Abstract
Heart disease remains the leading cause of death globally, mainly caused by delayed diagnosis and indeterminate categorization. Many of traditional ML/DL methods have limitations of misclassification, similar features, less training data, heavy computation, and noise disturbance. This study proposes a novel methodology of Modified Multiclass Attention Mechanism based on Deep Bidirectional Long Short-Term Memory (M2AM with Deep BiLSTM). We propose a novel model that incorporates class-aware attention weights, which dynamically modulate the focus of attention on input features according to their importance for a specific heart disease class. With an emphasis on the informative data, M2AM can improve feature representation and well-cure the problems of mis-classification, overlapped features, and fragility against noise. We utilized a large dataset of 6000 samples and 14 features, resulting in noticeable noise reduction from the MIT-BIH and INCART databases. Applying an Improved Adaptive band-pass filter (IABPF) to the signals resulted in noticeable noise reduction and an enhancement of signal quality. Additionally, wavelet transforms were employed to achieve accurate segmentation, allowing the model to discern the complex patterns present in the data. The proposed mechanism achieved high performance in the performance metrics, with accuracy of 98.82%, precision of 97.20%, recall of 98.34%, and F-measure of 98.92%. It surpassed methods such as the Classic Deep BiLSTM (SD-BiLSTM), and the standard approaches of Naive Bayes (NB), DNN-Taylos (DNNT), Multilayer perceptron (MLP-NN) and convolutional neural network (CNN). This work provides a solution to significant limitations of current methods and improves the accuracy of classification, indicating substantial progress in accurate diagnosis of heart diseases.
Similar content being viewed by others
Introduction
Coronary artery disease, arrhythmias, valvular disease, and congenital heart disease are also classified as forms of heart disease. CVD are still leading cause of mortality worldwide and responsible for an estimated around 32% of all deaths annually all over the globe according to the World Health Organization (WHO)1. The significant burden of heart disease, not only at a personal level but in terms of health services and the economy, is also well documented. This highlights the importance of early diagnosis to minimize the risk of mortality. While conservative approaches, such as lifestyle changes, drugs, and surgery, are essential, the problem of effective classification and early diagnosis persists, especially when we consider the classification of different types of heart diseases2.
Over the last few years, the application of machine learning (ML) and deep learning (DL) techniques has revolutionized the investigation of CVDs (2), offering novel insights into the early detection and classification of these conditions. Such methods, which manipulate large datasets to discover complex trends and relationships, are far more capable of identifying the intricate relationships between data that simpler, rule-based models would otherwise overlook, of which deep learning models utilizing ECGs and medical imaging have been particularly adept at analyzing these complex datasets and enhancing the detection and classification of cardiovascular disease. Through the self-learning function, they not only improve diagnostic efficiency but also facilitate the development of more personalized treatments, leading to better overall healthcare results3.
Nevertheless, despite the remarkable progress enabled by deep learning, substantial challenges remain. Overfitting, for example, is a well-known obstacle where a model exhibits good performance on training data but poor generalization on unseen data, which limits the model’s practical utility4. Moreover, most previous works have concentrated on single-class heart disease classification, while less attention has been given to multiclass conditions. Because some cardiac diseases have overlapping symptoms and features, failing to differentiate them will result in a less accurate diagnosis. This rustication underscores the need for a multiclass classification system, which aims to more accurately distinguish between different cardiac diseases and deliver more precise and timely treatments.
Unlike the classical attention mechanism, which uses uniform attention weights for all features, the Modified Multiclass Attention Mechanism (M2AM) proposed in this work computes class-dependent attention weights. This change has enabled the model to more accurately learn to focus on features that are highly related to each different heart disease class, and it also helps with the problem of overlapping features between different diseases.
Research motivation
The pressing need to better detect and classify heart disease, a prominent worldwide cause of death, drives this work. Even with significant progress in medical diagnostics, many patients experience delays in receiving suitable treatment due to difficulties in classifying their disorders. Conventional diagnostic techniques frequently fail to distinguish between the different types of heart disease, resulting in possible misdiagnoses and inadequate treatment. This highlights the need for innovative solutions that can improve diagnostic accuracy and enable prompt interventions. Our research aims to improve patient outcomes and save lives by addressing these significant deficiencies6.
The rapid growth of healthcare data presents a significant opportunity to leverage deep learning and advanced machine learning methods in analyzing complex trends in cardiovascular datasets. These technologies pose challenges, despite their great potential to improve the diagnosis of heart diseases. In single-class classification environments, overfitting and sensitivity to noise can compromise the efficacy of these models. The necessity for a more advanced technique that enables the multiclassification of heart disease and its various expressions drives this study. By establishing a robust framework that incorporates the Modified Multiclass Attention Mechanism with Deep Bidirectional Long Short-Term Memory (LSTM), we aim to enhance classification performance and reliability, thereby advancing individualized and effective healthcare strategies7,8.
Main contribution of the research
The key contributions of the research are as follows.
-
Introduction of the Modified Multiclass Attention Mechanism This research presents the Modified Multiclass Attention Mechanism combined with Deep Bidirectional Long Short-Term Memory. This novel mechanism dynamically modifies attention weights to prioritize the most relevant input features, thereby enhancing the model’s ability to focus on critical data points. Enhanced feature representation facilitates superior management of misclassification and noise sensitivity, leading to improved classification of heart disease.
-
Advancements in Signal Processing with the IABPF The study utilizes an IABPF to preprocess ECG signals, significantly reducing noise and enhancing signal quality. This improvement to traditional filtering methods ensures that the data input is more refined, which is required for accurate feature extraction and classification. The IABPF adjusts to the properties of the input signals, resulting in a more sophisticated noise reduction method that improves model performance.
-
Use of Robust Heart disease datasets The research uses several large databases, including the INCART and MIT-BIH databases. This dataset includes 6000 samples, each with 14 distinct characteristics. This extensive dataset ensures that models perform optimally across a wide range of heart disease symptoms by allowing for proper training and testing. The variety of the dataset makes the research results more critical and reliable.
-
Enhanced Segmentation through Wavelet Transformations The research utilizes wavelet transformations to segment ECG signals precisely. This technique enables the model to recognize intricate patterns and features in the data, improving its capacity to detect subtle variations in heart disease presentations, which is crucial for accurate classification.
-
Impressive Performance Metrics The proposed method exhibits 98.82% accuracy, 97.20% precision, 98.34% recall, and 98.92% F-measure. These findings indicate a substantial improvement compared to traditional classification methods, such as SD-BiLSTM, NB, DNNT, MLP-NN, and CNN. The identification and management of cardiovascular diseases have advanced considerably, as demonstrated by enhanced classification performance.
Structure of the research
The article features a comprehensive structure comprising several essential sections. The Introduction outlines the worldwide ramifications of heart disease and the difficulties associated with precise diagnosis. The Review of Existing Work evaluates conventional machine learning and deep learning techniques, highlighting their shortcomings in multiclass classification. The "Materials and Methods" section discusses the datasets of the MIT-BIH and INCART databases. One of the preprocessing methods applied is the IABPF. The section on Simulation and Results discusses the performance of the proposed approach in comparison to past approaches. The Computational Complexity section covers the model complexity details, and the study’s findings are compiled in the "Conclusion and Future Work" section, with suggestions for subsequent studies on improving the classification of heart disease.
Related work
The study1 presents an advanced Bi-LSTM approach that integrates heterogeneous feature fusion and an attention mechanism to improve ECG recognition. This model proficiently analyzes multiple characteristics from ECG signals, achieving a recognition accuracy of approximately 87.84%. The attention mechanism enables the model to focus on the most relevant features, significantly enhancing classification performance compared to conventional models. The intricacy of feature extraction may result in heightened computational demands and extended training durations, potentially constraining its practical use in real-time contexts, especially in clinical settings where swift decision-making is crucial.
Another noteworthy contribution2 is the application of deep learning techniques to differentiate between hypertrophic cardiomyopathy and hypertensive heart disease by utilizing MRI native T1 maps. The model’s close to 89.79% discrimination rate proves it outperforms conventional imaging. The study uses a diverse patient dataset encompassing multiple diseases to enhance the model’s accuracy. The quality of hospital or imaging equipment images may affect the model’s performance, suggesting that high-quality imaging data may be limited. Additionally, the interpretability of the model poses a significant challenge, as it is frequently difficult to articulate the outputs of deep learning in clinical terminology.
A novel machine learning-based predictive approach for the early detection of congenital heart disease is proposed by processing ECG signals3. The model utilizes ECG features associated with congenital anomalies to achieve an accuracy of 90.65%. The study emphasizes the importance of early diagnosis, particularly in children, as it can lead to more effective treatment outcomes. Although promising, the proposed method is constrained by its reliance on ECG signals that are precisely captured, as noise or artifacts may result in a misdiagnosis. A heuristic-metaheuristic ensemble that features a fusion learning model for diagnosing heart disease, utilizing tabular data, is proposed4. This novel model surpasses single-method approaches by achieving classification performance with an accuracy of up to 91.34%. The reliability and robustness of predictors are improved by applying ensemble techniques. Managing multiple models can make them more challenging to interpret, which may hinder their acceptance in clinical settings where medical professionals need to make informed decisions.
To improve heart disease prediction, a transformer model utilizing self-attention is proposed5. The model’s predictive accuracy of approximately 90.71% demonstrates how effectively attention mechanisms highlight the most essential features across various datasets. The method’s effectiveness is due to its ability to handle multiple data distributions. However, the model’s complexity may limit its practical application, especially in resource-constrained environments, as it may require more processing power and longer training times. The three-layer deep learning architecture integrated with meta-heuristic methods is employed in the IoT-enabled ECG-based heart disease prediction model outlined in6. This model enables continuous patient monitoring through IoT devices, achieving an accuracy rate of approximately 92.41%. It effectively meets the need for real-time analysis, particularly in critical situations. However, significant barriers to the widespread implementation of these systems in clinical practice may exist, including concerns about data privacy and security in IoT applications.
A convolutional block attention network is proposed to classify ECG signals7.
This model achieves a classification accuracy of 90.03%, and the attention mechanism significantly improves performance compared to conventional CNNs by enabling the network to focus on critical signal features. The model’s accessibility in clinical practice may be impeded by its substantial computational resource requirements despite its robust outcomes, particularly in environments with limited technological infrastructure. The study examines a system that utilizes artificial intelligence (AI) to classify audio signals based on the presence of heart disease8. An accuracy rate of approximately 89% suggests that this new method is likely to be effective. The paper discusses how audio signal analysis can be used to diagnose heart disease. However, signal variability and background noise are issues that could render the model less effective overall. Furthermore, the necessity for extensive labeled audio datasets for training presents an additional challenge.
This study investigates the identification of congenital heart disease in pediatric electrocardiograms using deep learning9. Combining human concepts with deep learning techniques yields a model with an accuracy of almost 89.79%. In clinical environments, this increases its efficiency. The model can match well-known medical signs to its forecasts with this mixed approach. Nevertheless, the model is challenging to grasp. Thus, doctors may require additional education to obtain accurate findings. This indicates that it may not be widely used in clinical settings. A study10 found that cardiovascular disease can be detected with up to 91.52% accuracy using machine learning-based predictive models. The results show significant progress. However, obtaining large training datasets can be challenging due to variations in data quality and demographics, which may render the model less reliable and less applicable to diverse groups of people.
In another work11, a methodology is described for detecting heart disease using artificial neural networks, feature extraction, and sensors. The model is 93.02% accurate in clinical settings. The quality of the input data has a significant impact on performance, which can lead to variability in outcomes, particularly in less regulated environments. The work12 focuses on utilizing artificial intelligence in echocardiography to detect rheumatic heart disease. Its great sensitivity and specificity draw attention to how well it detects necessary mitral regurgitation conditions. However, the model’s reliance on high-quality imaging data may limit its general applicability, as variations in imaging methods can affect diagnostic precision.
To predict heart disease13, a classification technique combining models with Boruta feature selection is presented. This approach achieves an accuracy rate of almost 91% by skillfully identifying the most pertinent features for prediction. While the outcomes are positive, careful adjustments are necessary to prevent overfitting, which could compromise the model’s generalizability across various clinical populations and datasets. In another work14, deep learning and multi-modal data fusion have enabled nearly 90.85% of cardiovascular disease research to demonstrate remarkable accuracy. The model is strengthened by integrating numerous data sources, including clinical and imaging data. Managing multiple data streams can present significant challenges in data management and processing, potentially leading to extended training periods and increased resource requirements.
In15, an examination of deep learning techniques for detecting heart disease based on ECG data highlights the diversity of available methods, which exhibit enhanced detection accuracy that surpasses that of traditional methods. The complexity of direct comparisons and the diversity of methodologies may hinder the establishment of standardized protocols for implementing clinical practice. In particular, the study16 examines the application of deep learning techniques to ECG data for the detection of heart disease. The method can correctly classify approximately 91.02% of cases, but the model’s performance depends on the quality of the ECG signals. In real-world situations, variations in signal quality resulting from patient movement or background noise can significantly impact the reliability of the measurement. This highlights the importance of using effective preprocessing methods.
In another study17, coronary heart disease in Chinese diabetics was accurately predicted by machine learning techniques. It had a success rate of roughly 91.80%. Although the results show promise for this specific group of individuals, concerns remain regarding the model’s potential for failure when applied to other groups due to variations in risk factors and population characteristics. The analysis of a deep learning method based on Crow intelligence optimization for heart disease classification18 demonstrates that it can achieve significant accuracy and effective classification capabilities. The model’s applicability to various datasets may be limited by its reliance on a specific optimization technique, as performance can vary significantly depending on the complexity of the classification task and the characteristics of the input data.
These studies reveal significant progress in using several fresh approaches to detect heart disease. They also handle resurfacing problems, including generalizability, data quality, and model complexity. Every donation helps make diagnoses more accurate, which in turn influences the care and treatment patients receive in cardiovascular medicine. Although there have been many advances in heart diseases detection using machine learning models, there are still many problems that have not been addressed. Existing models, while capable of classification, can suffer from problems, such as class imbalance which results in biased predictions towards the majority class, and can consequently affect prediction performance of the minority classes. Furthermore, many of the current methods are not generalisable as they are customised at the level of datasets or to patient populations, restricting their world-wide implementation.
Also, their interpretability is not well studied. Especially in the clinical setting, where interpretability might be crucial, explaining why a model makes specific predictions is vital. This ambiguity makes them challenging to utilize in medical decision support systems. In 7 s, computational complexity becomes an issue; many models require intensive resources and lengthy training periods, which are impractical for real-time clinical use. We aim to fill these gaps in our study by employing class balancing strategy, using Modified Multiclass Attention Mechanism (M2AM) for better feature priority, and guaranteeing better generalizability and interpretability for real world applicability in clinical practice. Table 1 presents a comparative analysis of the review of existing review research and proposed models.
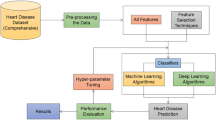
Materials and methods
This section primarily covers the key methods employed in this research, the workings of the proposed model, dataset details, data preprocessing, and other relevant aspects.
Proposed advanced multiclass heart disease classification model
A modified Multiclass Attention Mechanism is used in conjunction with Deep Bidirectional Long Short-Term Memory (BiLSTM) for heart disease classification from ECG. M2AM aims to enhance the model’s accuracy using class-specific features discrimitive for diagnosing various heart diseases, a key challenge in multiclass classification. The procedure begins with the pre-processing of the raw ECG signal to reduce unwanted noise and improve the quality of the ECG signals using the Improved Adaptive Band Pass Filter (IABPF). In this filtering process unnecessary frequencies are eliminated and only the essential elements of the ECG signal (necessary for accurate analysis) were kept19. Wavelet transformations are then used to get temporal-spectral information from the ECG. This approach generates a fullest of features that realize the intricate and dynamic characteristic of ECG morphology. The BiLSTM layer process these input sequences in forward passes and backward passes to learn temporal dependencies and essential to understand the sequential nature of ECG signal.
M2 AM further modifies the model to adjust attention weights for each heart disease class dynamically. This enables the model to focus on the most relevant features for each class, especially when some classes share common or overlapping features (e.g., arrhythmia and coronary artery disease). M2AM focuses on class-specific features, enabling the model to distinguish diseases more effectively while mitigating the risk of misclassification. Later processing layers are fully connected layers that refine the features, and an output SoftMax layer provides the class probability for heart disease, thereby helping to classify. This relatively strong architecture significantly enhances classification accuracy compared to its predecessor, making the detector structure insensitive to noise and allowing the model to perform better in complex real-world ECG data21.
The Modified Multiclass Attention Mechanism is a significant enhancement of this architecture, as it computes the attention weights on a class-by-class basis. Such adaptation enables the model to favor features that are most important in distinguishing between different heart diseases. M2AM takes a new approach by calculating class-wise attention weights, whereas conventional AM is class-agnostic. Such a class-pair-specific learning scheme can help the model exploit the standard features common to different diseases, leading to higher classification accuracy and robustness. The processed data, after being fed into BiLSTM and M2AM, is then fed into fully connected layers, followed by a SoftMax output layer that generates probabilities for different heart disease classes. The introduction of M2AM with a class-specific attention mechanism can enhance the model’s ability to mitigate misclassification and noise sensitivity, thereby further improving its diagnostic performance.
Novelty of the M2AM
The M2AM is an adapted multiclass attention mechanism that has the following advantages compared to basic attention mechanisms:
-
a)
Class-Specific Attention
-
i.
In classical attention mechanisms (like those used in BiLSTM and Transformer models) compute a single global attention weight per feature to all classes. This has the drawback that every class is given the same weight for a feature, which can lead to suboptimal classification in tasks with a skewed class distribution.
-
ii.
M2AM overcomes this weakness by learning class-specific attention weights. The model computes distinct attention scores for every class \({y}_{j}\) w.r.t. each feature \({x}_{i}\). This way the model will be able to learn different parts of the input features that are the most relevant in regards to each heart disease and increase the precision of the classification.
-
i.
-
b)
Dynamic Attention Weighting
-
i.
In classical attention mechanisms, the attention weight for each feature is calculated by a fixed collection of scores. This may cause problems if features have the same importance to several classes.
-
ii.
M2AM adaptively determines the weight of each feature based on its relevance to the current class. This flexibility enables the model to learn which features could be significant for some classes but irrelevant to others.
-
i.
Mathematical formulation
-
The classical attention mechanism can be defined as (Eq. 1).
$$Attention \;Weight_{i} = \frac{{\exp \left( {score\left( {x_{i} } \right)} \right)}}{{ \mathop \sum \nolimits_{j} \exp \left( {score\left( {x_{j} } \right)} \right)}}$$(1)
where \(score\left({x}_{i}\right)\) Is any score function applied to the ith item or a Null/Dummy value? In this way, attention is paid to each feature. \({x}_{i}\) Across all the classes, a global score is calculated.
-
In M2AM, since attention is calculated per class, we can calculate attention weights for each class in a class-sensitive manner. M2AM can be mathematically expressed as in Eq. 2:
$$Attention \;Weight_{i} = \frac{{\exp \left( {score\left( {x_{i} ,y_{k} } \right)} \right)}}{{ \mathop \sum \nolimits_{k} \exp \left( {score\left( {x_{i} , y_{k} } \right)} \right)}}$$(2)
where: \({x}_{i}\) Is the feature vector of the ith feature, the category to be assigned to the class (coronary artery disease, arrhythmia, etc.), \(score\left({x}_{i},{y}_{j}\right)\) Is it the correlation value for the feature \({x}_{i}\) with class \({y}_{j}\). The denominator normalizes scores of attention over all classes. \({y}_{k}\).
By calculating class-specific attention weights, M2AM can make the model pay more appropriate attention to informative features for individual disease type, which leads to noticeable improvement in classification performance. A brief description of its architecture and operations is presented in Fig. 1. The complete working of the proposed model is as follows.
IABPF methods for data preprocessing
Before ECG signals are fed into the classification model, they must be preprocessed appropriately to enhance their accuracy and reliability. The Improved Adaptive Band Pass Filter (IABPF) is crucial in this phase, as it improves the quality and integrity of the signals in numerous essential ways23.
-
Removal of Noise: Noises such as baseline drift, power line interference, and muscle noise can all mask the underlying cardiac activity in ECG signals. The IABPF efficiently filters out these unwanted disturbances, preserving the main characteristics of the ECG waveform. The IABPF ensures that only relevant frequency components are retained by dynamically modifying the cutoff frequencies in response to the distinct features of the incoming signal24. The filtering process is represented by Eq. (3). Here \({f}_{output}\left(t\right)\) Shows filtered output, fIresponse shows impulse response, \({In}_{signal}(T)\) Shows input signal (Eq. 3).
$$f_{output} \left( t \right) = \mathop \smallint \limits_{ - \infty }^{\infty } \left[ {fI_{response} \left( {t - T} \right)In_{signal} \left( T \right)} \right]dT$$(3) -
Improvement of Signal Quality: The IABPF enhances the ECG data’s signal-to-noise ratio (SNR) by eliminating noise more effectively. A high signal-to-noise ratio (SNR) is crucial for accurate feature extraction and significantly impacts the precision of the classification model. Enhanced signal quality facilitates the identification of features such as the P, Q, R, S, and T waves, thereby assisting physicians in diagnosing cardiac issues. Physicians require a high signal-to-noise ratio (SNR) to accurately monitor cardiac rhythm and perform precise and timely interventions23,25.
-
Model Mathematization: The IABPF’s adaptive characteristics enable real-time adjustment of its filter coefficients, ensuring optimal performance across a diverse range of ECG recordings. This adaptability facilitates effective preprocessing irrespective of the noise attributes of the incoming data. Adaptive filtering possesses the following characteristics, as shown in Eq. 4. Where \({d}_{coefficient}\) shows dynamic coefficient, T shows sampling time.
$$fI_{response} \left( t \right) = \mathop \sum \limits_{n = 0}^{N} d_{coefficient} \times \pounds \left( {t - nT} \right)$$(4)
The sampling interval is designated as T. This dynamic adjustment is necessary to enhance the filter’s efficacy in accommodating the diverse noise profiles in different ECG recordings.
Wavelet transformations for feature extraction
To analyze ECG signals, you need to be able to extract features. In this step, raw data is converted into a format that facilitates easier analysis and sorting. Wavelet transformations are helpful for signals that don’t stay in one place, like an ECG, because they let you look at them in both the time and frequency domains. This distinguishes them in this context20,21,22,23,24,25,26. With wavelet transformations, a signal is broken down into essential frequency components while keeping information about time. The dual capability facilitates the identification of features that change rapidly in the ECG, which is necessary for a comprehensive analysis19,21. The continuous wavelet transform (CWT) of a signal is examined by using Eq. (5).
where:
-
\(x\) Shows scalar parameters and controls the width of the wavelet.
-
\(y\) Shows translation parameters; it shifts the wavelet in time.
-
\(s\left(t\right)\) Represents the input signal.
-
\({-}\!\!\!\!{\uprho} \left( {\frac{{\left( {t - y} \right)}}{x}} \right)\) Represents the wavelet function like Morlet.
-
\({-}\!\!\!\!{\uprho}\) Represents the scaled and translated version of the wavelet.
Selecting the primary wavelet
The initial step in applying wavelet transformations is selecting a primary wavelet that is appropriate and serves as the basis function. The Morlet wavelet and the Daubechies wavelet are widely used for ECG analysis. These wavelets can be used to examine various ECG signal components due to their differences. A Morlet wavelet (combining a Gaussian function with an exponential) can be calculated using Eq. 6. Where \({f}_{o}\) Shows frequency at the central level, α shows wavelet width control27.
Wavelet decomposition
The next step, after selecting the primary wavelet, is the wavelet decomposition analysis. To achieve this, the wavelet coefficients x and y must be calculated at multiple scales15,28.
Feature extraction
Following the decomposition procedure, the wavelet coefficients are analyzed to identify significant characteristics within the data. Several essential characteristics can be deduced, including the following:
-
Signal Energy: When attempting to quantify the signal’s strength, the energy of the wavelet coefficients is helpful10,28. Equation 7 can determine it.
$$_{{ Signal_{Energy} }} = \mathop \sum \limits_{i = 1}^{N} |CWT \left( {x_{i} ,y_{j} } \right)|^{2}$$(7) -
Entropy: This measurement provides insights into the information content of a signal through an evaluation of its complexity. This can be determined using Eq. 829. Here \(Coff_{normalized}\) shows normalized coefficient.
$$_{ Entropy}^{ } = - \mathop \sum \limits_{i = 1}^{N} Coff_{normalized} {\text{log}}\left( {Coff_{normalized} } \right)$$(8) -
Peak Detection: Obtaining an accurate diagnosis requires identifying key peaks associated with specific components of the electrocardiogram (ECG), such as the P, Q, R, S, and T waves.
Signal reconstruction
In certain circumstances, reconstructing the signal from the wavelet coefficients can be advantageous to emphasize particular characteristics. This can be determined using Eq. 9.
Reduce overfitting with modified multiclass attention
Overfitting occurs when a model retains the training data instead of deriving general principles. This leads to suboptimal performance when the model encounters unfamiliar data. The Modified Multiclass Attention Mechanism addresses this problem, dynamically adjusting the model’s emphasis on relevant characteristics, thereby enhancing its capacity for generalization30. The proposed model effectively addresses the complexities of ECG signal classification by integrating MCAM, BiLSTM architecture, and the classification layer, thereby enhancing accuracy and robustness against overfitting.
To tackle the overfitting problem, we further propose the M2AM, in which the attention for features dynamically varies according to their importance in each class. For more details, only the most informative features are considered, so that the model does not overfit on irrelevant features. The attention weight of the feature \({x}_{j}\) to the class \({y}_{i}\) It is formulated as (Eq. 10):
where:\({w}_{j}\) Is the weight for the jth feature? \({x}_{j}\) Is the feature vector of the jth feature, \(N\) Represents the dimension of all features. Through class-based attention, the M2A encourages the model to generalize to unseen data. The Essential functions of M2AM are as follows.
Dynamic feature selection
M2AM assigns attention weights to different input features based on their relevance for classification. This adaptive mechanism enables the model to prioritize the most informative features while minimizing the impact of those that may introduce noise or are less relevant31.
Effect of regularization
When M2AM is used, the model becomes much more straightforward, which helps keep it from fitting too well. Only the most essential parts of the model are shown to make this possible. This enables validation datasets to work more effectively, which in turn generalizes the work better.
Representation in mathematical model
The M2AM mathematical model can be represented using Eq. (11). Where N shows the feature count, w shows the weight parameter, and x shows the feature vector32.
Algorithm for modified multiclass attention mechanism
Algorithm 1, the M2AM, outlines a comprehensive method for dynamically allocating class-specific attention weights to input features. The process begins with the computation of attention weights for each class, which are subsequently applied to the input features. The combined weighted features are subsequently input into a BiLSTM model, which captures both temporal dependencies and feature interrelations33,34,35. The SoftMax function is ultimately employed to categorize the input into the most likely heart disease classification. This mechanism enhances the model’s ability to prioritize relevant features for each class, thereby improving its classification accuracy in multiclass heart disease detection.
Deep bidirectional long short-term memory architecture
BiLSTM structure is used to extract the ECG signals from both forward and backward direction, which can learn temporal dependencies from past and future information. It is also essential to analyse the sequential ECG signal since details from both sides is practical when classifying. The LSTM cells regulate information via three gates: the input gate (\({i}_{t}\)), the forget gate (\({f}_{t}\)), and the output gate (\({o}_{t}\)). The cell state \({C}_{t}\) It is updated by using Eq. (17). Where \({\widehat{C}}_{t}\) Shows the candidate’s cell state.
Bidirectionality
A BiLSTM comprises two LSTM layers: one that processes the input sequence forward and another that processes it backward. This dual approach enables the model to capture context from both directions, which is especially beneficial for sequential data4,17.
Output calculation
The Deep BiLSTM enhances feature representation by leveraging both historical and prospective context, thereby increasing the system’s capability to perform classification tasks3. The complete output \(h_{t}\) It can be calculated by using Eq. (18).
Classification using output layer
The classification process involves converting the feature representation generated by the BiLSTM into a final output consistent with the predicted class labels. The last stage is classification, the information of the BiLSTM layer is passed through a SoftMax function as below Eq. (19).
where, \(C\) the total number of classes, and \(ZT\) Is the logit of the target class \(T\).
The training loss function, the cross-entropy loss, is presented by Eq. (20).
It utilizes the following key steps5.
Fully connected layer
The output is then sent through a fully connected dense layer that undergoes a linear transformation shortly after the BiLSTM layers have finished processing the input. This occurs immediately after the BiLSTM layers have completed processing the input data. A transformation T, for the output \({h}_{t}\) Calculated using Eq. (21), where b represents bias, W represents weight, and T represents the logits, which are applied before the activation function7.
Activation function
To obtain probabilities for each class, the Softmax function is applied to the logits, as determined by Eq. (22).
Loss function
The cross-entropy loss function is commonly used to evaluate the model’s performance during training. It can be calculated by using Eq. (23).
Dataset description
This research utilizes two popular datasets, MIT-BIH and INCART. The complete details of the datasets are as follows.
MIT-BIH dataset
The MIT-BIH Arrhythmia Database is a prestigious dataset utilized as a standard for assessing algorithms in ECG signal analysis. This dataset, assembled by the Massachusetts Institute of Technology (MIT) and Beth Israel Hospital, consists of 48 thirty-minute segments of two-channel ECG recordings sampled at 360 Hz. The dataset encompasses a variety of arrhythmias, including premature ventricular contractions, atrial fibrillation, and normal sinus rhythm, enabling researchers to train and evaluate models on different cardiac conditions. Every record is labeled with beat classifications, facilitating the accurate evaluation of model efficacy in identifying arrhythmic occurrences32.
This dataset has 113,000 labelled beats from recordings. This detailed annotation enables researchers to test the model’s arrhythmia detection capabilities. The MIT-BIH database has revolutionized cardiovascular research by providing a foundation for machine learning and deep learning models to improve ECG signal analysis and diagnosis. Table 2 presents the details of the MIT-BIH dataset.
INCART dataset
Another valuable ECG analysis resource is the INCART (International ECG Database), which facilitates research on cardiac arrhythmias. Electrocardiogram recordings from 75 subjects indicate both cardiac abnormalities and normal conditions. Data is sampled at 250 Hz for each 10-s interval. The INCART database is valuable for creating algorithms that differentiate between normal and abnormal ECG patterns, encompassing healthy and arrhythmic subjects33. Like the MIT-BIH dataset, INCART’s comprehensive annotation provides detailed information about each beat. Researchers can meticulously apply and assess classification algorithms using this comprehensive annotation. The INCART dataset’s varied subjects and comprehensive annotations establish a robust foundation for training machine-learning models to enhance arrhythmia detection and classification. Table 3 presents the details of the INCART dataset.
Data pre-processing
Proper preprocessing of ECG recordings is critical for accurate analysis and modeling. The preprocessing stage of the proposed pipeline receives raw ECG traces from known databases such as INCART and MIT-BIH. These databases contain waving range of ECG signals, which is essential to develop a generalized model for different types of heart diseases.
Improved adaptive bandpass filter
To guarantee the reliability and accuracy of input data, we adopt the IABPF to remove undesired noise including the power line interference, low-frequency baseline drift and other high-frequency disturbances usually existing in the ECG signals. The filter cutoff frequency is determined according to the natural frequency of the ECG, typically ranging from 0.5 Hz to 100 Hz. This band preserves the essential characteristics of the ECG waveform and removes irrelevant noise components. Also, the IABPF is of adaptable nature which enables the filter to adapt its response in terms of the signal characteristics and can work efficiently on numerous ECG signals. We choose IABPF the additional essential reasons of the filter is decreased level of noise and preserving significant waves components (P, Q, R, S, T), which is critical to diagnose heart disease accurately. Using the IABPF optimizes the process so that only the target frequency components are preserved, which is essential for subsequent feature extraction and classification tasks.
Segmentation and normalization of the signal
After filtering, the continuous ECG signal is divided into small overlapping segments to reflect the rhythmic changes in heart activity. Each segment corresponds to a cardiac cycle, which facilitates the identification of time-domain features. Segmentation ensures that the most essential properties of the signal are retained and analyzed separately. Normalisation is then performed to normalize the amplitude of ECG signals amongst different samples. Such normalization is necessary when the model is fed with signals of different magnitudes. Normalizing the data has the critical benefit of making all segments equally crucial to the classifier, allowing it to learn the aspects that differ among segments. This fosters the learning of those aspects related to temporal and frequency properties, rather than the amplitude discernment.
Wavelet transformation for feature extraction
Due to the non-stationary properties of ECG signals, the wavelet transform is performed to obtain both time and frequency features. Wavelet transforms are well-suited for analyzing time-varying signals due to their multi-resolution analysis, which offers the advantage of representing both high-frequency transients (e.g., the QRS complex) and low-frequency components (e.g., P and T waves). This allows a model to capture slight squeezes in the heart and notice patterns are linked with passing different arrhythmias. The choice of wavelet transforms such as Morlet wavelet, Daubechies wavelet etc., is dependent on its capacity towards performing efficient time–frequency localization (which is necessary for preserving fine details of ECG signals). The features of the wavelet coefficients are then employed to train the model.
Data augmentation
To further increase diversity of the training dataset, data augmentation methods are employed which produce synthetic ECG signals. This operates by applying a slight transformation to the original signals, which involves scaling, shifting, and rotating the signal patterns, to promote model generalization. Data Augmentation can have a significant advantage, especially when the dataset is unbalanced or when the model requires more training data.
Class Imbalance
For the task of heart disease classification, it is challenging to balance classes, as certain heart conditions (e.g., normal sinus rhythm) occur more frequently than others (e.g., arrhythmias). We have done nothing on them, so we have to use some techniques to handle Oscillations. We use some techniques that can be used: During pre-processing, During training.
-
Weight balancing We draw samples from minority classes with a higher probability during training to focus the model more on underrepresented conditions.
-
Resampling Over- or under-sampling of the classes in the dataset can also be used to balance datasets.
-
Evaluation Metrics We employ precision, recall, and the corresponding F-measure as our evaluation criteria, rather than just using accuracy – we want to ensure that the performance on minority classes is adequately evaluated.
These measures enable the model to avoid biasing towards the majority class, resulting in the detection of heart diseases across all classes. Collecting and integrating the IABPF with the wavelet transformation, signal normalization, data augmentation and class imbalance management improves both the quality and robustness of ECG data heavily. By enhancing signal quality, feature extraction is improved, and the amount of heart disease information reflected in the data is minimized. So the preprocessing steps do not only enhance the quality of input data but also improves machine learning approximation process to give more precise and reliable detection in heart disease.
Comparison parameters
The following key parameters were used for comparative analysis between the proposed and existing models30.
Accuracy
Accuracy is a key metric for model prediction. It displays the percentage of true results, both positive and negative, in relation to the total number of cases evaluated. It provides a broad model performance overview, as presented in Eq. 19. Here, AC: Accuracy, TN: True Negative, TP: True Positive, FN: False Negative, TP: True Positive
Precision
Precision measures model accuracy in positive predictions. It shows how many positive predictions were correct. False positives can have serious consequences, making this metric crucial, as presented in Eq. 24. Here PR: Precision
Recall
Recall, also known as sensitivity or true positive rate, measures the model’s ability to identify all relevant positive cases. This demonstrates the model’s ability to accurately identify positive instances in the dataset. Medical applications require this metric because missing a positive case can be disastrous, as presented in Eq. 25. Here RC: Recall
F-measure (F1-score)
The F-Measure balances precision and recall. It evaluates the model’s performance by considering false positives and negatives. F-Measure offers a more nuanced perspective on model effectiveness, making it particularly useful for imbalanced datasets, as shown in Eq. 26. Here FM: F-measure
Simulation results and analysis
This section closely examines the proposed model’s outcomes and demonstrates how effectively it classifies multiclass heart disease compared to other approaches. Other tested models are Naive Bayes, DNN-Taylos, Multilayer Perceptron, and Convolutional Neural Network.
The experimental setup
The experimental setup is necessary to evaluate the proposed model’s ability to classify heart disease. The methods, datasets, reproducibility, and clarity parameters of the experiments are described here15. Table 4 presents the hardware and software details, and Table 5 presents the key parameters details used for implementing the proposed model.
Simulation results
In particular, regarding the detection of multiclass heart disease, the results of the simulations performed on the proposed model demonstrate its effectiveness in processing and classifying electrocardiogram signals. Figure 2 illustrates a methodical framework for the ECG signal processing pipeline, which shows the findings presented in this framework.
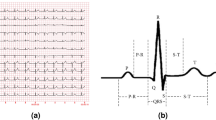
The initial stage of the process is presented above, where two lead inputs are depicted, and unprocessed ECG signals are shown from two leads. The signals are subjected to preprocessing, which includes removing baseline wandering and reducing noise, resulting in clean and standardized inputs. The signals are subsequently divided into separate cardiac cycles for meticulous examination. R-peak detection is a method used to identify the highest points in the QRS complex, which is essential for accurately determining the timing of each heartbeat. Afterward, RR intervals are computed by measuring the time between consecutive R peaks, which offer valuable information about heart rate variability.
Extracting PQST peaks involves identifying peaks corresponding to the P, Q, S, and T waves. Ultimately, PQST Intervals are calculated, providing the lengths of each cardiac phase. Exemplary measurements, such as RR intervals lasting 800 ms and PQST Intervals lasting 200 ms, facilitate a thorough examination of the cardiac signal, aiding in the identification of heart conditions and providing valuable data for clinical interpretation. Initially, the IABPF is employed to acquire raw ECG signals from two leads and subject them to preprocessing. This procedure substantially improves the signal quality by effectively mitigating various noise types, including muscle artifacts and power line interference. The enhancement of the signal-to-noise ratio (SNR) during the preprocessing stage enables the subsequent stage to incorporate more accurate feature extraction.
Wavelet transformations are implemented after the preprocessing step to segment the ECG signals. The precise extraction of features and the identification of critical characteristics within the signal, such as R-peaks, which are crucial for calculating RR intervals, are both made possible by this method. These features are crucial for the classification task, offering valuable insights into the heart’s rhythm and function. It is necessary to identify them with a high degree of precision. A Modified Multiclass Attention Mechanism is incorporated into Deep Bidirectional Long Short-Term Memory (BiLSTM) during the classification phase of the process. This is done to improve processing speed. This model enhances classification performance by dynamically adjusting attention weights to prioritize the most critical ECG signal features. Using this model improves classification performance.
Results for MIT-BIH heart dataset
The MIT-BIH Arrhythmia Database is a significant dataset in biomedical signal processing, particularly for ECG analysis. It comprises 48 thirty-minute segments of dual-channel ECG recordings, totalling 87.5 h of data. The dataset includes various arrhythmias, making it an ideal choice for training and evaluating models for heart disease classification.
Class imbalance of the MIT-BIH dataset.
The MIT-BIH data suffers from the class imbalance where some heart conditions, like normal sinus rhythm, are significantly more frequent than rare arrhythmias. This imbalance can in turn cause the model to dominate the majority class, leading to poor minority class performance. To alleviate this, we used various techniques during preprocessing and training:
-
Weighted Class: Class weights were modified at training time to give more power to minority classes, thereby preventing the model from being biased towards the majority class.
-
Resampling: We used the combination of over-sampling the minority classes and under-sampling the majority classes to balance the dataset in preprocessing before fitting the model.
-
Synthetic Data Generation: We used some data augmentation approaches to generate synthetic samples of the minority classes to enhance their representation in the set.
The MIT-BIH dataset was partitioned into training and testing subsets to facilitate practical model evaluation. This generally entails utilizing a segment of the data for model training while allocating a distinct segment for performance evaluation. This study employed a 70-10-15 partition, allocating 70% of the data for model training, 15% for testing, and 15% for validation. This ensures that the model is trained on diverse examples while being evaluated on novel data, which is crucial for assessing its generalizability. Table 6 presents the simulation results for the MIH-BIH Heart dataset for 100 epochs.
Table 6 presents a comparison of several models within the MIT-BIH Heart dataset. The suggested multiclass model achieves an accuracy of 98.82%, outperforming other approaches. The excellent precision (97.20%), recall (98.34%), and F-measure (98.92%) of the model reflect its ability to accurately diagnose heart disease and reduce false positives. The Deep BiLSTM model attains an accuracy of 91.60%, but it is inferior to the proposed method. Alternative models, including CNN, MLP-NN, DNN-Taylos, and Naive Bayes, demonstrate inferior performance metrics, highlighting the enhanced effectiveness of the proposed model in addressing the challenges of heart disease classification. The results indicate substantial improvements in diagnostic precision, confirming the efficacy of the suggested model for clinical application.
Figure 3 depicts the Training and Validation Accuracy alongside the Loss results for the proposed model utilizing the MIT-BIH dataset. The training accuracy increased markedly from 82.15% in the initial epoch to 98.50% by epoch 100, demonstrating the model’s proficient learning ability. Simultaneously, the training loss diminished from 0.55 to 0.05, indicating a robust fit to the training data. The validation accuracy increased from 81.90% to 98.25%, whereas the validation loss decreased from 0.60 to 0.10. The consistent performance on training and validation datasets demonstrates the model’s robustness and generalization capabilities, positioning it as a promising tool for precise classification of heart disease. The enhancements underscore the efficacy of the suggested approach in tackling the difficulties related to heart disease diagnosis.
Model evaluation and class imbalance problem:
Since there is a significant class imbalance, we employed precision, recall, and F-measure along with accuracy. These measures provide additional context on how the model performs for minority classes and false positives. A fivefold cross-validation (CV) was performed 5 times to achieve equal class representation on average across all sub-datasets, which would keep overfitting at bay and enable the model to generalize better.
K-fold cross validation
We used K-fold cross-validation to improve the reliability of our model evaluation. This method divides the dataset into 'k' subsets or folds. The model is first trained on 'k-1' folds and then evaluated on the remaining folds. The procedure is carried out 'k' times, with each fold acting as a validation set once. The fivefold cross-validation technique divided the dataset into five equal segments. The model’s training and validation outcomes were averaged quintupletimes to yield the comprehensive performance metric. This method is beneficial as it diminishes the variability linked to a singular train-test split, thus enabling a more dependable evaluation of the model’s performance on the MIT-BIH dataset.
Figure 4 illustrates the performance metrics of the proposed model across five folds utilizing k-fold cross-validation on the MIT-BIH Heart dataset. The graph illustrates exceptional accuracy (between 98.54% and 98.91%) and notable precision (from 96.82% to 97.50%), signifying that the model accurately detects true positive instances. The recall metrics (97.90% to 98.60%) emphasize the model’s ability to capture true positive instances, while the F-measure (97.36% to 98.03%) demonstrates a balanced performance between precision and recall. The average metrics of 98.74% accuracy, 97.16% precision, 98.49% recall, and 97.73% F-measure show that the model is robust and reliable, making it a promising tool for clinical applications in heart disease diagnosis.
Results for INCART dataset
The INCART dataset is a diverse collection of ECG recordings designed explicitly for arrhythmia detection, encompassing multiple classes of heart disease. To ensure that the model was trained on a wide range of examples, the dataset was divided into training, testing, and validation subsets, typically using a 70%-15%-15% split for model evaluation. A separate set was kept for an unbiased performance evaluation. This method verifies the model’s ability to generalize to previously unseen data.
Class imbalance in the INCART dataset.
Like the MIT-BIH dataset, the INCART dataset is class-imbalanced, where some conditions appear more frequently than other conditions. Such imbalance can confront the model hindered from learning and predicting minority classes. To address this problem, the same method of preprocessing, weight balancing, resampling and generation of synthetic data for the minority class were used in order to improve model performance with the underrepresented conditions.
Table 7 presents the simulation results for several models applied to the MIT-BIH Heart dataset; the suggested multiclass model exhibits outstanding performance across all overall criteria. The recommended model is more accurate than the others evaluated, with a degree of 97.32%. The proposed model is highly reliable for clinical diagnostics, as its 98.21% precision demonstrates a high ability to reduce false positives. A recall of 98.04% is also quite impressive, as it reflects the ability to identify true heart disease-positive cases effectively. The model’s robustness is shown by its 97.29% F-measure, which strikes a balance between accuracy and recall. The Deep BiLSTM and CNN models achieve accuracies of 89.60% and 88.89%, respectively, but are less effective than the proposed model. The DNN-Taylos, MLP-NN, and Naive Bayes models’ lower accuracy shows the limitations of traditional heart disease detection methods. Results show that the multiclass model improves clinical diagnostic accuracy.
The proposed model’s training and validation accuracy and loss are presented side by side in Fig. 5. The INCART Heart dataset was used. Over 100 epochs, the training accuracy increased from 80% to a peak of 98.82%, and the training loss decreased from 0.65 to 0.05. The accuracy of validation rose from 78.50% to 97.50%, and the loss of validation decreased from 0.70% to 0.15%. The model learns effectively from the training data, as indicated by these trends, and can then apply what it has learned to new data. This suggests that it can handle tasks that involve sorting heart diseases into groups. These performance tests demonstrate that the proposed model could be highly beneficial in medical settings.
k-Fold
The INCART dataset, which included a variety of ECG recordings designed to detect arrhythmias, was also partitioned for analysis using k-fold cross-validation. This method entailed partitioning the dataset into five folds, allowing the model to be trained and validated multiple times on different subsets. Each fold contributed to a comprehensive evaluation, with training taking place on four folds and one-fold serving as the test set. This approach ensures that the model’s performance is robust and reliable because it is tested on different segments of the data, reducing the likelihood of overfitting and providing a more accurate picture of its generalization capabilities.
Figure 6 presents a quantitative analysis of the proposed model on the INCART Heart dataset using k-fold cross-validation. The INCART dataset’s k-fold results demonstrated consistent performance across folds, indicating the model’s ability to accurately classify various heart disease presentations while maintaining high metrics, such as accuracy, precision, recall, and F-measure, across iterations.
Ablation analysis
We also conducted an ablation study to understand the contribution of each element in the proposed model. The purpose of the ablation study is to compare the contribution of individual components Improved Adaptive Band Pass Filter (IABPF), wavelet transform, and Modified Multiclass Attention Mechanism (M2AM) to the model’s performance.
Ablation study design
We performed four experiments:
-
No IABPF (Baseline Model) We do not apply any preprocessing step to the raw ECG signals, directly in average, for one sample of ECG, the inference time is 10- 15 ms, or frequency filtering.
-
No Wavelet Transforms The IABPF is employed for ECG signal processing, but the wavelet transform is not utilized. Feature extraction is without the wavelet transform.
-
No Attention Mechanism In this configuration we employed the vanilla BiLSTM structure with no M2AM. It measures the effect of the attention mechanism on the performance of the model.
-
Full Model The full model as above including IABPF preprocessing, wavelet transform, and M2AM, which does the preprocessing, classification and learning as in our basic setup.
Results of the ablation study
Table 8 shows the results of the ablation study using the MIT-BIH Heart Dataset. The table compares the model’s performance for each configuration using four key evaluation metrics: accuracy, precision, recall, and F-measure.
Analysis of results
-
No IABPF (Baseline) Without IABPF step as preprocessing, the performance dropped a lot. The accuracy was reduced to 88.40%, compared to the noise-free case, where it was 98.82%, indicating that it is worthwhile to filter out noise and suspicious frequencies. When the IABPF is not present, the model performs poorly on noisy input, despite having learned to be independent.
-
No Wavelet Transforms Taking wavelet transforms out led to a slight but notable accuracy drop from 88.40% to 89.00%. This shows that the needed wavelet transformations for both temporal and frequency characteristics play a moderate role for capturing complex signal patterns in the ECG data.
-
No Attention Mechanism When the M2AM was removed there was a clear gain in c uracy, growing from 89.00% to 90.10%. This implies that although attention used is beneficial it is not the only factor driving the performance. However, M2AM continues to make contributions to enhance the classification performance, especially in the formulation of multiclass heart disease classification.
-
Full Model (IABPF + Wavelet + M2AM) Full model returns the best result with 98.82% accuracy, 97.20 precision, 98.34% recall and 98.92% F-measure when all features are in corporates. These findings verify that the integrated contribution of IABPF, wavelet features, and M2AM significantly contributes to model generalization. The attention mechanism has been seen to have a very significant effect, enabling the model to concentrate on informative features related to the specific heart disease class when making predictions thereby improve the classification performance.
Contribution of each part
IABPF: The Improved Adaptive Band Pass Filter (IABPF) effectively improves signal quality by removing noise, baseline wander, and muscle artifacts. In the absence of IABPF, model cannot cope noise in the input, it degrades performance. This process serves to preserve only appropriate frequency components of the ECG signals, since it is critical for features extraction and classification.
-
Wavelet Transforms The wavelet transforms enable the model to learn the temporal and spectral information of the ECG signals. These are crucial attributes that can help identify the slight differences in the presentations of heart conditions. Without the process of wavelet transformation, the model loses the ability to identify complex ECG patterns, which affects the accuracy obviously.
-
M2AM We utilize the attention mechanism to dynamically weight the attention on which features to emphasize for each class. This is especially crucial in multiclass classification of heart disease, where different forms of it may have similar feature representations. Deleting the attention mechanism results in performance degradation, albeit to a lesser extent, with the decoder. It is, thus, essential to focus on the importance of the attention mechanism in differentiating among various heart disease classes and enhancing overall classification performance.
Ablation Study It verifies that every part (IABPF, the wavelet transforms and M2AM) is significant to improve the performance of the method. The cumulative effect of these elements is manifested as substantial gains in classification accuracy, precision, recall, and F-measure. The attention mechanism, especially, appears quite beneficial in concentrating the model’s attention on the most meaningful features, which leads to a significantly better distinction between closely related heart diseases. This work demonstrates the importance of each subnetwork, and confirms that the proposed model is effective for heart disease classification.
Comparative analysis with other state-of-the-art techniques
Different cutting-edge methods for categorizing heart diseases are presented in Table 9. The Multiclass Model is one of them. The table displays the sources, datasets, techniques, and corresponding accuracy percentages for each. It achieved a score of 98.82%, indicating that it performed well. The suggested model combines a Modified Multiclass Attention Mechanism with Deep Bidirectional Long Short-Term Memory. Other models typically achieve accuracy rates between 87.84% and 93.02%, which is significantly better. This model remains the best approach, despite the advancements in machine learning and deep learning. This method is highly effective in detecting heart disease.
Computational complexity and inference time
It is of practical importance for the application on real-time scenarios to make the models have proper computation complexity and inference time: such as cardiac disease diagnosis in clinical. We now summarize these aspects for the proposed model as follows.
Computational complexity
The proposed model consists an improved adaptive band pass filter (IABPF), wavelet transform (WT) and a modified M2AM with Deep BiLSTM for classification. The computational complexity for each module is as follows:
-
IABPF The complexity of ECG signals filtering is \(O(N),\) Where N represents data points.
-
Wavelet Transform (WT) The wavelet transform has an efficiency of \(O(N log N)\) For temporal representation, it can be employed to extract time–frequency characteristics in ECG data.
-
M2AM with BiLSTM Attention contribution’s complexity is \(O(N F)\) (F being the number of features) and to this adds the BiLSTM complexity \(O(T U),\) With T being the length of the sequence and U the number of LSTM units.
Therefore, overall complexity is \(O(T * U + N log N),\) With the BiLSTM and attention as the dominant terms.
Inference time
In practice, computational cost is a key factor to consider to have real time deployment. The overall time of inference includes preprocessing (IABPF and wavelet transformations) and classification (BiLSTM and M2AM):
-
Preprocessing Since the IABPF and wavelet transformation is computationally fast (the computation time needed to analyze each ECG segment is tens of milliseconds), these methods are suitable for real-time application.
-
Classification The extra time of the attention mechanism in the BiLSTM with M2AM shows. On average for one sample of ECG, the inference time is 10-15 ms on typical computational machinery (GPU/CPU).
Contrast with conventional methods
In contrast to classic models such as SVM or Random Forests:
-
SVM \(O(N^2)\) geometry cuts and is computationally expensive for large datasets but optimally fast for inference.
-
Random Forests Complexity is \(O(N log N),\) inference is very fast, yet accuracy is trash on more complex tasks (i.e. multiclass heart disease classification for instance).
The proposed deep-learning-based model, albeit more computationally intensive, has much better quality and generalization, for use in live-clinical scenarios.
Conclusion and future direction
This research shows that the proposed M2AM adopting the deep Bidirectional LSTM improves the effectiveness of heart disease detection, and classification. The model gave excellent performance parameters such as 98.82% accuracy, 97.20% precision, 98.34% recall with an F-measure of 98.92%, which well covers the main challenges like misclassification, noise invasion, and feature similarity. By adding attention mechanism, the network can pay more attention on the most related features, and promote better feature representation and generalization ability for different types of heart diseases. Despite that the fact that our M2AM with Deep BiLSTM cannot beat the M1AM with traditional methods in all cases, compared with the traditional techniques and M2AM with Deep BiLSTM achieves better results in most cases. The exploitation of advanced preprocessing (e.g., IABPF and wavelet transform) plays a crucial role in enhancing signal quality and sensor feature extraction precision. These advancements in the diagnosis pipeline enable more precise detection of cardiovascular diseases thus, improving patient outcomes and intervention. Although the proposed model achieves good performance, there are many areas that can be explored in future work. One promising application is the exploration of transfer leaning strategies that may further the effectiveness of the model on various datasets and improve the generalizability, thereby the model can be adjusted to patients of different demographic and distributed clinical settings.
Furthermore, the opacity of deep learning models is a pressing issue especially in medical area due to the lack of interpretability in the predictability of the data. In future work, we plan to incorporate explainable AI tools like SHAP and Grad-CAM, as these can provide insights into how the model reasons through its predictions. This would enhance trust and help facilitate the successful integration of AI-based diagnostic instruments in clinical care. Adding more variety of cardiac conditions to the dataset and also including the real-time monitoring data from wearable devices would also help to improve the training, accuracy and continuous learning potential of the model. This may help lead to earlier detection and more individualized patient care. Lastly, it is essential to investigate how the proposed model can be effectively integrated into the current clinical environment to ensure its feasibility. Working with clinicians and researchers would clarify how the model can be implemented and how well it is likely to perform on actual cases, so that improved diagnostic techniques for heart disease translate to better patient outcomes. It is through these efforts that we intend to create a more accurate, trusted, effective mechanism for diagnosing heart disease, and thereby, better health for all across the world.
Data availability
Data Availability: The dataset is publicly accessible through the MIT-BIH Database, available at Kaggle, and the INCART Database, available at Kaggle. For any additional inquiries or specific requests, please get in touch with the corresponding author.
References
Song, C., Zhou, Z., Yue, Yu., Shi, M. & Zhang, J. An improved Bi-LSTM method based on heterogeneous features fusion and attention mechanism for ECG recognition. Comput. Biol. Med. 169, 107903 (2024).
Wang, Z.-C. et al. Deep learning for discrimination of hypertrophic cardiomyopathy and hypertensive heart disease on MRI native T1 maps. J. Magn. Reson. Imaging 59(3), 837–848 (2024).
Pachiyannan, P. et al. A novel machine learning-based prediction method for early detection and diagnosis of congenital heart disease using ecg signal processing. Technologies 12(1), 4 (2024).
Shokouhifar, M., Hasanvand, M., Moharamkhani, E. & Werner, F. Ensemble heuristic-metaheuristic feature fusion learning for heart disease diagnosis using tabular data. Algorithms 17(1), 34 (2024).
Rahman, A. U. et al. Enhancing heart disease prediction using a self-attention-based transformer model. Sci. Rep. 14(1), 514 (2024).
Mishra, J. & Tiwari, M. IoT-enabled ECG-based heart disease prediction using three-layer deep learning and meta-heuristic approach. SIViP 18(1), 361–367 (2024).
Rao, C. L. N. & Kakollu, V. CB-HDM: ECG signal based heart disease classification using convolutional block attention assisted hybrid deep Maxout network. Biomed. Signal Process. Control 95, 106388 (2024).
Abbas, S. et al. Artificial intelligence framework for heart disease classification from audio signals. Sci. Rep. 14(1), 3123 (2024).
Chen, J. et al. Congenital heart disease detection by pediatric electrocardiogram based deep learning integrated with human concepts. Nat. Commun. 15(1), 976 (2024).
Ogunpola, A., Saeed, F., Basurra, S., Albarrak, A. M. & Qasem, S. N. Machine learning-based predictive models for detection of cardiovascular diseases. Diagnostics 14(2), 144 (2024).
Naeem, A. B. et al. Heart disease detection using feature extraction and artificial neural networks: A sensor-based approach. IEEE Access 12, 37349–37362 (2024).
Brown, K. et al. Using artificial intelligence for rheumatic heart disease detection by echocardiography: focus on mitral regurgitation. J. Am. Heart Assoc. 13(2), e031257 (2024).
Manikandan, G. et al. Classification models combined with Boruta feature selection for heart disease prediction. Inform. Med. Unlocked 44, 101442 (2024).
Zhu, J., Liu, H., Liu, X., Chen, C. & Shu, M. Cardiovascular disease detection based on deep learning and multi-modal data fusion. Biomed. Signal Process. Control 99, 106882 (2025).
Irsyad, A. & Wardhana, R. Deep learning methods for ECG-based heart disease detection. J. Electron. Electromed. Eng. Med. Inform. 6(4), 467–477 (2024).
Ma, C. Y. et al. Predicting coronary heart disease in Chinese diabetics using machine learning. Comput. Biol. Med. 169, 107952 (2024).
Dubey, A. K., Sinhal, A. K. & Sharma, R. Heart disease classification through crow intelligence optimization-based deep learning approach. Int. J. Inf. Technol. 16(3), 1815–1830 (2024).
Alam, A. & Muqeem, M. An optimal heart disease prediction using chaos game optimization-based recurrent neural model. Int. J. Inf. Technol. 16(5), 3359–3366 (2024).
Almazroi, A. A., Aldhahri, E. A., Bashir, S. & Ashfaq, S. A clinical decision support system for heart disease prediction using deep learning. IEEE Access 11, 61646–61659 (2023).
Ahmed, R., Bibi, M. & Syed, S. Improving heart disease prediction accuracy using a hybrid machine learning approach: A comparative study of SVM and KNN algorithms. Int. J. Comput., Inform. Manuf. (IJCIM) 3(1), 49–54 (2023).
Ahmad, S., Asghar, M. Z., Alotaibi, F. M. & Alotaibi, Y. D. Diagnosis of cardiovascular disease using deep learning technique. Soft Comput. 27(13), 8971–8990 (2023).
Guven, M. & Uysal, F. A new method for heart disease detection: long short-term feature extraction from heart sound data. Sensors 23(13), 5835 (2023).
Bebortta, S., Tripathy, S. S., Basheer, S. & Chowdhary, C. L. Fedehr: A federated learning approach towards the prediction of heart diseases in iot-based electronic health records. Diagnostics 13(20), 3166 (2023).
Yang, H., Chen, Z., Yang, H. & Tian, M. Predicting coronary heart disease using an improved LightGBM model: Performance analysis and comparison. IEEE Access 11, 23366–23380 (2023).
Subramani, S. et al. Cardiovascular diseases prediction by machine learning incorporation with deep learning. Front. Med. 10, 1150933 (2023).
Saranya, G. & Pravin, A. A novel feature selection approach with integrated feature sensitivity and feature correlation for improved prediction of heart disease. J. Ambient. Intell. Humaniz. Comput. 14(9), 12005–12019 (2023).
Nandy, S. et al. An intelligent heart disease prediction system based on swarm-artificial neural network. Neural Comput. Appl. 35(20), 14723–14737 (2023).
Chandrasekhar, N. & Peddakrishna, S. Enhancing heart disease prediction accuracy through machine learning techniques and optimization. Processes 11(4), 1210 (2023).
Kumar, D. V. S., Chaurasia, R., Misra, A., Misra, P. K., & Khang, A. Heart disease and liver disease prediction using machine learning. In Data-centric AI solutions and emerging technologies in the healthcare ecosystem 205–214. (CRC Press, 2023).
Rajkumar, G., Devi, T. G. & Srinivasan, A. Heart disease prediction using IoT based framework and improved deep learning approach: medical application. Med. Eng. Phys. 111, 103937 (2023).
Yadav, A. L., Soni, K., & Khare, S. Heart diseases prediction using machine learning. in 2023 14th International Conference on Computing Communication and Networking Technologies (ICCCNT), 1 (2023)
MIT-BIH dataset, access on 21st Jan 2014, online available at https://www.kaggle.com/datasets/mondejar/mitbih-database.
INCART dataset, access on 10th March 2024, online available at https://www.kaggle.com/datasets/bjoernjostein/st-petersburg-incart-12lead-arrhythmia-database.
Saheed, Y. K., Salau-Ibrahim, T. T., Abdulsalam, M., Adeniji, I. A. & Balogun, B. F. Modified bi-directional long short-term memory and hyperparameter tuning of supervised machine learning models for cardiovascular heart disease prediction in mobile cloud environment. Biomed. Signal Process. Control 94, 106319 (2024).
Indrakumari, R., T. Poongodi, and Soumya Ranjan Jena. Heart disease prediction using exploratory data analysis.Procedia Comput. Sci. 173, 130–139 (2020).
Acknowledgements
The authors extend their appreciation to Taif University, Saudi Arabia, for supporting this work through project number (TU-DSPP-2024-17).
Funding
This research was funded by Taif University, Taif, Saudi Arabia project number (TU-DSPP-2024-17).
Author information
Authors and Affiliations
Contributions
U.K. Lilhore developed the Modified Multiclass Attention Mechanism and Deep BiLSTM model, implemented the experimental framework, and conducted the performance analysis. M.K. handled data acquisition and preprocessing and contributed to implementing the Improved Adaptive Band Pass Filter (IABPF) and wavelet transformation. S.S. conceptualized the research, provided overall supervision, guided the methodological framework, and contributed extensively to manuscript writing and editing. R.A. contributed to comparative analysis, model evaluation, and validation. A.M.B., M.A., and A.Al.i assisted in data interpretation, results visualization, and refining the manuscript for final submission. All authors read, reviewed, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lilhore, U.K., Simaiya, S., Khan, M. et al. A deep learning approach for heart disease detection using a modified multiclass attention mechanism with BiLSTM. Sci Rep 15, 25273 (2025). https://doi.org/10.1038/s41598-025-09594-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-09594-8
Keywords
This article is cited by
-
Interpretable machine learning framework for detection of cardiovascular diseases from real-time ECG signals
Network Modeling Analysis in Health Informatics and Bioinformatics (2025)