Abstract
Pseudomonas aeruginosa (PA) bloodstream infections (BSIs) are severe, life-threatening events. PA success involves a complex interplay between virulence, antimicrobial resistance and epidemicity. This study examines the impact of COVID-19 pandemic on PA-BSI characteristics. Seventy-eight bacteraemic PA isolates were collected in two periods: pre-COVID 2015–2018 (48 PA) and post-COVID 2022–2023 (30 PA). An increase in non-susceptibility rates was found in post-COVID period, noteworthy for gentamicin, netilmicin and multidrug-resistance, although extensively drug resistant/difficult-to-treat (XDR/DTR) isolates were only detected in pre-COVID period. All isolates were cefiderocol-susceptible, but resistance to novel cephalosporin-inhibitor combinations and colistin was identified in pre-COVID period. Carbapenem-non-susceptible PA persisted around 20%, associated with OprD alterations and one pre-COVID-isolate harboured blaVIM-2. High clonal diversity was found: 71 pulsotypes and 52 sequence types (most prevalent: ST17, ST244 and ST274). A shift in serotypes was detected between pre-COVID-PA (O:1) and post-COVID-PA (O:6). Eight virulotypes were detected, highlighting one exlA-hypervirulent post-COVID-PA, and exoU+/exoS- genotype in 17% pre-COVID-PA but none post-COVID-PA. Biofilm production was significantly higher in pre-COVID-PA. This work shed light on the evolving nature of PA infections amidst the COVID-19 pandemic, providing valuable insights for clinical management and reinforcing the importance of judicious antibiotic use in the context of public health crises.
Similar content being viewed by others
Introduction
Bloodstream infections (BSIs) are serious clinical events with life-threatening consequences. BSIs have high morbidity worldwide and an estimated overall crude mortality rate of 15%-30%. Pseudomonas aeruginosa (PA) is the third most commonly identified BSI cause among Gram-negative microorganisms, and accounts for 3 to 8% of all BSIs. In Europe, the prevalence of PA-BSI is 6.1%, being 7% in Spain1. The death rate due to PA-BSI is severe (ranging from 32 to 73%) with an estimated attributable mortality of approximately 30%, the highest one among causative agents2,3,4,5,6. In this regard, factors related to the host, the organism, and the treatment (such as neutropenia, a respiratory source of bacteraemia, bacterial intrinsic virulence, antibiotic resistance or the timely administration of adequate antimicrobial therapy, among others) may increase mortality2,7,8.
PA-BSI represents an important clinical problem, which is aggravated by the high rate of PA antibiotic resistance. Beyond having an intrinsic resistance to several antibiotics, PA can develop other resistances through chromosomal mutations or by acquisition of horizontally transferable antibiotic resistance determinants, such as carbapenemases, that limit treatment options9,10. In addition, the multiple pathogenicity factors of PA, such as biofilm production and secretion of adhesins, toxins, proteases and pigments, favour the PA success in infecting the host cell and evading the host immune system. Lipopolysaccharide (LPS) is also an important virulence factor in PA, exerting direct endotoxic effects, with O:1, O:6, and O:11 being the most frequent serotypes in many clinical settings10,11. Moreover, the presence of exoU gene that encodes a cytotoxic phospholipase secreted by the type III secretion system (T3SS), determines a greater impact in bacterial virulence and is associated with early deaths in BSIs, but not with overall mortality6. Indeed, the interconnections between antimicrobial resistance and virulence traits, and how they may impact the outcome of bacterial infections is a subject of growing interest.
The COVID-19 pandemic has not only posed an unprecedented global health crisis, but has also inadvertently cast a spotlight on an equally pressing issue—antibiotic resistance. As healthcare systems worldwide grappled with the surge in COVID-19 cases, extensive and improper antibiotic usage became the norm, often as a precautionary measure to mitigate secondary bacterial infections. Furthermore, there was a huge increase in the use of biocides that could also encourage indirect pressure on antibiotic resistance12,13. On the other hand, the implementation of enhanced infection prevention and control measures, alongside improved hand hygiene practices, was initially anticipated to have a positive impact on the rates of various healthcare-associated infections. Consequently, it remains uncertain whether the incidence of PA bacteraemia increased, decreased, or remained unchanged after the pandemic, with conflicting findings reported in the literature13,14,15,16.
In this work, the aim was to delve into the intricate interplay between the COVID-19 pandemic, and the emergence of antibiotic resistance of PA-BSI in a non-outbreak scenario. The objective was to provide insights into the molecular epidemiology of this microorganism in clinical blood samples, crucial not only for guiding treatment strategies but also for reinforcing the importance of judicious antibiotic use in the context of public health crises.
Results
Clinical and epidemiological data
A total of seventy-eight PA-BSI were collected from 78 episodes from 76 patients, in two different periods: pre-COVID period 2015–2018 (48 isolates), and post-COVID period 2022–2023 (30 isolates). The patient’s clinical and demographic characteristics are included in Table 1. The average age of the patients was 72.4 year-old (range 15–95), and the majority were men (63%). The most frequently suspected clinical sources of infection were abdominal focus (23%) and UTI (22%) (Fig. 1; Table 1). The BSIs were classified as hospital acquired (34 isolates, 44%), health care-associated (34 isolates, 44%) or community acquired (8 isolates, 10%) (Table 1). Most BSIs of pre-COVID-episodes were hospital acquired (48%) and health care-associated (42%), as well as post-COVID-BSI episodes (37% and 47%, respectively). PA-BSI were isolated in different services, highlighting oncology/haematology (23.1%) and infectious diseases (19.2%) (Fig. 1; Table 1). Crude mortality was 53% in the 78 episodes (54% in pre-COVID and 50% in post-COVID-period), and with an attributable mortality (died within 30 days of BSIs) of 24% due to the PA-BSI. The distribution of isolates according to hospital ward or sources of infection in each period did not show notable differences (Fig. 1).
Clinical features of the 78 episodes. (a) Source of BSI. (b) Admision departments where the PA-BSI were isolated. The pre-COVID and post-COVID strains are differentiated in blue and red, respectively. UTI (urinary tract infection), STI (soft tissue infection), ICU (intensive care unit), IM (internal medicine), ID (infectious disease).
Only three patients co-harboured COVID and PA-BSI, and the origin of the infection was respiratory (S1 Table). One of these three patients died very likely due to the PA-BSI.
Clonal relatedness: Pulsed-Field-Gel-Electrophoresis (PFGE) and Multilocus-sequence-typing (MLST)
Clonal diversity was evaluated in all isolates by PFGE and MLST. SpeI-PFGE classified the 78 PA-BSI in 71 different pulsotypes, revealing a remarkable clonal diversity (Fig. 2). Six pulsotypes (four with pre-COVID-PA and two with post-COVID-PA) were shared between two or three strains each. The two pairs of PA isolated from the same patients were found to be clonally identical (S1 Table). None of the pulsotypes was detected in both periods, pointing out that there was not any clonal relationship between pre- and post-COVID isolates.
Dendrogram constructed using PFGE patterns of the 78 PA-BSI of this study. PFGE patterns were analyzed by GelJ v2 program, using the Dice coefficient. The MLST, the serotype, the antimicrobial resistance phenotype and the virulotype data have been included. The pre-COVID and post-COVID isolates are differentiated with blue and red circles, respectively.
A total of 52 sequence types (STs) were identified among the 78 isolates, 34 in the pre-COVID period and 25 in the post-COVID. Five of them resulted from a new allele combination, which were submitted to the P. aeruginosa MLST database, and assigned as ST3145, ST3146, ST3989, ST4373 and ST4381. Seven STs (ST244, ST274, ST252, ST381, ST395, ST487 and ST611) were detected in PA-BSI from both periods. The most prevalent STs were ST17, ST244 and ST274 (5 isolates each), although there were not detected equally in both periods (Fig. 3). On the other hand, 40 STs were found in only one isolate. These results confirmed the remarkable clonal diversity, and isolates with indistinguishable PFGE pattern had the same ST. Among the worldwide top ten PA high-risk clones (HRC)10, five isolates belonged to ST244, the Ps1015 isolate was ST298, and the Ps1056 isolate belonged to the HRC ST175.
Ten different serotypes were assigned using monovalent specific sera, and 21 PA-BSI were non-typable isolates (27%) (including polyagglutinable and no agglutinable ones) (Fig. 4). Serotype O:6 was the most prevalent (17% of PA-BSI), followed by O:1 (15%) (S1 Table). Regarding serotyping in the pre- and post-COVID-PA, a decrease in the detection of serotype O:1 (23% vs. 3%) was observed, in contrast, the prevalence of serotypes O:2 (4% vs. 17%), O:5 (8% vs. 17%), O:6 (10% vs. 27%) and O:9 (5% vs. 10%) was higher in the post-COVID period (Fig. 4).
Antibiotic susceptibility and resistance mechanisms: OprD, MBLs and integrons
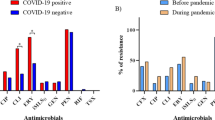
The Fig. 5 shows the susceptibility analysis of the 78 PA-BSI to the 17 antipseudomonal agents. Non-susceptibility rates were higher in the post-COVID-PA, being statistically significant higher for gentamicin and netilmicin. The highest rates of non-susceptibility in the pre-COVID-PA were for aztreonam and ciprofloxacin (33.3% and 22.9%, respectively), and in the post-COVID-PA for aztreonam, netilmicin and gentamicin (43.3%, 43.3% and 40%, respectively). All 78 isolates were susceptible to cefiderocol; however, resistance to the new antimicrobial combinations, ceftazidime-avibactam and ceftolozane-tazobactam, as well as to colistin, were found in the pre-COVID-PA (Fig. 5a).
Comparative analysis of (a) non-susceptibility to 17 antipseudomonal agents tested in the 78 PA-BSI of pre-COVID (blue) and post-COVID (red) periods, and (b) resistance phenotypes detected in both periods: XDR (purple), MDR (light red) and non-MDR (light yellow).*p < 0.05. Abbreviations: PIP: piperacillin, TZP: piperacillin-tazobactam, CAZ: ceftazidime, FEP: cefepime, ATM: aztreonam, IPM: imipenem, MEM: meropenem, DOR: doripenem, GEN: gentamicin, TOB: tobramycin, AMK: amikacin, NET: netilmicin, CIP: ciprofloxacin, FDC: cefiderocol, CZA: ceftazidime-avibactam, C/T: ceftolozane-tazobactam, CST: colistin, XDR: extensively drug resistant, MDR: multidrug-resistant.
Depending on their resistance phenotype, PA-BSI were classified as multiS (susceptible to all tested antipseudomonal agents), modR (non-susceptible to ≥ 1 agent in ≤ 2 antimicrobial categories), MDR (non-susceptible to ≥ 1 agent in ≥ 3 categories) or XDR (non-susceptible to ≥ 1 agent in all but ≤ 2 categories)17,18. The strains that were non-susceptible to all of the following antibiotics: piperacillin–tazobactam, ceftazidime, cefepime, aztreonam, meropenem, imipenem, and ciprofloxacin, were considered difficult-to-treat resistant (DTR)19.
Regarding the distribution of resistance profiles, most of the strains were non-MDR (76.9%), which included multiS and modR isolates. The presence of non-MDR isolates was higher in the pre-COVID period (87.5% vs. 60%), and a remarkable increase of MDR isolates was detected in the post-COVID period (40% vs 6.3%). However, three XDR isolates (6.25%), also classified as DTR, were detected only in the pre-COVID period (Fig. 5b).
The carbapenem-non-susceptibility was quite extended and maintained in the time (18–20%). However, metallo-beta-lactamase (MBL) phenotype was only detected in the Ps1031 strain (1.3%), isolated in the pre-COVID period, ST395, and harboured the blaVIM-2 gene. None other carbapenemase encoding gene was detected in the remaining isolates.
Alterations in the oprD gene were analysed in the 13 carbapenem-resistant PA (Table 2). Four isolates had a premature stop codon reducing OprD length to 63 or to 276 codons. Other four isolates contained deleted base pairs in their oprD gene, and the insertion sequence (IS) element, ISPa104, was found at codon 378 in PA Ps1019. This IS was first characterized and described in this study, and was included in ISFinder Database (https://isfinder.biotoul.fr/). Regarding the remaining four isolates, one isolate harboured the wild type OprD, and other two isolates and the VIM-positive PA showed non OprD inactivating mutations, including amino acid substitutions plus the deletion of two amino acids in the loop 7 region (loop L7-short).
Integrons were only detected in two isolates: the class 1 integron intI1 + aadB + qacEΔ1-sul1 (In7, assigned by the INTEGRALL Database [http://integrall.bio.ua.pt/]) in the ST175-Ps1056 isolate, and the intI1 + blaVIM-2 + qacEΔ1-sul1 (In56) in the MBL-producing Ps1031 isolate.
Virulence determinants
Eight virulotypes were assigned among the 78 PA-BSI, according to the presence/absence of 14 virulence and quorum sensing genes, being the virulotype V1 (exoS, exoY, exoT, lasA, lasB, exoA, aprA, rhlAB, rhlI, rhlR, lasI and lasR positive) the most frequent one (80.8%) (Table 3). Regarding the amplification of T3SS effector exoU and exoS genes, the following genotypes were obtained (total PA/pre-COVID-PA/post-COVID-PA): 68 PA exoU-/exoS+ genotype (87/83/93%), 8 exoU+/exoS- (10/17/0%), one exoU+/exoS+ (1/0/3%) and one PA exoU-/exoS- (1/0/3%) (Fig. 6). Interestingly, one strain (Ps1457, ST273) exhibited the rare co-occurrence of both exoS and exoU genes (V7). Noteworthy was the identification of a post-COVID-PA (Ps1450) devoid of all T3SS genes (exoU, exoS, exoT and exoY) and exoA gene, that harboured the exlA gene associated with hyper-virulence in PA (V8) (Table 3; Fig. 6). Further highlighting the unique genetic makeup, and its potential hyper-virulent characteristics of this Ps1450 isolate belonging to ST2211.
The presence of lasA, lasB, aprA, lasI, and lasR genes was observed in all isolates, although a subset of five isolates revealed the presence of insertion sequences disrupting the lasR gene. Specifically, ISPPu21 was identified in two isolates (Ps1030 and Ps1184, ST252), ISPsp7 in Ps1321 (ST381), ISPa41 in Ps1447 (ST499), and ISPa104 in Ps1019 (ST3145) (Table 3). Notably, in Ps1019, the new ISPa104 was also found truncating rhlR and oprD genes (S1 Table). Of particular interest was the absence of rhlR/rhlI and rhlAB genes in the Ps1056 isolate (ST175) (Table 3).
Biofilm, elastase and pigment production
Compared to P. aeruginosa PAO1, the biofilm biomass was higher in 16 PA (20.5%) isolates, lower in 53 (68%) and similar in 9 PA (11.5%). Interestingly, there was a statistically significant difference in the PA-BSI biofilm production between both periods, being higher in the pre-COVID-PA (Fig. 7a). Furthermore, there were also differences in the biofilm production considering the PA-BSI resistance phenotype. Thus, XDR isolates were the less producers, and susceptible isolates the highest biofilm producers (Fig. 7a; S1 Table). Very low biofilm producers were also the rhlR-affected PA (22.5% in Ps1056 and 27.9% in Ps1019).
Comparative analysis of pigments, elastase and biofilm production in pre-COVID (blue) and post-COVID (red) periods. (a) Biofilm biomass, (b) pyocyanin, (c) pyorubin, (d) pyoverdine and (e) elastase activity are represented in isolates from pre-COVID (blue) and post-COVID (red) periods. *p < 0.05. Black rhombous represents ST175 isolate and black squares XDR/DTR isolates.
Elastase activity and pigment production were assessed in all isolates compared to PAO1. Figure 7b–e illustrates the percentage of pyocyanin, pyorubin and pyoverdine production, and elastase activity for PA of each period. Notably, the unique isolate belonging to high risk clone ST175 (Ps1056) showed the lowest values for both pigment production and elastase activity. Regarding pyorubin, pre-COVID isolates demonstrated significantly higher production than post-COVID-PA. For other pigment productions and elastase activity, no statistically significant differences were observed, neither based on resistance pattern nor virulotype, although, generally, pre-COVID strains exhibited higher production levels.
In brief, in this study the BSIs was mainly hospital-acquired or health care-associated. Crude mortality was 53% in the 78 episodes, with an attributable mortality of 24% due to the PA-BSI. During the study period, the antimicrobial non-susceptibility increased and virulence decreased in the PA-BSI. MDR isolates remarkably increased in the post-COVID period, but the unique XDR/DTR isolates were detected only in the pre-COVID period. The biofilm production, elastase activity and pigment production were higher in pre-COVID PA-BSI than in post-COVID ones. None post-COVID-PA harboured the exoU gene, but singular isolates that exhibited the hypervirulent exlA gene or the co-occurrence of exoU and exoS genes were isolated.
Discussion
PA is one of the most problematic Gram negative pathogens that cause hospital infections, especially due to its extraordinary ability to acquire resistance and its variety of virulence traits. In this study, we analysed the prevalence and characteristics of PA-BSIs in two different periods: pre-COVID (2015–2018) and post-COVID (2022–2023). Similar to other studies4,8, most of the BSIs were classified as hospital acquired or health-care associated, highlighting the importance of the contact with the health-care system as a factor to suffer PA-BSI, either for inpatients or outpatients with recurrent contact with the health care system. Additionally, PA-BSI is an important cause of mortality, and in our study, the detected mortality (24%) was within the range previously reported (20–51.2%)4,6,20. Nevertheless, in this study, no correlation between mortality and resistance, virulence (exoU/exoS, or pigments and biofilm production) or origin of the infection was established.
Supporting the non-clonal epidemic PA population structure and the non-outbreak scenario in PA-BSI from our hospital, no clonal relationship was found between PA from both periods. Among the worldwide top ten PA-HRC10, PA-BSI belonging to ST175, ST244 and ST298 were detected in this study. Notably, the HRC ST111, ST175, and ST235 disseminated in hospitals worldwide and linked to drug resistance, were absent, with the exception of a single ST175 isolate identified during the pre-COVID period. These findings align with McCarthy et al.21, that similarly detected none HRC complexes (HRCCs) in BSI isolates in a non-outbreak context. Otherwise, other “international clones” have gained relevance in the last few years, which could be also considered a HRC, such as ST395 or ST274, that our collection contained three and five PA, respectively. Indeed, the unique detected MBL-producing PA belonged to ST395 clone. The ST395 and ST274 spread has been documented within hospitals worldwide, as well as in XDR isolates in Spain22,23.
The association between high genetic diversity and antimicrobial susceptibility is extensively documented. As consistently reported, XDR/MDR isolates showed predominantly a clonal population structure being also associated with HRCC, while clonal diversity and absence of HRCC is notably more pronounced for non-MDR isolates3,18,24. Given these findings, it could be hypothesized that the lack of genotypic correlation among isolates may suggest an environmental origin of infection, wherein various successful clonal types contribute to invasive infections.
The rising prevalence of MDR and XDR PA in clinical settings poses a growing global threat within hospital environments. However, the isolates examined in this study exhibited notable susceptibility, with 77% identified as non-MDR, 19.2% classified as MDR, and merely 3.8% as XDR/DTR. In contrast to our findings, other investigations on PA-BSI in Spain3,8 and other countries25,26 reported higher percentages of MDR/XDR isolates.
In our work, XDR/DTR isolates were exclusively identified in the pre-COVID period, while the prevalence of MDR escalated notably from 6.3% to 40% in the post-COVID period. This result stands in opposition to findings by Ioannou et al.16 who observed lower rates of MDR, XDR, and DTR PA after the onset of the COVID-19 pandemic. Notably, their study was performed after a focused antimicrobial stewardship intervention, particularly targeting carbapenem use. Similarly, Sastre-Femenia et al.27 reported a decline in MDR/XDR rates for post-COVID-PA, although this reduction was less prominent in BSIs. Other authors reported that antibiotic susceptibility patterns for PA remained relatively stable14. The fact is that it remains unclear how the incidence of PA bacteraemia changed during the COVID-19 pandemic, with diverse studies yielding seemingly contradictory conclusions14,28.
While the factors underlying PA epidemiological changes are likely multifactorial and complex; when the pre-COVID to the post-COVID periods were compared, no changes in the single-room organization system of the Hospital Universitario San Pedro (Logroño, Spain) (with 630 beds and 85% of them in single rooms) were performed. However, Programs to Optimize the Antimicrobial Use, that work in order to optimize the appropriateness of antimicrobial prescriptions, control the emergence of resistance, and guarantee the use of cost-effective treatments, were progressively implemented from January 2020 in the hospital and from September 2021 in the primary care, although they properly worked after pandemic. During COVID-19 pandemic, the extensive and improper use of antibiotics in the hospital increased the global consumption, but in the primary care setting, a decrease was observed. The Spanish lockdown (from 14 March to 21 June 2020) and less accessibility to health centers during COVID-19 pandemic could have helped to reduce the transmission of infections, and to reduce unnecessary antibiotic prescriptions. These facts involved fewer antibiotic consumptions. The global antibiotic consumption in the hospital decreased in the post-COVID period in comparison to the pre-COVID rates, however, the antibiotic consumption in primary care started to raise in the post-COVID period, although it did not reach 2019 levels. Furthermore, during pandemic, the huge increase in the use of biocides could also encourage indirect pressure on antibiotic resistance. For all these reasons, the elevated resistance rates observed in post-COVID isolates may stem from the inappropriate utilization of antibiotics during the COVID-19 pandemic, the resistance selection in the community as well as the probable indirect resistance selection promoted by the intensified use of biocides. Regarding carbapenem-resistant PA (CR-PA), the prevalence is around 17% in Spain1 and in our work, the CR-PA remained stable among 20% during both periods, but lower than the observed in other studies3,8,25,26. In CR-PA, the most common mechanism of resistance to carbapenems is the loss or alteration of the OprD porin, rather than the production of carbapenemases. Multiple studies conducted in Spain have shown that a significant proportion of CR-PA exhibit these OprD inactivating mutations, contrasting with the minority of cases with carbapenemase production8,27,29. In line with this fact, in our study, only 6% of the CR-PA harboured a MBL gene (blaVIM-2), and the OprD deficiencies or alterations were the mechanism most frequently involved in imipenem-resistant PA.
The pathogenesis of PA is multifactorial as suggested their notable number of virulence determinants. Eight virulotypes were observed, and virulotype V1 was the most prevalent (81%), encompassing all virulence genes except exoU and exlA. Our findings are therefore in agreement with previous studies showing that exoU+ PA (range 10%-40%) are less frequent than exoS+ ones (60%–90%) among clinical PA30. The presence of exoU is associated with increased mortality rates, being this gene one of the most important virulence determinants3. Furthermore, Peña et al.30 found that the exoU genotype was significantly less frequent among MDR strains. This trend is evident in our results, where post-COVID strains exhibited a higher percentage of MDR and fewer exoU-positive strains.
Among the different periods, 83% of strains in the pre-COVID era harboured the exoS gene, with 17% containing exoU. In the post-COVID period, 93% carried the exoS gene, while none exhibited the presence of exoU; however a strain harboured both genes and another PA none of them. Pre-COVID-PA seem to be more pathogenic than post-COVID-PA associated with HRC and a higher percentage of exoU-positive strains. However, in the post-COVID period, Ps1450 strain belonging to ST2211, lacked all T3SS and exoA genes, but contained the toxin ExlA. The ExlBA is a pore-forming toxin that induces plasma membrane rupture in host cells. Since its first description in 2014, exlA-positive strains have been identified, causing several types of infectious diseases, and detected from different origins worldwide31,32. In particular, Ps1450 has similar characteristics than the exlA-positive AZPAE15042 strain (ST2211; O:11) whose high cytotoxicity has been previously proven32,33.
However, no connection could be established between mortality and PA resistance or exoU/exlA virulence genes, although it should be consider the number of samples, and that most of them were susceptible. Another potential limitation of our study is that the presence of T3SS genes was documented but whether the corresponding cytotoxins are actually expressed and subsequently secreted was not evaluated.
Pigments not only confer a distinctive blue-green colour to PA colonies, but also play a crucial role in the physiology and pathogenesis of this bacterium. Pigment production in PA-BSI of this study was decreased in MDR isolates, although this difference was not statistically significate. Furthermore, the only HRC ST175 isolate, was the fewer producer of pigments, in accordance with Mulet et al.18, who observed that MDR/XDR HRC were associated with defective pigment production. Interestingly, MDR PA isolates demonstrated lower biofilm production than non-MDR ones. Biofilm production was significantly higher in the pre-COVID period supporting the idea that pre-COVID isolates seem more virulent than post-COVID ones.
In conclusion, our study reveals a remarkably diverse PA population, characterized by a moderate prevalence of MDR strains, albeit with a majority exhibiting high susceptibility, and a wide variety of virulence traits, that caused severe BSI resulting in a crude mortality rate exceeding 50%. This distinctive scenario, occurring in a non-outbreak context with limited presence of high-risk clones requires ongoing monitoring. Moreover, the study period coincided with the COVID-19 pandemic, emphasizing the dynamic genomic landscape of PA over time and highlighting the potential influence of external factors, such as the ongoing pandemic, on the genetic landscape of this opportunistic pathogen.
The results obtained in this work shed light on the evolving nature of PA providing valuable insights for clinical management and infection control, and underscores the significance of prudent antibiotic usage, especially in the context of public health crises like the COVID-19 pandemic.
Material and methods
Clinical isolates
A total of seventy-eight PA isolates were recovered from 76 patients in the Hospital Universitario San Pedro (Logroño, Spain) in two different periods: pre-COVID period from July 2015 to February 2018 (48 isolates), and the post-COVID period from January 2022 to February 2023 (30 isolates). For each period, isolates included in the study corresponded to consecutive, non-duplicated (one per patient or episode), and bloodstream PA isolations. After excluding certain isolates due to issues in the collection or preservation processes, 78 PA isolates were finally included in this study (81.5% of the total).
All isolates were recovered from blood samples and detailed clinical data were retrospectively collected. The BSIs were classified as hospital acquired, community acquired or health care associated according to Friedman criteria34.
Antimicrobial susceptibility testing
The antibiotic susceptibility test was performed against 16 antipseudomonal agents (piperacillin, piperacillin-tazobactam, ceftazidime-avibactam, ceftolozane-tazobactam, ceftazidime, cefepime, cefiderocol, aztreonam, imipenem, meropenem, doripenem, gentamicin, tobramycin, netilmicin, amikacin and ciprofloxacin) using the disk diffusion method35. Colistin resistance was also screened by colistin broth disk elution method35.
MBL, extended-spectrum-beta-lactamase and class A carbapenemase phenotypes were determined by double-disc synergy tests36. Strains with positive MBL or class A carbapenemase phenotypes were screened for their genes by multiplex PCR and subsequent sequencing37,38.
Molecular typing and serotyping
PFGE and MLST were carried out in all isolates in order to evaluate clonal relatedness. PFGE was performed with SpeI enzyme for DNA restriction, and the PFGE patterns were analysed by the Java program GelJ using the Dice coefficient39,40. A 90% similarity cut-off was selected to consider a clonal relationship among strains. MLST was performed by PCR and sequencing of seven housekeeping genes41. STs were assigned at the PubMLST database (http://pubmlst.org/paeruginosa/). To classify PA, the ST235, ST111, ST233, ST244, ST357, ST308, ST175, ST277, ST654 and ST298 were considered as the worldwide top ten PA-HRC10.
Serotype identification was performed by slide agglutination with monovalent antisera specific for 16 different P. aeruginosa O serotypes (Bio-Rad, Marnes-la-Coquette, France).
Characterization of porin oprD gene and integron structures
OprD protein alterations were analysed in all carbapenem resistant isolates by PCR, sequencing and comparison with the sequence of the P. aeruginosa PAO1 reference strain41.
The presence of genes encoding type 1, 2, and 3 integrases (intI1, intI2, intI3), and 3′-conserved segment of class 1 integrons (qacEΔ1 + sul1) was studied by PCR. The variable regions of integrase-positive isolates were further amplified by PCR and sequenced, as previously described41.
Virulence marker genes
The following genes involved in virulence and quorum sensing (QS) were analysed by PCR: exoU, exoS, exoY, exoT, lasA, lasB, exoA, aprA, rhlAB, rhlI, rhlR, lasI, lasR and exlA31,41. Different virulotypes were assigned according to the presence/absence of these genes.
Biofilm formation assay
Biofilm assay was performed in 96-well microtiter plates using an initial 1 × 106 CFU/mL inoculum. Biofilm formation was allowed to occur for 24 h at 37 °C, and crystal violet (CV) staining assay was carried out to analyse total biofilm biomass42. Measures were performed on a plate reader (POLAR star Omega microplate reader, BMG Labtech). All experiments were performed in triplicate and including P. aeruginosa PAO1 as control strain.
Elastase and pigment production
The LasB elastolytic activity was measured using Elastin-Congo red as substrate43. The chloroform-extract method was used for pyocyanin, pyorubin and pyoverdin pigments extraction and quantification44. All experiments were performed in triplicate and including P. aeruginosa PAO1 as control strain.
Data analysis
GraphPad Prism (version 7.04) and R-commander program (version 4.2.2) were used for graphical representation and statistical analysis. p-values were the result of Fisher’s exact tests, t-tests, Kruskal–Wallis or Mann–Whitney tests, as appropriate. A p-value of ≤ 0.05 was considered statistically significant.
Ethics approval
Ethical approval for this study was obtained from the Ethical Committee of Clinical Research of La Rioja in 29/03/2019 (CEImLAR P.I. 357). Informed consent was obtained from all participants and/or their legal guardians. All experiments were carried out according to relevant guidelines and regulations.
Data availability
The new insertion sequence ISPa104 was included in ISFinder Database (https://isfinder.biotoul.fr/).
References
European Centre for Disease Prevention and Control. Antimicrobial resistance in the EU/EEA (EARS-Net) - Annual Epidemiological Report 2022; Stockholm: ECDC; 2023.
Hattemer, A. et al. Bacterial and clinical characteristics of health care- and community-acquired bloodstream infections due to Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57, 3969–3975. https://doi.org/10.1128/AAC.02467-12 (2013).
Recio, R. et al. Pathogenic characteristics of Pseudomonas aeruginosa bacteraemia isolates in a high-endemicity setting for ST175 and ST235 high-risk clones. Eur. J. Clin. Microbiol. Infect. Dis. 39, 671–678. https://doi.org/10.1007/s10096-019-03780-z (2020).
Callejas-Díaz, A. et al. Impact of Pseudomonas aeruginosa bacteraemia in a tertiary hospital: Mortality and prognostic factors. Med. Clin. 152, 83–89. https://doi.org/10.1016/j.medcli.2018.04.020 (2019).
Pont, S., Janet-Maitre, M., Faudry, E., Cretin, F. & Attrée, I. Molecular mechanisms involved in Pseudomonas aeruginosa. Bacteremia. Adv. Exp. Med. Biol. 1386, 325–345. https://doi.org/10.1007/978-3-031-08491-1_12 (2022).
Gupte, A., Jyot, J., Ravi, M. & Ramphal, R. High pyocyanin production and non-motility of Pseudomonas aeruginosa isolates are correlated with septic shock or death in bacteremic patients. PLoS ONE 16(6), e0253259. https://doi.org/10.1371/journal.pone.0253259 (2021).
Hoang, S. et al. Risk factors for colonization and infection by Pseudomonas aeruginosa in patients hospitalized in intensive care units in France. PLoS ONE 13(3), e0193300. https://doi.org/10.1371/journal.pone.0193300 (2018).
Peña, C. et al. Prospective multicenter study of the impact of carbapenem resistance on mortality in Pseudomonas aeruginosa bloodstream infections. Antimicrob. Agents Chemother. 56, 1265–1272. https://doi.org/10.1128/AAC.05991-11 (2012).
Laborda, P., Hernando-Amado, S., Martínez, J. L. & Sanz-García, F. Antibiotic resistance in Pseudomonas. Adv. Exp. Med. Biol. 1386, 117–143. https://doi.org/10.1007/978-3-031-08491-1_5 (2022).
Oliver, A. et al. Pseudomonas aeruginosa antimicrobial susceptibility profiles, resistance mechanisms and international clonal lineages: Update from ESGARS-ESCMID/ISARPAE Group. Clin. Microbiol. Infect. 30(4), 469–480. https://doi.org/10.1016/j.cmi.2023.12.026 (2024).
Del Barrio-Tofiño, E. et al. Association between Pseudomonas aeruginosa O-antigen serotypes, resistance profiles and high-risk clones: Results from a Spanish nationwide survey. J. Antimicrob. Chemother. 74(11), 3217–3220. https://doi.org/10.1093/jac/dkz346 (2019).
Imoto, W. et al. Impact of coronavirus disease 2019 on infectious disease treatment and infection control at a tertiary hospital in Japan. J. Infect. Chemother. 28(5), 616–622. https://doi.org/10.1016/j.jiac.2022.01.008 (2022).
Sulayyim, H. J. A., Ismail, R., Hamid, A. A. & Ghafar, N. A. Antibiotic resistance during COVID-19: A systematic review. Int. J. Environ. Res. Public Health 19(19), 11931. https://doi.org/10.3390/ijerph191911931 (2022).
Ng, Q. X. et al. Trends in Pseudomonas aeruginosa (P. aeruginosa) bacteremia during the COVID-19 pandemic: A systematic review. Antibiotics (Basel) 12(2), 409. https://doi.org/10.3390/antibiotics12020409 (2023).
Alcántar-Curiel, M. D. et al. Gram-negative ESKAPE bacteria bloodstream infections in patients during the COVID-19 pandemic. PeerJ 11, e15007. https://doi.org/10.7717/peerj.15007 (2023).
Ioannou, P., Alexakis, K., Maraki, S. & Kofteridis, D. P. Pseudomonas bacteremia in a tertiary hospital and factors associated with mortality. Antibiotics (Basel) 12(4), 670. https://doi.org/10.3390/antibiotics12040670 (2023).
Magiorakos, A. P. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18(3), 268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x (2012).
Mulet, X. et al. Biological markers of Pseudomonas aeruginosa epidemic high-risk clones. Antimicrob. Agents Chemother. 57(11), 5527–5535. https://doi.org/10.1128/AAC.01481-13 (2013).
Kadri, S. S. et al. Difficult-to-treat resistance in gram-negative bacteremia at 173 US hospitals: Retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin. Infect. Dis. 67(12), 1803–1814. https://doi.org/10.1093/cid/ciy378 (2018).
Herrera, S. et al. Pseudomonas aeruginosa Bloodstream Infection, resistance, and mortality: Do solid organ transplant recipients do better or worse?. Antibiotics (Basel) 12, 380. https://doi.org/10.3390/antibiotics12020380 (2023).
McCarthy, K. L., Wailan, A. M., Jennison, A. V., Kidd, T. J. & Paterson, D. L. P. aeruginosa blood stream infection isolates: A “full house” of virulence genes in isolates associated with rapid patient death and patient survival. Microb. Pathog. 119, 81–85. https://doi.org/10.1016/j.micpath.2018.03.062 (2018).
Petitjean, M. et al. Genomic characterization of a local epidemic Pseudomonas aeruginosa reveals specific features of the widespread clone ST395. Microb. Genom. 3(10), e000129. https://doi.org/10.1099/mgen.0.000129 (2017).
Chichón, G. et al. Spread of Pseudomonas aeruginosa ST274 Clone in Different Niches: Resistome, Virulome, and Phylogenetic Relationship. Antibiotics (Basel) 12(11), 1561. https://doi.org/10.3390/antibiotics12111561 (2023).
Treepong, P. et al. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin. Microbiol. Infect. 24(3), 258–266. https://doi.org/10.1016/j.cmi.2017.06.018 (2018).
Nemec, A., Krizova, L., Maixnerova, M. & Musilek, M. Multidrug-resistant epidemic clones among bloodstream isolates of Pseudomonas aeruginosa in the Czech Republic. Res. Microbiol. 161(3), 234–242. https://doi.org/10.1016/j.resmic.2010.02.002 (2010).
Fehlberg, L. C. et al. Beta-lactam resistance mechanisms in Pseudomonas aeruginosa strains causing bloodstream infections: Comparative results between Brazilian and American isolates. Microb. Drug Resist. 18(4), 402–407. https://doi.org/10.1089/mdr.2011.0174 (2012).
Sastre-Femenia, M. À. et al. Pseudomonas aeruginosa antibiotic susceptibility profiles, genomic epidemiology and resistance mechanisms: A nation-wide five-year time lapse analysis. Lancet Reg. Health Eur. 19(34), 100736. https://doi.org/10.1016/j.lanepe.2023.100736 (2023).
Langford, B. J. et al. Antibiotic resistance associated with the COVID-19 pandemic: A systematic review and meta-analysis. Clin. Microbiol. Infect. 29(3), 302–309. https://doi.org/10.1016/j.cmi.2022.12.006 (2023).
Estepa, V. et al. Characterisation of carbapenem-resistance mechanisms in clinical Pseudomonas aeruginosa isolates recovered in a Spanish hospital. Enferm. Infecc. Microbiol. Clin. 35(3), 141–147. https://doi.org/10.1016/j.eimc.2015.12.014 (2017).
Peña, C. et al. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin. Infect. Dis. 60(4), 539–548. https://doi.org/10.1093/cid/ciu866 (2015).
Ruiz-Roldán, L. et al. Antimicrobial resistance and virulence of Pseudomonas spp. among healthy animals: concern about exolysin ExlA detection. Sci. Rep. 10(1), 11667. https://doi.org/10.1038/s41598-020-68575-1 (2020).
Bouillot, S. et al. Inflammasome activation by Pseudomonas aeruginosa’s ExlA pore-forming toxin is detrimental for the host. Cell Microbiol. 22(11), e13251. https://doi.org/10.1111/cmi.13251 (2020).
Reboud, E. et al. Phenotype and toxicity of the recently discovered exlA-positive Pseudomonas aeruginosa strains collected worldwide. Environ. Microbiol. 18(10), 3425–3439. https://doi.org/10.1111/1462-2920.13262 (2016).
Friedman, N. D. et al. Health care–associated bloodstream infections in adults: A reason to change the accepted definition of community-acquired infections. Ann. Intern. Med. 137(10), 791–797. https://doi.org/10.7326/0003-4819-137-10-200211190-00007 (2002).
CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 34th ed. CLSI supplement M100. (Wayne, PA: Clinical and Laboratory Standards Institute; 2022).
Estepa, V., Rojo-Bezares, B., Torres, C. & Sáenz, Y. Genetic lineages and antimicrobial resistance in Pseudomonas spp. isolates recovered from food samples. Foodborne Pathog. Dis. 12, 486–491. https://doi.org/10.1089/fpd.2014.1928 (2015).
Ellington, M. J., Kistler, J., Livermore, D. M. & Woodford, N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 59, 321–322. https://doi.org/10.1093/jac/dkl481 (2007).
Hong, S. S. et al. Multiplex PCR for rapid detection of genes encoding class A carbapenemases. Ann. Lab. Med. 32, 359–361. https://doi.org/10.3343/alm.2012.32.5.359 (2012).
Rojo-Bezares, B. et al. A novel class 1 integron array carrying blaVIM-2 genes and a new insertion sequence in a Pseudomonas aeruginosa strain isolated from a Spanish hospital. J. Med. Microbiol. 60, 1053–1054. https://doi.org/10.1099/jmm.0.030973-0 (2011).
Heras, J. et al. GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinform. 16, 270. https://doi.org/10.1186/s12859-015-0703-0 (2015).
Ruiz-Roldán, L. et al. Pseudomonas aeruginosa Isolates from Spanish children: Occurrence in faecal samples, antimicrobial resistance, virulence, and molecular typing. Biomed. Res. Int. 2018, 8060178. https://doi.org/10.1155/2018/8060178 (2018).
Peeters, E., Nelis, H. J. & Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 72(2), 157–165. https://doi.org/10.1016/j.mimet.2007.11.010 (2008).
Pearson, J. P., Pesci, E. C. & Iglewski, B. H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179, 5756–5767. https://doi.org/10.1128/jb.179.18.5756-5767.1997 (1997).
Anantharajah, A. et al. SalicylideneAcylhydrazides and Hydroxyquinolines act as inhibitors of type three secretion systems in Pseudomonas aeruginosa by distinct mechanisms. Antimicrob. Agents Chemother. 61, e02566-e2616. https://doi.org/10.1128/AAC.02566-16 (2017).
Acknowledgements
The authors want to thank the students Celia Gil, Ángela de la Rosa, Andrea Lázaro, and Carmen González for their experimental collaboration in this work. We would like to acknowledge the help and predisposition of Dr. Mercedes Sanz Franco of the Infectious Diseases Department in the Hospital Universitario San Pedro.
Funding
This study has been funded by Instituto de Salud Carlos III (ISCIII), through the project Fondo de Investigaciones Sanitarias PI20/00356 (Acción Estratégica en Salud 2020) (Co-funded by European Regional Development Fund (FEDER) "A way to make Europe"). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
ML and YS, conceptualization; ML, CAA, CL, BRB and YS, methodology; ML, BRB, CL and YS, formal analysis; ML, CAA, JMAG and YS, statistical analysis, and interpretation of data; CAA and JMAG, analysis and interpretation of clinical data; YS, funding acquisition; ML and YS, writing—original draft; All authors participated in reviewing of the manuscript. All authors have read, reviewed, and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
López, M., Alonso, C.A., Azcona-Gutiérrez, J.M. et al. Impact of the COVID-19 pandemic on the epidemiology and molecular features of Pseudomonas aeruginosa bloodstream infections. Sci Rep 15, 24853 (2025). https://doi.org/10.1038/s41598-025-09492-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-09492-z