-xylonolactone. This conversion is considered a critical step in xylose utilization in certain bacteria, such as Gluconobacter oxydans40. In Armillaria, D-xylose 1-dehydrogenase activity may be essential for survival and growth in xylose-rich environments. Additionally, D-xylose can be further metabolized into pyruvate through a series of enzymatic reactions, suggesting that the increased branching observed in GR24-treated A. gallica may be accompanied by enhanced metabolic activity. Another upregulated oxidoreductase-encoding gene, ARMGADRAFT_1006485, was identified as D-arabinitol 2-dehydrogenase, an enzyme that catalyzes the oxidation of D-arabinitol with the assistance of NAD+ or NADP+. In Zygosaccharomyces rouxii, D-arabinitol 2-dehydrogenase has been shown to participate in D-arabinitol biosynthesis alongside ribulokinase41. The ability of D-arabinitol 2-dehydrogenase in Armillaria to convert D-arabinitol into nucleotide derivatives such as ribitol and glycolaldehyde suggests enhanced glucose and energy metabolism during rhizomorph branching in A. gallica 541.
Several genes related to galacturan 1,4-alpha-galacturonidase, D-galacturonate reductase, and L-glyceraldehyde reductase were upregulated in A. gallica 541, ultimately contributing to its glycerolipid metabolism. Galacturan 1,4-alpha-galacturonidase plays a crucial role in breaking down polygalacturonic acid in the cell wall, leading to cell wall disintegration and collapse, as observed in Beta vulgaris L42. In A. gallica 541, the activity of this enzyme may induce structural modifications in the cell wall, potentially affecting nutrient exchange and cellular signaling. These changes could increase Armillaria’s ability to infect and colonize its host. The reorganization of the cell wall may also influence fungal growth rate and morphology, which aligns with the increased rhizomorph branching observed in A. gallica 541 following GR24 treatment. D-galacturonate reductase is a key enzyme in the D-galacturonate metabolic pathway. Studies have shown that deletion of the gene encoding NADPH-dependent D-galacturonate reductase in Trichoderma reesei resulted in inhibited fungal growth, highlighting its essential role in fungal development43. In A. gallica 541 treated with GR24, genes related to fungal growth and development were upregulated, leading to increased metabolic activity, particularly in sugar metabolism and glycerol metabolism. This finding is consistent with reports that GR24 increases the dry weight of Armillaria hyphae, promotes mycelium formation, and enlarges the diameter of mycelial spheres44.
The ability of SLs to promote fungal growth has been widely documented in AM fungi, where they have been shown to increase spore germination45stimulate mycelial branching46regulate mitochondrial metabolism47and facilitate symbiotic relationships with plants by activating mitochondrial function48 and inducing the expression of ROS-related genes49. For instance, GR24 treatment rapidly increased NADH concentration, NADH dehydrogenase activity, and ATP levels in Gigaspora rosea cells within minutes, demonstrating the strong and immediate impact of SLs on mitochondrial energy metabolism in fungi48. In this study, GR24 may have influenced the metabolic processes of A. gallica 541 by upregulating genes related to cytochrome P450, NAD(P)-dependent energy metabolism, and carbon source exchange, ultimately leading to increased rhizomorph branching. Additionally, the GR24-induced improvement of Armillaria growth and development offers new insights for optimizing the cultivation of Armillaria and G. elata. In G. elata, an increase in carotenoid cleavage dioxygenases (CCDs) and ABC transporters (PDRs), key genes involved in the biosynthesis and transport of aurillactone, has been identified as a critical signal for establishing symbiosis between G. elata and Armillaria15. The observed effects of GR24 on Armillaria growth and rhizomorph branching provide a potential strategy for improving the efficiency of symbiotic establishment with medicinal plants such as G. elata and P. umbellatus. Improving these symbiotic interactions could contribute to increased yields of G. elata and P. umbellatus while also providing a valuable reference for further research into the mechanisms underlying Armillaria-host symbiosis.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequence data that support the findings of this study have been deposited in the National Center for Biotechnology Information with the primary accession PRJNA1155725.
References
Heinzelmann, R. et al. Latest advances and future perspectives in Armillaria research. Can. J. Plant. Pathol. 41(1), 1–23 (2019).
Sipos, G. et al. Genome expansion and lineage-specific genetic innovations in the forest pathogenic fungi Armillaria. Nat. Ecol. Evol. 1(12), 1931–1941 (2017).
Wang, Y. et al. Effect of symbiotic fungi-Armillaria gallica on the yield of Gastrodia Elata bl. And insight into the response of soil microbial community. Front. Microbiol. 14, 1233555 (2023).
Xing, Y. M. et al. Armillaria mellea symbiosis drives metabolomic and transcriptomic changes in Polyporus umbellatus sclerotia. Front. Microbiol. 12, 792530 (2022).
Devkota, P. & Hammerschmidt, R. The infection process of Armillaria mellea and Armillaria solidipes. Physiol. Mol. Plant. P. 112, 101543 (2020).
Porter, D. L. et al. The melanized layer of Armillaria Ostoyae rhizomorphs: its protective role and functions. J. Mech. Behav. Biomed. 125, 104934 (2022).
Mihail, J. D. & Bruhn, J. N. Dynamics of bioluminescence by armillaria gallica, A. mellea and A. tabescens. Mycologia 99(3), 341–350 (2007).
Sipos, G., Anderson, J. B. & Nagy, L. G. Armillaria. Curr. Biol. 28(7), R297–R298 (2018).
Morrison, D. J. Rhizomorph growth habit, saprophytic ability and virulence of 15 Armillaria species. For. Pathol. 34(1), 15–26 (2004).
Yu, E. et al. An exploration of mechanism of high quality and yield of Gastrodia Elata bl. F. glauca by the isolation, identification and evaluation of Armillaria. BMC Plant. Biol. 22(1), 621 (2022).
Aliche, E. B., Screpanti, C., De Mesmaeker, A., Munnik, T. & Bouwmeester, H. J. Science and application of Strigolactones. New. Phytol. 227(4), 1001–1011 (2020).
Siame, B. A., Weerasuriya, Y., Wood, K., Ejeta, G. & Butler, L. G. Isolation of strigol, a germination stimulant for Striga asiatica, from host plants. J. Agric. Food Chem. 41(9), 1486–1491 (1993).
Akiyama, K., Ogasawara, S., Ito, S. & Hayashi, H. Structural requirements of Strigolactones for hyphal branching in AM fungi. Plant. Cell. Physiol. 51(7), 1104–1117 (2010).
Omoarelojie, L. O. & Van Staden, J. Plant-endophytic fungi interactions: A Strigolactone perspective. S Afr. J. Bot. 134, 280–284 (2020).
Yuan, Y. et al. The Gastrodia Elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 9(1), 1615 (2018).
Liu, L. et al. Different symbiotic species of Armillaria affect the yield and active compound contents of Polyporus umbellatus. Microorganisms 13(2), 228 (2025).
Sipos, G. et al. Genome expansion and lineage-specific genetic innovations in the forest pathogenic fungi Armillaria. Nat. Ecol. Evol. 1, 1931–1941 (2017).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Li, B., Liu, L., Shan, T. T. & Guo., S. Selection of reference genes for real-time quantitative PCR of Armillaria mellea. Microbiol. China. 49(2), 473–482 (2022).
Yuan, J. S., Reed, A., Chen, F. & Stewart, C. N. Statistical analysis of real-time PCR data. BMC Bioinform. 7, 1–12 (2006).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677 (2025).
Hua, Z. et al. The Armillaria response to Gastrodia elata is partially mediated by strigolactone-induced changes in reactive oxygen species. Microbiol. Res. 278, 127536 (2024).
Fiorilli, V., Novero, M. & Lanfranco, L. Evaluation of the effect of strigolactones and synthetic analogs on fungi. In Strigolactones: Methods and Protocols. 75–89 (2021).
Besserer, A. et al. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 148(1), 402–413 (2008).
Ho-Plágaro, T. et al. A dual regulatory role for the arbuscular mycorrhizal master regulator RAM1 in tomato. J. Exp. Bot. 75(16), 5021–5036 (2024).
Yang, X. et al. Knocking out of carotenoid catabolic genes in rice fails to boost carotenoid accumulation, but reveals a mutation in Strigolactone biosynthesis. Plant Cell Rep. 36, 1533–1545 (2017).
Črešnar, B. & Petrič, Š. Cytochrome P450 enzymes in the fungal Kingdom. BBA-Proteins Proteom. 1814(1), 29–35 (2011).
Zhang, X., Guo, J., Cheng, F. & Li, S. Cytochrome P450 enzymes in fungal natural product biosynthesis. Nat. Prod. Rep. 38(6), 1072–1099 (2021).
Shin, J., Kim, J. E., Lee, Y. W. & Son, H. Fungal cytochrome P450s and the P450 complement (CYPome) of Fusarium graminearum. Toxins 10(3), 112 (2018).
Park, J. et al. Fungal cytochrome P450 database. BMC Genom. 9, 1–11 (2008).
Nett, R. S. et al. Elucidation of Gibberellin biosynthesis in bacteria reveals convergent evolution. Nat. Chem. Biol. 13(1), 69–74 (2017).
Davière, J. M. & Achard, P. Gibberellin signaling in plants. Development 140(6), 1147–1151 (2013).
Cai, G. et al. Characterization of the transcriptional responses of Armillaria gallica 012m to GA3. Arch. Microbiol. 205(9), 308 (2023).
Tudzynski, B., Hedden, P., Carrera, E. & Gaskin, P. The P450-4 gene of Gibberella Fujikuroi encodes ent-kaurene oxidase in the Gibberellin biosynthesis pathway. AEM 67(8), 3514–3522 (2001).
Zhang, F. L. & Feng, T. Diterpenes specially produced by fungi: structures, biological activities, and biosynthesis (2010–2020). J. Fungi. 8(3), 244 (2022).
Dauda, W. P. et al. Robust profiling of cytochrome P450s (P450ome) in notable Aspergillus spp. Life 12(3), 451 (2022).
Fan, T. et al. Transcriptomics combined with metabolomics unveiled the key genes and metabolites of mycelium growth in Morchella importuna. Front. Microbiol. 14, 1079353 (2023).
Marttinen, E. M., Decker, E. L., Heinonen, P., Reski, R. & Valkonen, J. P. Putative NAD (P)-binding Rossmann fold protein is involved in chitosan-induced peroxidase activity and Lipoxygenase expression in Physcomitrium patens. Mol. Plant. Microbe in. 36(11), 682–692 (2023).
Moummou, H., Kallberg, Y., Tonfack, L. B., Persson, B. & van Der Rest, B. The plant short-chain dehydrogenase (SDR) superfamily: genome-wide inventory and diversification patterns. BMC Plant. Biol. 12, 1–17 (2012).
Dai, L. et al. Multi strategy in production of high titer gluconic acid by the fermentation of concentrated cellulosic hydrolysate with Gluconobacter oxydans. Ind. Crop Prod. 189, 115748 (2022).
Li, X. et al. High-level production of d-arabitol by Zygosaccharomyces rouxii from glucose: metabolic engineering and process optimization. Bioresour Technol. 367, 128251 (2023).
Li, X. et al. Genomic and transcriptomic-based analysis of agronomic traits in sugar beet (Beta vulgaris L.) pure line IMA1. Front. Plant. Sci. 13, 1028885 (2022).
Paasikallio, T., Huuskonen, A. & Wiebe, M. G. Scaling up and scaling down the production of galactaric acid from pectin using Trichoderma Reesei. Microb. Cell. Fact. 16, 1–11 (2017).
Xu, C. B. et al. Effects of Strigolactones on the growth of Armillaria mellea. Edible Fungi China. 41(6), 6 (2022).
Kountche, B. A. et al. Effect of the Strigolactone analogs Methyl phenlactonoates on spore germination and root colonization of arbuscular mycorrhizal fungi. Heliyon 4(11) (2018).
Mori, N., Nishiuma, K., Sugiyama, T., Hayashi, H. & Akiyama, K. Carlactone-type Strigolactones and their synthetic analogues as inducers of hyphal branching in arbuscular mycorrhizal fungi. Phytochemistry 130, 90–98 (2016).
Besserer, A. et al. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4(7), e226 (2006).
Belmondo, S. et al. Identification of genes involved in fungal responses to strigolactones using mutants from fungal pathogens. Curr. Genet. 63, 201–213 (2017).
Acknowledgements
This research was financially supported by grants from CAMS Innovation Fund for Medical Sciences(2021-I2M-1-031, 2022-I2M-2-001), and Fundamental Research Funds for the Central Universities, Peking Union Medical College (3332023052).
Author information
Authors and Affiliations
Contributions
Liu Liu performed the research and wrote the main manuscript text,Li Bing reviewed and edited the manuscript,Li Shoujian made the data analysis, Liu Youyan and Zhou Lingfeng revised the article, and Guo Shunxing contributed to refining the ideas and finalizing this paper. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, L., Li, B., Li, S. et al. Transcriptomics reveals the mechanism of branching production in response to GR24 in A. gallica 541. Sci Rep 15, 43471 (2025). https://doi.org/10.1038/s41598-025-09015-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-09015-w