Abstract
One of the most powerful tools for identifying genomic regions associated with various phenotypes is GWAS. Identifying genes influencing milk production traits in Iranian Holstein dairy cows is crucial to understanding the genetic mechanisms underlying these traits and improving future milk production. Therefore, using a single-step GWAS, this study aimed to identify genomic regions, genes, and pathways associated with milk yield (MY), milk fat percentage (FP), milk protein percentage (PP), and somatic cell count (SCC) traits in the Iranian Holstein cattle population. In this study, 210 animals were genotyped using 30K (150 animals from Herd 1) and 50K (60 animals from Herd 2) SNP arrays. Genotypes were then imputed to whole-genome sequence level using the 1000 Bull Genomes Project reference panel, resulting in 6,583,595 high-confidence imputed SNPs forGWAS analysis. Genomic regions associated with milk production traits included 184 significant SNP markers (milk yield, milk fat, milk protein, and somatic cell count, with 86, 18, 22, and 58 significant SNP markers, respectively) based on a significance threshold of P value < 1 × 10⁻⁸ across 10 chromosomes (2, 5, 7, 17, 19, 21, 24, 26, and 28). For the traits FP, PP, MY, and SCS, 5, 6, 9, and 7 candidate genes were identified near the significant SNPs, respectively. Key genes with important biological roles included ATE1, FGFR2, ALDH1A3, CHSY1, GABRG3, FBXO36, PID1, TRIP12, CD52, WDTC1, MATN1, CIDEA, LYZ, CPM, FBXO42, MAML3, SGMS2, HADH, CYP2U1, SCLT1 and THRSP. Therefore, the ATE1, FGFR2, and LYZ genes is not only a key marker for udder health and milk quality but also a promising candidate for genomic selection and therapeutic applications aimed at improving disease resistance in dairy herds. Our research led to the discovery of novel SNPs linked to milk production traits, which could be valuable for future livestock breeding programs.
Similar content being viewed by others
Introduction
The increasing global population and the essential role of milk in meeting nutritional needs have made enhancing the performance of domestic animals, particularly dairy cows, a top priority in breeding goals and programs worldwide1. This focus centers on improving key economic traits like milk production. Milk production and udder health are crucial economic factors that significantly impact the profitability of dairy operations2. Improvements in milk production traits directly benefit these operations, while enhancing resistance to mastitis can reduce the financial burden associated with treatment1,3.
Over time, substantial progress has been made in enhancing the production performance of dairy cows. However, mastitis remains a significant challenge. This infectious disease, caused by environmental and management factors combined with the animal's often weakened resistance and immunity (primarily acquired) to pathogens, leads to substantial economic losses in the dairy industry and raises concerns about the quality of dairy products globally1,2. The high costs associated with mastitis have led to increased attention to mastitis resistance as a vital breeding goal, considering economic and animal welfare aspects4. Direct recording of mastitis occurrences is not routine in most countries, and direct selection for mastitis resistance is uncommon1,5,6. Consequently, the somatic cell count (SCC) in milk, or its logarithmic transformation, measures mastitis due to its higher genetic variance, ease of recording, and strong positive correlation with mastitis incidence5. These complex traits, influenced by multiple genes, are affected by various factors, including management practices, environmental conditions, and the animal's physiological state. Control of these traits involves numerous genes and variants, each with minor effects on the observed phenotype1,6. Strong genetic selection and improved management and nutrition can lead to increased milk production and decreased mastitis prevalence. Research highlights a positive yet detrimental relationship (antagonism) between somatic cells and production traits, notably milk production1,2,7. The somatic cell count is a crucial indicator for assessing the quality and health of raw milk and is a factor in pricing. An elevated SCS negatively impacts raw milk's processing quality and overall quality due to changes in its composition, including fat, protein, lactose, and acidity levels1,2.
The advent of genome-wide panels of single nucleotide polymorphisms (SNPs) has revolutionized the field. SNPs are extensively valuable for detecting and localizing quantitative trait loci (QTLs) for complex traits across diverse species1,2,8. They have proven robust and practical tools for identifying accidental mutations linked to economically significant traits in livestock 3,9,10 and human diseases11,12. Numerous studies over the years have focused on identifying QTLs for various traits in dairy cattle, leading to the discovery of many QTLs across different chromosomes9,13. New sequencing technologies have opened new opportunities to identify markers associated with economically essential genes and milk production traits. Genome-wide association Studies (GWAS) have emerged as a highly efficient strategy for uncovering candidate genes and markers associated with quantitative traits3. The primary aim of GWAS is to pinpoint the most likely genomic locations that control these traits14. Moreover, genome-wide scanning studies contribute to a deeper understanding of the genes and polymorphisms linked to economic traits, ultimately shedding light on the underlying mechanisms of the traits under investigation1,15. In dairy cattle, the GWAS method has been instrumental in estimating SNPs influencing production traits like milk yield, fat yield, protein percentage1,3,4,16,17, and health traits such as mastitis, uterine health1,18, longevity within the herd19,20,21, and reproductive traits16,22,23,24,25.
While different studies have reported SNPs and genes affecting somatic cells and the occurrence of mastitis, these findings have often varied, with limited overlap in identified SNPs between studies. Several factors contribute to these discrepancies, including environmental conditions, the specific type of dairy management (industrial or semi-industrial), variations in native pathogens and the host's response, and the genetic background of the studied population. These factors significantly impact the relationship between genetic variants and genes across the genome and the resulting phenotype26,27. Notably, this is the first study conducted in Iranian Holstein cows using a GWAS approach to investigate milk production and mastitis traits. In this study, 150 animals from Herd 1 were genotyped using a 30K SNP array, and 60 animals from Herd 2 were genotyped using a 50K SNP array, totaling 210 animals. Genotype data were subsequently imputed to whole-genome sequence level using the 1000 Bull Genomes Project reference panel, resulting in 6,583,595 high-confidence imputed SNPs used for GWAS. Consequently, the primary objective of this study is to examine the association of genome-wide SNPs with somatic cell count, milk yield, milk fat (%), and milk protein (%) traits. This comprehensive analysis seeks to identify known and novel genes or genomic and chromosomal regions linked to the inheritance of these traits, individually or in combination, within the Iranian Holstein cattle population.
Results
Descriptive statistics and
Descriptive statistics for milk production traits and somatic cell count in the Iranian Holstein population are presented in Table 1, and the distribution of each milk production trait and somatic cell count is shown in Fig. 1. On average, Iranian Holstein cows had a milk yield of 38.32. The mean milk fat percentage, milk protein percentage, and somatic cell count were 3.304, 2.899, and 64.41, respectively. The coefficient of variation for milk production traits and somatic cell count indicated acceptable diversity for these traits in Iranian Holstein cows, with values of 19.87, 7.24, 15.33, and 133.69 for milk yield, protein percentage, fat percentage, and somatic cell count, respectively. The estimates of variance components and heritability for the four traits (milk yield, milk protein, milk fat, and somatic cell score) from single-trait animal models are shown in Table 2. Overall, the heritability values for milk yield, milk protein, milk fat, and somatic cell score were 53%, 52%, 43%, and 39%, respectively.
GWAS for somatic cell count and milk production
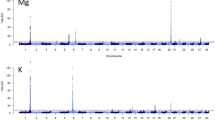
The results of the GWAS analysis for all studied traits (milk production, fat percentage, protein percentage, and somatic cell count) were reported based on the significance threshold of P value < 1 × 10⁻⁸ (supplementary 1). For the MY trait, 86 SNPs were identified on the following chromosomes: BTA2 (19), BTA5 (30), BTA17 (1), BTA21 (33), BTA24 (2), and BTA28 (1). And also, 18 SNPs were observed for the MF (milk fat) trait on BTA7 (11), BTA21 (6), and BTA26 (1). Furthermore, for the MP (milk protein) trait, 22 SNPs passed the significance threshold (P value < 10⁻⁸) and were located in the regions of BTA7 (11), BTA21 (9), BTA22 (1), and BTA26 (1). Moreover, the GWA for the somatic cell count (SCC) trait showed 58 marker-trait associations (P value < 1 × 10⁻⁸) locating on chromosomes BTA2 (1), BTA17 (42), BTA19 (12), and BTA21 (1). The Manhattan and Q-Q plot plots for the studied traits are illustrated in Fig. 2. The Manhattan plots clearly illustrate distinct genomic regions of association for each trait, with particularly strong signals on BTA5 and BTA21 for milk traits, and BTA17 for SCC, suggesting potential QTL hotspots. The Q-Q plots show a strong deviation from the expected distribution under the null hypothesis, further confirming the presence of true genetic associations and the robustness of the GWAS. Notably, several novel genes such as ATE1, FGFR2, LYZ, and MAML3 were identified near the top-associated SNPs, highlighting their potential roles in milk composition, udder health, and host defense mechanisms. These findings provide new insights into the genetic basis of production and health traits and offer promising targets for genomic selection and functional validation in dairy cattle.
QTL regions for somatic cell count and milk production
In Table 3 summarizes the important SNPs (P < 1 × 10⁻⁸) linked to milk production characteristics in Iranian Holstein cows that are situated close to identified QTLs. The results indicate that on chromosomes BTA7, BTA21, and BTA26, important QTLs associated with milk decenoic acid content (MFA-C10:1), milk capric acid content (MFA-C10:0), milk myristoleic acid content (MFA-C14:1), milk palmitoleic acid content (MFA-C16:1), milk lauroleic acid content (MFA-C12:1), milk myristic acid content (MFA-C14:0), milk palmitic acid content (MFA-C16:0), milk protein yield (PY), milk yield (MY), yield grade (YGRADE) were identified in proximity to the significant SNPs for the milk fat percentage trait. Near the significant SNPs associated with the milk yield trait on chromosomes BTA2, BTA5, BTA17, BTA21, BTA24, and BTA28, QTLs related to milk fat yield (FY), somatic cell count (SCC), bovine respiratory disease susceptibility (BRDS), milk yield (MY), milk protein yield (PY), body weight (BW), fat percentage (FATP), bovine tuberculosis susceptibility (BTBS), Clinical mastitis (CM), and Age at first calving (AGEFC) were observed (Table 3). Additionally, QTLs associated with specific somatic cell count (SCC), body weight (BW), milk protein percentage (PP), milk yield (MY), muscularity (MUSC), and average daily gain (ADG), traits were identified for the SCS trait. Regarding the milk protein percentage trait, several important QTLs, including milk protein yield (PY), milk decenoic acid content (MFA-C10:1), milk capric acid content (MFA-C10:0), milk myristoleic acid content (MFA-C14:1), milk palmitoleic acid content (MFA-C16:1), milk lauroleic acid content (MFA-C12:1), milk myristic acid content (MFA-C14:0), milk palmitic acid content (MFA-C16:0), calf size (CALFSZ), milk yield (MY), carcass weight (CWT), muscularity (MUSC), feed conversion ratio (FCR), average daily gain (ADG), and body weight (BY) were determined to be close to the significant SNPs on chromosomes BTA7, BTA21, BTA22, and BTA26.
Gene ontology for somatic cell count and milk production
Over 137 genes associating to milk production and somatic cell count traits were discovered using the gene ontology analysis (supplementary 2), 33 of them are essential genes (Table 4). For the milk fat percentage trait, five candidate genes were discovered around SNPs 26:41,368,775 (2), 21:5,475,347, and 21:5,525,195 (2), which influence the activity of the ATE1, FGFR2, ALDH1A3, CHSY1, and GABRG3 genes (Table 4). And also, for the milk protein percentage trait, six candidate genes were identified, affecting the activity of ATE1, FGFR2, ZNF346, FGFR4, TMEM40, and NTRK3 (Table 4). Furthermore, nine candidate genes were identified around SNPs 2:117,632,966 (2), 2:117,637,569, 24:33,558,520 (3), 24:42,643,763, and 5:19,359,629 (2) for the milk yield trait, affecting the activity of the FBXO36, PID1, TRIP12, CD52, WDTC1, MATN1, CIDEA, LYZ, and CPM genes (Table 4). Moreover, the 13 candidate genes were discovered around those SNPs that associated to somatic cell count trait, relating the activity of the FBXO42, MAML3, SGMS2, SCLT1, HADH, CYP2U1, DLK1, THRSP, ANKRD26, TMEM26, VEGFA, MED4, and VAV1 genes (Table 4).
Gene networks
The results of the gene network analysis for milk production traits, including milk yield (Fig. 3), milk protein percentage (Fig. 4), milk fat percentage (Fig. 5), somatic cell count (Fig. 6), and all traits are shown in Fig. 7. A densely co-expressed network was drawed by using Gene Mania (Fig. 7). This network consisted of 137 genes with 1764 interactions. Among these genes, CAND1, VEGFA, AFGLS2, FGFR2, NUP107, and MPPE1 genes have played roles in several intracellular transport processes. Therefore, the identified candidate genes in our study exhibited significant protein–protein interactions to each other or related genes.
Gene networks analysis for milk protein percentage trait in Holstein cows. Dark circles with and without slash represent candidate genes and associated genes, respectively. Arrows in pink, blue, red and bone color represent co-expression, pathway, physical interactions and shared protein domains, respectively.
Gene networks analysis for milk fat percentage trait in Holstein cows. Dark circles with and without slash represent candidate genes and associated genes, respectively. Arrows in pink, blue, red and bone color represent co-expression, pathway, physical interactions and shared protein domains, respectively.
Gene networks analysis for somatic cell count trait in Holstein cows. Dark circles with and without slash represent candidate genes and associated genes, respectively. Arrows in pink, blue, red and bone color represent co-expression, pathway, physical interactions and shared protein domains, respectively.
Discussion
Phenotypes of milk production traits are primarily quantitative and governed by polygenic mechanisms. Extensive research has been conducted on milk traits over the years. For instance, in 1944, a study confirmed significant QTLs associated with protein yield and fat yield traits, linked to beta-lactoglobulin and kappa-casein, respectively28. Subsequent studies identified numerous QTLs associated with milk traits across 30 different bovine chromosomes1,3,4,29,30,31,32.
Despite the numerous studies, the genetic mechanisms controlling these traits remain largely unclear. Therefore, further research to elucidate the genetic mechanisms governing these traits is precious. To this end, a GWAS was conducted on 210 Iranian Holstein cows, identifying several significant SNPs associated with milk production traits, including milk yield, milk fat, milk protein, and somatic cell count. In this study, significant milk yield SNPs were identified on chromosomes BTA2, BTA5, BTA17, BTA21, BTA24, and BTA28, consistent with previous research findings3,15,30,32,33. Eighteen marker-trait associations were found on chromosomes BTA7, BTA21, and BTA26 for milk fat percentage, corroborating earlier studies29,33,34. For milk protein percentage, 22 SNP markers were identified on BTA7, BTA21, BTA22, and BTA26, with some overlap with previous reports, which identified chromosomes 21 and 22 as the main contributors to this trait3,32,35,36. Several SNPs identified on chromosomes BTA2, BTA17, BTA19, and BTA21 for somatic cell count were also noted in prior research, though some significant SNPs discovered in this study had not been previously reported37,38,39,40,41.
Many genes were located alongside the identified markers, which may directly or indirectly influence the expression of genes associated with milk production traits. However, no reports have yet been published on the effects of some of these genes on milk production traits in cattle, indicating the need to expand our knowledge regarding the functions of these genes in bovines. On Chr26, tow genes (ATE1 and FGFR2) associated with milk fat percentage and milk protein percentage was identified. The ATE1 gene, identified in this study as significantly associated with somatic cell count in Iranian Holstein cows, plays a critical role in protein post-translational modification through arginylation, a process essential for regulating protein stability and degradation. This gene is known to be involved in various cellular functions, including stress response, apoptosis, and cell cycle control. Its identification as a candidate gene in the context of milk production suggests that ATE1 may influence immune and inflammatory responses in the mammary gland, potentially affecting mastitis susceptibility. This makes ATE1 a promising target for further functional studies and a valuable marker for improving udder health in genomic selection programs42. The FGFR2 (Fibroblast Growth Factor Receptor 2) gene emerged as a candidate associated with supernumerary teats (SNT) in the GWAS of Iranian Holstein cows, suggesting a potential role in mammary gland morphology and development. FGFR2 is a key component of the fibroblast growth factor signaling pathway, which regulates cell growth, differentiation, and tissue development. Previous studies have linked FGFR2 to mammary gland proliferation and its dysregulation to breast cancer development. Specifically, FGFR2 expression has been observed in the endometrial and trophoblastic epithelium, and its activation has been shown to influence epithelial integrity and fertility. These functions underscore FGFR2’s involvement in reproductive and mammary traits, making it a biologically plausible candidate gene for traits like supernumerary teats, which have implications for udder health, milkability, and the efficiency of mechanized milking systems42. ATE1 is a eukaryotic protein that plays a role in metabolism and apoptosis, reducing chromosomal aberrations through cell–cell contact43. A GWAS conducted by Fang et al.42 on Capra hircus demonstrated that the ATE1 gene is associated with udder size. Another gene identified in this study, FGFR2, has been linked to breast cancer44. Overexpression of growth hormone (GH) has been shown to promote mammary proliferation via FGFR2 and FGF742,45. On Chr24, the several genes (ALDH1A3, CHSY1, and GABRG3) were found alongside significant markers for milk fat percentage. The third enzyme from the aldehyde dehydrogenase 1 family, encoded by the ALDH1A3 gene, plays a detoxification and antioxidant role by converting retinaldehyde to retinoic acid44. In a GWAS conducted on Chinese Holstein cows, ALDH1A3 was associated with milk production traits, such as fat and protein content46. The CHSY1 gene has been previously shown to contribute to bone growth47, and this study demonstrates that it may also be linked to milk-related traits. Another essential gene identified is GABRG3, associated with teat size48. In other GWAS studies on cattle, GABRG3 has also been linked to carcass traits and feed efficiency49,50,51.
On Chr2, several genes associated with milk yield traits were identified, including the FBXO36 gene, which was linked to milk yield in this population. FBXO36, a member of the F-box protein family, plays a role in protein ubiquitination and is involved in critical cellular functions such as nutrient sensing, signal transduction, circadian rhythms, and the cell cycle, contributing to mastitis resistance in Holstein cows52,53,54. The function of this gene has been demonstrated in various cattle populations, showcasing its multifunctional role. These associations include specific diseases, infections, and biological functions related to adaptation55,56. Additionally, on the same chromosome, the PID1 gene plays a role in human lipid metabolism, reducing the sensitivity of adipocytes to insulin through the interaction of the phosphotyrosine-binding domain 1 with the lipoprotein receptor57. A GWAS study on cattle has identified the role of the PID1 gene in lipid metabolism and fatty acid synthesis58. TRIP12 is another gene that regulates the balance between protein synthesis and degradation and is involved in mammal muscle differentiation59. The exact role has been proposed for TRIP12 in intramuscular fat content in cattle58,60. Other critical genes on this chromosome include CD52, WDTC1, and MATN1. The CD52 gene encodes a glycoprotein that reduces T-cell activation61. The WDTC1 gene regulates fat-related gene transcription62. Reduced expression of MATN1 has been associated with impaired muscle growth63. On Chr24, the CIDEA gene was found alongside significant markers. Previous reports have highlighted its role in lipid synthesis in milk, which is influenced by the complex regulation of multiple gene expressions. CIDEA is a protein expressed in adipose tissue and associated with lipid droplets64. High expression of this gene in the mammary glands of lactating mice has been linked to lipid secretion65. Additionally, the CIDEA gene and several lipogenic enzymes are regulated post-partum in the mammary tissue of cattle66. On Chr5, the LYZ (Lysozyme) gene was identified by Salehin et al.67. They reported the significant effect of the LYZ gene on somatic cell count and milk production in cattle. The LYZ gene is of significant importance due to its strong antibacterial and immune-regulatory properties, particularly within the mammary gland of dairy animals. This gene encodes for lysozyme, an antimicrobial enzyme abundantly secreted in milk, saliva, and other bodily fluids, where it plays a crucial role in the innate immune system by breaking down bacterial cell walls. In the context of dairy production, LYZ is highly expressed in the mammary gland of buffaloes, contributing to their enhanced resistance to mastitis compared to cattle68. Therefore, the LYZ gene is not only a key marker for udder health and milk quality but also a promising candidate for genomic selection and therapeutic applications aimed at improving disease resistance in dairy herds. Another gene identified on this chromosome was CPM. The CPM protein plays a role in adipose tissue differentiation and has been identified as a candidate gene for milk fatty acids in Holstein cows69.
In Chr19, UCP1 gene was detected near significant SNPs with SCS trait. UCP1 gene is a mitochondrial carrier protein. Król et al.70 showed that the expression of UCP1 gene decreases during lactation in mice. Also, the effective function of UCP1 gene on milk protein percentage, milk fat percentage and milk yield has also been reported71. CYP2U1, SGMS2 and HADH genes cause the secretion of fat cells in milk because they play an important role in the metabolism of lipids and fatty acids72. In a GWAS experiment on cows, the role of these three genes (CYP2U1, SGMS2 and HADH) was reported as candidate genes for milk fat73. In Iranian Holstein cattle, SNP 17:28,549,748 in BTA17 was associated with SCS. According to Duchemin et al.74, this region contains the SCLT1 gene, which affects the fatty acid composition of milk from Holstein cows. The identified THRSP gene was located in the vicinity of the significant SNP associated with the SCS trait. THRSP gene in goat, with chest circumference and body weight75, with average daily weight gain, waist-eye area and back fat thickness in pig76 and in cattle with fatty acid composition milk74 and water holding capacity are correlated with meat tenderness77.
A new strategy in animal breeding programs, including for cattle, is using genomic information for economically important traits58. Identifying biological processes and genomic regions influencing milk production traits is essential for understanding the underlying genetic mechanisms. This study has identified novel genes as well as previously reported genes. In future breeding programs, the identified candidate gene variants can be utilized to improve milk production traits in dairy cattle. Additionally, validation studies involving gene expression analysis may be necessary in certain animal groups due to possible mutations in the identified candidate genes. This is essential for confirming the impact of these genes on the traits under investigation.
Conclusions
The genetic evaluation of milk production traits and somatic cell count in Holstein cows can be facilitated by combining genomic data in GWAS studies. We have identified several SNPs, important regions in various BTAs, and a list of candidate genes (both novel and known) that may contribute to variations in milk production traits and somatic cell count in Holstein cows. The genes ATE1, FGFR2, ALDH1A3, CHSY1, GABRG3, FBXO36, PID1, TRIP12, CD52, WDTC1, MATN1, CIDEA, LYZ, CPM, UCP1, MAML3, SGMS2, HADH, CYP2U1, SCLT1 and THRSP have been suggested as candidate genes for milk production traits and somatic cell count in Holstein cattle. These genes may be used for higher profit identification, causal mutations, and genomic predictions for milk production traits and somatic cell count in dairy cattle. This study demonstrated the feasibility of genetic evaluation for milk production traits and somatic cell count in the Iranian Holstein population, and it should be incorporated into the selection index for Iranian dairy cows.
Materials and methods
Phenotypic data
In the dairy farm of Ferdous Pars Agriculture Development, Iranian Holstein cows were selected. To conduct this study, 210 female cows (150 and 60 cattle, respectively, in herds 1 and 2) were selected for the study based on the breeding value of the milk production trait78. Animals were chosen using the two-tailed selection strategy outlined by Jiménez-Montero et al.79, which was based on estimated breeding values (EBVs) for milk yield. The EBVs were calculated by the National Animal Breeding Centre of Iran (Karaj, Iran) using a lactation model, as described in Eq. (1).80. The authors of the article confirm that the study was reported in accordance with the ARRIVE guidelines.
In this model, yij represents the milk yield, adjusted to a standard 305-day lactation period with twice-daily milking. The term μ denotes the overall population mean, hysi accounts for the fixed effect of the i herd-year-season group, aij represents the breeding value of the jth animal within the ith herd-year-season group, and eij captures the random residual error. The average accuracy of the estimated breeding values (EBVs) for milk yield was calculated to be 0.6180.
The following cases were also taken into consideration during the sampling in addition to those mentioned above: the sampling involved analyzing the livestock's pedigrees using the CFC V9.0 SP7 software81, and ensuring that both herds had a high diversity of livestock was done by choosing livestock with minimal kinship relationships80. A complete pedigree (The pedigree of the cows is given in Supplementary 3) and records were available for the selected animals, and it was ensured that the animals were not candidates for elimination. During the first to sixth lactation of 210 Holstein cows located on one Iranian farms with two herds, 75,228 phenotypic records were collected from May 2013 to December 2020. Among the traits studied were test-day milk yield (MY; kg/d), somatic cell count (SCC, converted according to Ali and Shook,82), milk protein percentage (PP, %), and milk fat percentage (FP, %). A summary of the phenotypic data is shown in Table 1.
Genotype imputation and quality control (QC)
One-hundred fifty (150) and Sixty )60( animals from herd 1 and 2 were genotyped by the GGP-LD v.4 SNP panel (with 30,108 SNPs) and the Affymetrix Axiom Bovine Array-50 K (with 51,987 SNPs), respectively.
Using the software PLINK 2.0 to control genotyping quality, four criteria were used. Those animals with over 5% missing genotypes were excluded, those with minor allele frequencies (MAFs) less than 5%, and SNPs that were not genotyped for more than 5% of animals and chi scores were less than 10–6 (Chi-square < 10−6) were excluded from the Hardy–Weinberg equilibrium test. To check imputation accuracy and identify and remove markers that had lower accuracy and stepwise imputation, Minimac3 2.0.1 software was used83.
The 210 cows (150 from Herd 1 and 60 from Herd 2) were genotyped using two SNP panels: the GGP-LD v.4 (30,108 SNPs) and the Affymetrix Axiom Bovine Array-50 K (51,987 SNPs). These animals comprised the target population84. Genotypes were then imputed to whole-genome sequence level using a reference population of 234 animals from the 1000 Bull Genomes Project. This reference panel included key progenitors from four major breeds: Holstein–Friesian (n = 129), Fleckvieh (n = 43), Jersey (n = 15), and Angus (n = 47), each genotyped using the BovineHD BeadChip and whole-genome sequencing data85. Quality control was applied to both SNP chip and sequence data, resulting in 578,505 SNPs from the BovineHD chip and 12,063,146 SNPs from the sequence data after filtering. Genotype phasing was conducted using Eagle v2.3, and imputation was performed with Minimac3 for both reference and target populations78. After removing imputed SNPs with an accuracy (R2) below 0.30, 6,583,595 high-confidence SNPs were retained and used in the genome-wide association analysis.
GWAS for somatic cells count and milk production
A mixed linear model in EMMAX was used to association studies between imputed genotypes and milk production and somatic cell count traits86. EMMAX adjust for both population stratification and relatedness in the association study. The mixed model used for this study was as follow Eq. (1):
where X is a n × q matrix of fixed effects including overall mean, covariates and the testing SNP; y is a n × 1 vector of the phenotypic measurement, b is a q × 1 vector denoting the coefficients of fixed effects; Z is a n × t incidence matrix which relates phenotypes to the corresponding random polygenic effect; u is a t × 1 vector of the random polygenic effect and e is a n × 1 vector of the residual effects. Furthermore, Var(u)=\({\sigma }_{g}^{2}K\) and var(e)=\({\sigma }_{e}^{2}I\) that I is identity matrix and K is a kinship matrix among all imputed sequence genotypes.
In GWAS, a Bonferroni-corrected genomic threshold of 1 × 10–8 (P < 0.05 / total number of SNPs) for association study is known. We used the R 4.3.2 software to draw the Manhattan plot using the qqman package87.
Gene annotation
Our study used Ensembl annotations of the UMD3.1 genome version (http://www.ensembl.org/biomart/martview) to identify candidate genes surrounding (within one megabase) SNPs that passed the threshold of P < 1 × 10–8. An analysis of gene ontologies was performed using DAVID Bioinformatics Resources version 6.7 (http://david.abcc.ncifcrf.gov/). Also, to identify those QTLs that fall within 1 Mb of SNPs that meet the threshold of P < 1 × 10–8, the QTLdb of cattle was used (https://www.animalgenome.org/cgibin/QTLdb/BT/index). The GeneMANIA (http://genemania.org/) was then used to draw gene networks.
Data availability
The datasets generated and analyzed during the current study are available in the Figshare repository [https://doi.org/10.6084/m9.figshare.28604060].
References
Buaban, S., Lengnudum, K., Boonkum, W. & Phakdeedindan, P. Genome-wide association study on milk production and somatic cell score for Thai dairy cattle using weighted single-step approach with random regression test-day model. J. Dairy Sci. 105(1), 468–494 (2022).
Snelling, W. M. et al. Breeding and genetics symposium: networks and pathways to guide genomic selection–. J. Anim. Sci. 91(2), 537–552 (2013).
Li, H. et al. Genome-wide association study of milk production traits in a crossbred dairy sheep population using three statistical models. Anim. Genet. 51(4), 624–628 (2020).
Jiang, L. et al. Genome wide association studies for milk production traits in Chinese Holstein population. PLoS ONE 5(10), e13661 (2010).
Heringstad, B., Sehested, E. & Steine, T. Correlated selection responses in somatic cell score from selection against clinical mastitis. J. Dairy Sci. 91(11), 4437–4439 (2008).
Rupp, R. & Boichard, D. Genetics of resistance to mastitis in dairy cattle. Vet. Res. 34(5), 671–688 (2003).
Carlen, E., Strandberg, E. & Roth, A. Genetic parameters for clinical mastitis, somatic cell score, and production in the first three lactations of Swedish Holstein cows. J. Dairy Sci. 87(9), 3062–3070 (2004).
Daw, E. W., Heath, S. C. & Lu, Y. Single-nucleotide polymorphism versus microsatellite markers in a combined linkage and segregation analysis of a quantitative trait. BMC Genet. 6(Suppl 1), S32 (2005).
Freebern, E. et al. GWAS and fine-mapping of livability and six disease traits in Holstein cattle. BMC Genom. 21, 1–11 (2020).
Horin, P., Osickova, J., Necesankova, M., Matiasovic, J., Musilova, P., Kubickova, S., & Rubes, J. Single nucleotide polymorphisms of interleukin-1 beta related genes and their associations with infection in the horse. In Animal Genomics for Animal Health Vol. 132 347-351 Karger Publishers. (2008)
Craig, D. W. & Stephan, D. A. Applications of whole-genome high-density SNP genotyping. Expert Rev. Mol. Diagn. 5(2), 159–170 (2005).
Scott, Laura J., et al. 2007 "A association study of type diabetes in nts." science 5 (2) 159-170.
Abdalla, I. M. et al. Identification of candidate genes and functional pathways associated with body size traits in chinese holstein cattle based on GWAS analysis. Animals 13(6), 992 (2023).
Zepeda-Batista, J. L. et al. Discovering of genomic variations associated to growth traits by GWAS in Braunvieh cattle. Genes 12(11), 1666 (2021).
Otto, P. I. et al. Single-step genome-wide association studies (GWAS) and post-GWAS analyses to identify genomic regions and candidate genes for milk yield in Brazilian Girolando cattle. J. Dairy Sci. 103(11), 10347–10360 (2020).
Nayeri, S. et al. Genome-wide association for milk production and female fertility traits in Canadian dairy Holstein cattle. BMC Genet. 17, 1–11 (2016).
van den Berg, I., Boichard, D. & Lund, M. S. Comparing power and precision of within-breed and multibreed genome-wide association studies of production traits using whole-genome sequence data for 5 French and Danish dairy cattle breeds. J. Dairy Sci. 99(11), 8932–8945 (2016).
Sahana, G. et al. Genome-wide association study using high-density single nucleotide polymorphism arrays and whole-genome sequences for clinical mastitis traits in dairy cattle. J. Dairy Sci. 97(11), 7258–7275 (2014).
Steri, R., Moioli, B., Catillo, G., Galli, A. & Buttazzoni, L. Genome-wide association study for longevity in the Holstein cattle population. Animal 13(7), 1350–1357 (2019).
Zhang, H. et al. Genetic parameters and genome-wide association studies of eight longevity traits representing either full or partial lifespan in Chinese Holsteins. Front. Genet. 12, 634986 (2021).
Zhang, Q., Guldbrandtsen, B., Thomasen, J. R., Lund, M. S. & Sahana, G. Genome-wide association study for longevity with whole-genome sequencing in 3 cattle breeds. J. Dairy Sci. 99(9), 7289–7298 (2016).
Abdollahi-Arpanahi, R., Carvalho, M. R., Ribeiro, E. S. & Peñagaricano, F. Association of lipid-related genes implicated in conceptus elongation with female fertility traits in dairy cattle. J. Dairy Sci. 102(11), 10020–10029 (2019).
Frischknecht, M. et al. Intergenomics Consortium Fries R Russ I, Sölkner J, Bieber A Genome-wide association studies of fertility and calving traits in Brown Swiss cattle using imputed whole-genome sequences. BMC Genom. 18(1), 3 (2017).
Liu, A. et al. Variance components and correlations of female fertility traits in Chinese Holstein population. J. Anim. Sci. & Biotechnol. 8, 1–9 (2017).
Lu, X. et al. Genome-wide association study on reproduction-related body-shape traits of Chinese Holstein cows. Animals 11(7), 1927 (2021).
Wang, X. et al. Genome-wide association study in Chinese Holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. 16(1), 9 (2015).
Chen Xing, C. X., Cheng ZhangRui, C. Z., Zhang ShuJun, Z. S., Werling, D., & Wathes, D. C. Combining genome wide association studies and differential gene expression data analyses identifies candidate genes affecting mastitis caused by two different pathogens in the dairy cow (2015)
Bovenhuis H, Weller JI. Mapping and analysis of dairy cattle quantitative trait loci by maximum likelihood methodology using milk protein genes as genetic markers. Genetics 137, 267–280 (1994).
Iung, L. H. S. et al. Genome-wide association study for milk production traits in a Brazilian Holstein population. J. Dairy Sci. 102(6), 5305–5314 (2019).
Križanac, A. M., Reimer, C., Heise, J., Liu, Z., Pryce, J., Bennewitz, J., et al. Sequence-based GWAS in 180 000 German Holstein cattle reveals new candidate genes for milk production traits. bioRxiv 2023–12. (2023)
Chen SY, Gloria LS, Pedrosa VB, Doucette J, Boerman JP, Brito LF. Unraveling the genomic background of resilience based on variability in milk yield and milk production levels in North American Holstein cattle through genome-wide association study and Mendelian randomization analyses. J. Dairy Sci. 107, 1035–1053 (2024).
Liu L, Zhou J, Chen CJ, Zhang J, Wen W, Tian J, et al. GWAS-based identification of new loci for milk yield, fat, and protein in Holstein cattle. Animals 10, 2048 (2020).
Silva, A. A. et al. GWAS and gene networks for milk-related traits from test-day multiple lactations in Portuguese Holstein cattle. J. Appl. Genet. 61(3), 465–476 (2020).
Atashi, H. et al. Single-step genome-wide association for selected milk fatty acids in Dual-Purpose Belgian Blue cows. J. Dairy Sci. 106(9), 6299–6315 (2023).
Shamsollahi, M. & Zhang, S. Genome wide association study associated with milk protein composition. Anim. Sci. Res. 34(1), 31–44 (2024).
Bakhshalizadeh, S., Zerehdaran, S., Hasanpur, K., & Javadmanesh, A. Identification of potential candidate genes associated with milk protein differences in Holstein cows: a meta-analysis integrating GWAS and RNA-Seq transcriptome. Canadian Journal of Animal Science (2024)
Ilie, D. E. et al. Genome-wide association studies for milk somatic cell score in romanian dairy cattle. Genes 12(10), 1495 (2021).
Sutera, A. M. et al. Genome-wide association study identifies new candidate markers for somatic cells score in a local dairy sheep. Front. Genet. 12, 643531 (2021).
Wang, P. et al. Genome-wide association analysis of milk production, somatic cell score, and body conformation traits in Holstein cows. Front. Vet. Sci. 9, 932034 (2022).
Strillacci MG, Punturiero C, Milanesi R, Bernini F, Mason T, Bagnato A. Antibiotic treatments and somatic cell count as phenotype to map QTL for mastitis susceptibility in Holstein cattle breed. Ital. J. Anim. Sci. 22, 190–199 (2023).
Lu X, Jiang H, Arbab AAI, Wang B, Liu D, Abdalla IM, et al. Investigating genetic characteristics of Chinese Holstein cow’s milk somatic cell score by genetic parameter estimation and genome-wide association. Agriculture 13, 267 (2023).
Fang X, Gu B, Chen M, Sun R, Zhang J, Zhao L, Zhao Y. Genome-wide association study of the reproductive traits of the Dazu black goat (Capra hircus) using whole-genome resequencing. Genes 14, 1960 (2023).
Jiang C, Moorthy BT, Patel DM, Kumar A, Morgan WM, Alfonso B, et al. Regulation of mitochondrial respiratory chain complex levels, organization, and function by arginyltransferase 1. Front. Cell Dev. Biol. 8, 603688 (2020).
Bao Y, Wang L, Shi L, Yun F, Liu X, Chen Y, et al. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell. Mol. Biol. Lett. 24, 38 (2019).
Spencer TE, Bazer FW. Uterine and placental factors regulating conceptus growth in domestic animals. J. Anim. Sci. 82(Suppl 13), E4–E13 (2004).
Bao Z, Lin J, Ye L, Zhang Q, Chen J, Yang Q, Yu Q. Modulation of mammary gland development and milk production by growth hormone expression in GH transgenic goats. Front. Physiol. 7, 278 (2016).
Yu H, Yu S, Guo J, Cheng G, Mei C, Zan L. Genome-wide association study reveals novel loci associated with body conformation traits in Qinchuan cattle. Animals 13, 3628 (2023).
Tao L, He XY, Wang FY, Pan LX, Wang XY, Gan SQ, et al. Identification of genes associated with litter size combining genomic approaches in Luzhong mutton sheep. Anim. Genet. 52, 545–549 (2021).
Olivieri BF, Mercadante MEZ, Cyrillo JNDSG, Branco RH, Bonilha SFM, de Albuquerque LG, et al. Genomic regions associated with feed efficiency indicator traits in an experimental Nellore cattle population. PLoS One 11, e0164390 (2016).
Maiorano AM, Cardoso DF, Carvalheiro R, Júnior GAF, de Albuquerque LG, de Oliveira HN. Signatures of selection in Nelore cattle revealed by whole-genome sequencing data. Genomics 114, 110304 (2022).
Ceballos, F.C., Hazelhurst, S. & Ramsay, M. Assessing runs of Homozygosity: a comparison of SNP Array and whole genome sequence low coverage data. BMC Genomics 19, 106 (2018).
Cuesta, L. M., Liron, J. P., Nieto Farias, M. V., Dolcini, G. L. & Ceriani, M. C. Effect of bovine leukemia virus (BLV) infection on bovine mammary epithelial cells RNA-seq transcriptome profile. PLoS ONE 15(6), e0234939 (2020).
Johnston, D. et al. Elucidation of the host bronchial lymph node miRNA transcriptome response to bovine respiratory syncytial virus. Front. Genet. 12, 633125 (2021).
Gondaira, S. et al. Innate immune response in bovine neutrophils stimulated with mycoplasma bovis. Vet. Res. 52(1), 11 (2021).
Kurz, J. P. et al. A genome-wide association study for mastitis resistance in phenotypically well-characterized Holstein dairy cattle using a selective genotyping approach. Immunogenetics 71, 35–47 (2019).
Laodim, T. et al. Genetic factors influencing milk and fat yields in tropically adapted dairy cattle: Insights from quantitative trait loci analysis and gene associations. Anim. Biosci. 37(4), 576 (2024).
Shi, C. M. et al. Knockdown of NYGGF4 (PID1) rescues insulin resistance and mitochondrial dysfunction induced by FCCP in 3T3-L1 adipocytes. Mitochondrion 12(6), 600–606 (2012).
Machado, P. C. et al. Genome-wide association analysis reveals novel loci related with visual score traits in nellore cattle raised in pasture-based systems. Animals 12(24), 3526 (2022).
Egerman, M. A. & Glass, D. J. Signaling pathways controlling skeletal muscle mass. Crit. Rev. Biochem. Mol. Biol. 49(1), 59–68 (2014).
Silva, D. B. et al. Spliced genes in muscle from Nelore Cattle and their association with carcass and meat quality. Sci. Rep. 10(1), 14701 (2020).
Furukawa, A., Wisel, S. A. & Tang, Q. Impact of immune-modulatory drugs on regulatory T cell. Transplantation 100(11), 2288–2300 (2016).
Ducos, E. et al. Remarkable evolutionary conservation of antiobesity ADIPOSE/WDTC1 homologs in animals and plants. Genetics 207(1), 153–162 (2017).
García-Contreras, C. et al. Impact of genotype, body weight and sex on the prenatal muscle transcriptome of Iberian pigs. PLoS ONE 15(1), e0227861 (2020).
Sun, X. et al. High expression of cell death-inducing DFFA-like effector a (CIDEA) promotes milk fat content in dairy cows with clinical ketosis. J. Dairy Sci. 102(2), 1682–1692 (2019).
Wang, W. et al. Cidea is an essential transcriptional coactivator regulating mammary gland secretion of milk lipids. Nat. Med. 18(2), 235–243 (2012).
Bionaz, M. et al. Old and new stories: Revelations from functional analysis of the bovine mammary transcriptome during the lactation cycle. PLoS ONE 7(3), e33268 (2012).
Salehin, M., Ghosh, A. K., Mallick, P. K. & Bhattacharya, T. K. Molecular characterization, polymorphism and association study of lysozyme gene with milk production and somatic cell trait in Bos indicus× Bos taurus cattle. Animal 3(5), 623–631 (2009).
Su, J. et al. Comparative evolutionary and molecular genetics based study of Buffalo lysozyme gene family to elucidate their antibacterial function. Int. J. Biol. Macromol. 15(234), 123646 (2023).
Shi, L. et al. Identification of genetic effects and potential causal polymorphisms of CPM gene impacting milk fatty acid traits in Chinese Holstein. Anim. Genet. 51(4), 491–501 (2020).
Król, E., Martin, S. A., Huhtaniemi, I. T., Douglas, A. & Speakman, J. R. Negative correlation between milk production and brown adipose tissue gene expression in lactating mice. J. Exp. Biol. 214(24), 4160–4170 (2011).
Zhou, H. et al. Haplotypic variation in the UCP1 gene is associated with milk traits in dairy cows. J. Dairy Res. 84(1), 68–75 (2017).
Grilz-Seger, G. et al. High-resolution population structure and runs of homozygosity reveal the genetic architecture of complex traits in the Lipizzan horse. BMC Genom. 20, 1–17 (2019).
Illa, S. K., Mukherjee, S., Nath, S. & Mukherjee, A. Genome-wide scanning for signatures of selection revealed the putative genomic regions and candidate genes controlling milk composition and coat color traits in Sahiwal cattle. Front. Genet. 12, 699422 (2021).
Duchemin, S. I., Bovenhuis, H., Megens, H. J., Van Arendonk, J. A. M. & Visker, M. H. P. W. Fine-mapping of BTA17 using imputed sequences for associations with de novo synthesized fatty acids in bovine milk. J. Dairy Sci. 100(11), 9125–9135 (2017).
An, X. et al. Polymorphism identification in the goat THRSP gene and association analysis with growth traits. Arch. Anim. Breed. 55(1), 78–83 (2012).
Wang, X. et al. Polymorphisms in 5 proximal regulating region of THRSP gene are associated with fat production in pigs. 3 Biotech 10(1), 9 (2020).
Oh, D. Y. et al. Identification of exonic nucleotide variants of the thyroid hormone responsive protein gene associated with carcass traits and Fatty Acid composition in korean cattle. Asia.-Aust. J. Anim. Sci. 27(10), 1373 (2014).
JalilSarghale, A. et al. Genome-wide association studies for methane emission and ruminal volatile fatty acids using Holstein cattle sequence data. BMC Genet. 21(1), 1–4 (2020).
Jiménez-Montero, J. A., Gonzalez-Recio, O. & Alenda, R. Genotyping strategies for genomic selection in small dairy cattle populations. Animal 6, 1216–1224 (2012).
Abdollahi-Arpanahi, R., Razmkabir, M., SayadNezhad, M. & Eghbal, A. Determination of the number of test day records is required to replace lactation model with random regression model?. Iran. J. Anim. Sci. 48, 391–398 (2017).
Sargolzaei, M., Schenkel, F. S., Jansen, G. B. & Schaeffer, L. R. Extent of linkage disequilibrium in Holstein cattle in North America. J. Dairy Sci. 91(5), 2106–2117 (2008).
Ali, A. K. A. & Shook, G. E. An optimum transformation for somatic cell concentration in milk. J. Dairy Sci. 63, 487–490 (1980).
Fuchsberger, C., Abecasis, G. R. & Hinds, D. A. Minimac2: Faster genotype imputation. Bioinformatics 31(5), 782–784 (2014).
Daetwyler, H. D. et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46(8), 858–865 (2014).
Chen, S. Y. et al. Identifying pleiotropic variants and candidate genes for fertility and reproduction traits in Holstein cattle via association studies based on imputed whole-genome sequence genotypes. BMC Genom. 23(1), 1–22 (2022).
Hoze, C. et al. A splice site mutation in CENPU is associated with recessive embryonic lethality in Holstein cattle. J. Dairy Sci. 103(1), 607–612 (2020).
Turner SD. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv. 005165 (2014).
Acknowledgements
We would like to extend thanks to the Ferdows Pars Agricultural Holding Company, and National Animal Breeding Centre of Iran for giving us access to the animals and recording. Finally, we acknowledge the 1000 Bull Genomes Project for making their research data publicly available.
Funding
This research did not receive any specific funding.
Author information
Authors and Affiliations
Contributions
NM performed the experiments and data analysis and wrote the article draft; MS and AJS supervised the project and provided editorial input on the writing. NM, MS, AJS, MS, MKD and MK contributed to the data collection and supervised the analysis. All authors discussed the results and contributed to the final manuscript. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The samples collected from the studied animals were performed in accordance with animal ethics and approved by the Animal Use Committee of the University of Tehran and the National Animal Breeding Centre of Iran. In addition, permission for sampling was obtained from the farmers on site. Also, we have obtained the informed consent of the owner(s) to use the animals in our study in this area from the University of Tehran and the National Animal Breeding Center of Iran. The authors of the article confirm that the study was reported in accordance with the ARRIVE guidelines.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Maddahi, N., Sadeghi, M., Sarghale, A.J. et al. Identification of candidate gene networks affecting the number of somatic cells count and milk production in Iranian Holstein cows using Genome-wide association study. Sci Rep 15, 32168 (2025). https://doi.org/10.1038/s41598-025-09103-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-09103-x