Abstract
Inhibition of carbonic anhydrases (EC 4.2.1.1, hCAs) is known to be a potential target for treatment of several disorders like epilepsy, glaucoma, obesity, and cancer. The current research focuses on the synthesis of a series of 4’-amino-[1,1’-biphenyl]-4-sulfonamide derivatives (9a-e) through cross-coupling reactions in the presence of Cu(OAc)2 catalyst. The structural elucidation of synthesized derivatives was carried out through1H NMR, and13C NMR. These sulphonamide derivatives were screened for inhibitory potential towards various isozymes of carbonic anhydrases including hCA-II, hCA-IX, and hCA-XII. Enzyme inhibition assay exhibited that synthesized derivatives with IC50 ± SEM, 9e (0.38 ± 0.03µM), 9d (0.21 ± 0.03 µM) and 9b (0.69 ± 0.15 µM) had remarkable inhibition potency against hCA-II, hCA-IX and hCA-XII respectively. It was noted that 9d exhibited 8-fold more inhibitor potential as compared to standard inhibitor acetazolamide. For the most potent inhibitors, enzyme kinetic analysis was also carried out to find inhibition mode. Furthermore, the drug-ability of synthesized compounds was evaluated through SwissADME tools, and found that all the newly prepared derivatives successfully satisfied the drug-ability criteria. Molecular docking studies were conducted to identify the types of interactions between the synthesized ligands and the target proteins.
Similar content being viewed by others
Introduction
CAs (Carbonic anhydrases) zinc based metalloenzymes are naturally found in archaic prokaryotic and eukaryotic families1. CAs are categorized into eight different families, including α, β, γ, δ, ξ, η, θ, and ι families that catalyze carbon dioxide (CO2) hydration to proton (H+) and bicarbonate (HCO3)2,3,4,5. This reaction is very important for physiological processes like acid-base balance and also for maintaining the optimum extra/intracellular pH in the living organism3,6. CAs are involved in numerous normal physiological functions such as transporting CO2/HCO3 in homeostasis, secretion of electrolytes, and a lot of biosynthetic processes including lipogenesis, oncogenesis, gluconeogenesis, bone resorption, calcification, as well as ureagenesis7,8. Among these families, α-CAs are expressed in all vertebrates, and up to now, 15 isoforms have been reported in humans, which differ in catalytic activities and cell and tissue distribution9,10. Variation in the expression of CA isoforms may lead to pathological conditions such as epilepsy, neuropathic pain, glaucoma, edema, cancer, osteoporosis, obesity, and CNS disorders, thereby making the CAs more valuable and validated pharmacological targets for treating such dysfunctions11,12,13. Among human CA isoforms, an important role is played by hCA-II in angiogenesis through VEGFR (vascular endothelial growth factor receptor) signaling pathway. The progress and development of cancerous tumors is associated with the activation of both the genes CA-II and VEGFR in response to hypoxia14. Hypoxic solid tumors were found to be the result of overexpression of membrane-bounded isoforms such as CA-IX and CA-XII15. Additionally, the survival, pH maintenance and spreading of tumors is associated with isoforms (hCA-IX and hCA-XII). Therefore, CA-IX/XII isozymes inhibition may be the potential target for cancer treatment16,17. A variety of carboxylic acids substituted with aryl- or alkyl groups, sulfamoyl carbamates, sulfonamides, and sulfamides, have already been reported as known inhibitors of CA isoforms18. Sulfonamide derivatives, among all known inhibitors, were found to be the most potent, as deprotonated nitrogen of the sulfonamide group interacts with Zn++ in active pocket of CA, thereby blocking the catalytic property of the enzyme19. In addition, sulfonamides have a lot of pharmacological applications mainly anticancer, anti-inflammatory, antiplatelet, antioxidant, and antimicrobial, antidiabetic, and antimalarial activities20.
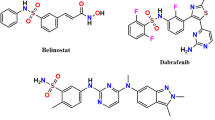
Acetazolamide (AAZ) (1) has been used as a standard CA inhibitor for more than 40 years11,18,21 Multiple heterocyclic aromatic sulfonamides22,23,24,25,26,27,28,29,30,31 such as brinzolamide (2), methazolamide (3), and indisulam (4), have been clinically employed against cancerous activities of carbonic anhydrases shown in Fig. 1.
Standard inhibitors potentially used against hCA isoforms. A free open access software [ChemDraw v20.1 available at. https://disk.yandex.com/d/sY_jZQuwkLDLIg] was used.
Moreover, numerous sulfonamide derivatives exhibit antidepressant, anti-oxidant, and anti-microbial activities32,33,34,35,36. In recent years, 4-((1-acetyl-5-oxo-2,5-dihydro-1 H-pyrrol-3-yl)amino)benzenesulfonamide (5) and 2-(4-hydroxy-6-methyl-2-oxopyridin-1(2 H)-yl)ethane-1-sulfonamide (6) have been found as selective potential inhibitors against hCA-I, and hCA-IX respectively37. The excellent inhibition activities of various sulfonamide compounds encouraged us to put more effort into synthesizing its aryl/heteroaryl derivatives. At the end of the 20th century, Chan Lam cross-coupling reactions catalyzed by Cu-salts were developed in organic synthesis. These cross-coupling reactions occur under mild conditions and are free from toxic environments. In addition, it provides an efficient way to form C-N bond (between aryl/hetero-arylated boronic acids and aliphatic/aromatic amines)38.
In the current study, various arylated and hetero-arylated derivatives of 4’-amino-[1,1’-biphenyl]−4-sulfonamide have been prepared through Chan Lam cross-coupling reactions. Furthermore, all the freshly prepared compounds have been analyzed for their inhibition potential against CA isoforms (hCA-IX, hCA-XII, hCA-II). CAs IX and XII, two distinct isoforms of carbonic anhydrase, are linked to cancer progression, metastasis, and suboptimal therapeutic responses. These isoforms have been recognized as alternative targets for chemotherapy aimed at hypoxic solid tumors. All the synthesized compounds exhibited remarkable activities as hCA inhibitors as compared to acetazolamide (standard drug). The ability of each compound to exert inhibitory control differs based on its structural characteristics. Furthermore, ligand-protein interaction/binding affinities of compounds have been analyzed through in silico studies or molecular docking.
Results and discussion
Chemistry
Chan Lam cross-coupling reactions catalyzed by Cu (II) are a facile and perfect methodology for N-arylations. Construction of the C-N bond through Chan Lam cross-coupling reactions has been accomplished by employing various transition metal catalysts such as copper and nickel39,40. These reactions are affected by the nature of the solvent, type of Cu salt, presence or absence of visible light, reaction time, kind of bases (N, N’ N’ N’-tetramethylethylenediamine(TMEDA), Et3N, pyridine). Chan Lam products’ yield varies depending on the nature of N-nucleophilic and electrophilic (Aryl/heteroaryl boronic acids) partners. C-N bond formation can occur under mild conditions in these reactions38.
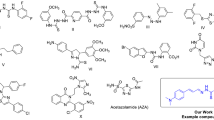
In the present research, an easy and efficient methodology was adopted for C-N cross-coupling catalyzed by Cu(II). We started working with commercially available 4’-amino-[1,1’-biphenyl]−4-sulfonamide (7) and reacted it with a variety of aryl/heteroaryl boronic acids (8). All the new products (9a-e) have been prepared by using Cu(OAc)2 catalyst, (TMEDA) as a base and distilled methanol (solvent) under a named reaction (Chan Lam cross-coupling reaction). The synthesized molecules provided good to excellent yields (65–81%) in Fig. 2. The solubility of compound (7) in methanol was the reason for the high yields.
Chan Lam cross-coupling reaction between 4’-amino-[1,1’-biphenyl]−4-sulfonamide and multiple aryl/hetero aryl boronic acids to form products (9a-e). A free open access software [ChemDraw v20.1 available at. https://disk.yandex.com/d/sY_jZQuwkLDLIg] was used.
Conditions: (i) 7 (0.8 mmol, 1.0 eq, 200 mg), 8 (1.6 mmol, 2.0 eq), Cu(OAc)2 (0.8 mmol, 1.0 eq, 160 mg), TMEDA (1.6 mmol, 2.0 eq, 180 mg), ambient oxygen, distilled water (2 mL), MeOH (10 mL), 25°C, 24 h.
Initially, coupling reactions were performed in ethyl acetate solvent, but this time, yields were moderate due to less solubility of (7) in ethyl acetate. However, yields were surprisingly enhanced by changing the solvent (methanol) while keeping all the other reaction conditions the same. Previously, Boykin and researchers41 performed Chan Lam reactions in methanol to obtain significant yields. Our reactions provided better results in methanol/water (10:1) than pure methanol under an oxygenated atmosphere. Aryl/heteroaryl boronic acids substituted with electron-withdrawing moieties provided product 9b with maximum yield (81%), while electron-donating moieties comparatively gave product 9c with less yield (65%) shown in Table 1. Additionally, it was noted that yields of reactions have a direct relation with reaction time. This has been illustrated by monitoring the reactions through thin layer chromatography (TLC) after intervals, and specifically, a reaction time of 24 h gave maximum yields.
Biological studies
Cell viability assay
The cell viability and cytotoxicity were assessed using the MTT assay42which involved testing against HEK-293 T normal cells and BHK-21 cell line. The evaluation of cell viability and safety profiles for the synthesized compounds, including 9a to 9e, was carried out. All compounds were analyzed at a final concentration of 100 µM, and the results were compared to those of the standard drug cisplatin. The findings indicated that most compounds exhibited noncytotoxic characteristics and were considered safe at the 100 µM level. These non-toxic compounds could be evaluated for further bioassays shown in Table 2.
Carbonic anhydrase Inhibition assay and Structure-Activity relationship (SAR)
All the newly synthesized analogs (9a-e) have been screened for their inhibition potential against multiple hCA isozymes including hCA-IX, hCA-XII, and hCA-II. In the current research work, a standard potent hCA inhibitor acetazolamide (AAZ) was also screened. The inhibitory activities of all the derivatives (9a-e) were compared with AAZ and the results are encapsulated in Table 3. A few molecules showed greater inhibition potential as compared to AAZ based on their structure. However, others depicted lesser potency when compared with AAZ.
Sulphonamide derivatives (9a-e) were found potent against hCA-II having IC50 domain 0.38–2.11 µM. 9e exhibited fantastic inhibition potency (IC50 = 0.38 ± 0.03 µM) greater than standard drug (IC50 1.92 ± 0.05 µM). Which has Cl at the meta position while F at para position of ring A as shown in Fig. 2. Additionally, 9b and 9d also showed outstanding inhibitions (IC50 = 1.03 ± 0.10, 1.01 ± 0.10 µM, respectively) in contrast with AAZ (standard compound). In the case of 9b, MeOCO, a moiety with a strong electron-withdrawing effect, is attached at the para position of ring A. Whereas 9d has a heterocyclic ring (4-pyridinyl). It was observed that 9a (has 4-Cl at the para position of ring A) expressed moderate inhibition potential (IC50 value = 1.03 ± 0.10 µM) in contrast with 9b because of less electron-withdrawing effect of Cl than F.
Regarding hCA-IX, the synthesized molecules (9a-e) were found pharmaceutically active as anti-hCA-IX with IC50 range 0.21–3.43 µM. The biological activity trend of (9a-e) observed as anti-hCA-IX is different from hCA-II. Excellent inhibitory activity against hCA-IX was exhibited by 9d (IC50 = 0.21 ± 0.03) with a pyridinyl moiety compared to AAZ (IC50 = 1.81 ± 0.10). However, 9b provided almost similar results (1.46 ± 0.08) in both cases. Ring A substituted by halo-groups assuming the structures of 9a and 9e showed moderate inhibition effects (IC50 = 2.79 ± 0.02, 3.43 ± 0.15), respectively.
Considering the hCA-XII inhibitions, 9b, 9d, and 9a provided excellent results (IC50 = 0.69 ± 0.15, 0.88 ± 0.09, and 1.05 ± 0.10, respectively) compared to AAZ (IC50 = 1.26 ± 0.07). The high potency of 9a was due to electron-withdrawing character of the methoxycarbonyl moiety. However, 9e showed moderate inhibition, slightly lower than AAZ (Fig. 3).
Comparison of IC50 ± SEM of 9e, 9d, and 9b with AAZ. A free open access software [ChemDraw v20.1 available at. https://disk.yandex.com/d/sY_jZQuwkLDLIg] was used.
Exceptionally, 9c (substituted with methyl groups at both the meta positions of ring A) was found inactive against all hCA isoforms (hCA-II, hCA-IX, and hCA-XII). To recapitulate, the compounds with electron-withdrawing substitutions exhibited moderate to excellent efficiency, whereas electrons with donating moieties substituted compounds showed no results against any isoform43.
Regarding the selectivity of action towards the target compared to off-target CAs (see Table 3), compounds 9a and 9d demonstrated significant inhibitory specificity for hCAII/IX and hCAII/XII. These synthesized derivatives produced a preferred inhibition of the target CA IX and XII over the main off-target hCA II as compared to AAZ.
Enzyme kinetics of carbonic anhydrases
Most potent compounds among the synthesized derivatives (9a-e) were selected for enzyme kinetic studies. The Lineweaver-Burk plot defined that 9e and 9b exhibited the competitive mode of inhibition for hCA-II and hCA-XII in Figs. 4 and 5, respectively. As presented in Figs. 6 and 7d revealed an uncompetitive mode of inhibition for hCA-XII. For enzyme kinetic analysis, the substrate and inhibitor concentrations are mentioned in Table 4.
Putative binding interaction of 9b in the active pocket of CA-XII (PDB ID# 4HT2). A free open access software [ChemDraw v20.1 available at. https://disk.yandex.com/d/sY_jZQuwkLDLIg] was used.
Molecular Docking studies
Molecular docking was carried out to investigate the binding mode of synthesized ligands within active sites of target enzyme proteins and further correlate with the experimental data. The compounds exhibited good binding affinities within the active pockets through conventional hydrogen bonding, carbon-hydrogen bond, π-π, and hydrophobic interactions. A correlation was found between the in vitro enzyme inhibition assay results and those of the in-silico studies. Especially, substances with the lowest IC50 values showed the highest affinity for binding with the target protein High-resolution crystal of hCA-II/hCA-IX, and hCA-XII with resolutions of 1.03, 1.8, and 1.45 Å were selected for molecular docking studies. Crystal structure of CA-II encompasses a co-crystalized ligand (N-[2-(3,4-dimethoxyphenyl)ethyl]−4-sulfamoylbenzamide) in its pocket. This co-crystalized ligand was re-docked to validate the docking procedure and a free binding energy of ΔG= −7.1135 Kcal/mole was noted (Fig. S11). Among all synthesized derivatives, 9e was most potent inhibitor of CA-II during in vitro studies (IC50 = 0.38 ± 0.03 µM). Molecular docking studies of 9e revealed a binding energy of −6.4521 Kcal/mole, and this compound established two hydrogen bonds with Thre1199, three Carbon-hydrogen bonds with His1094, Gly1132, and Leu1198 amino acid residues in the active pocket of targeted CA-II protein. Within the active pocket, the deprotonated nitrogen (N) of primary sulfonamide and the cofactor (Zn++) form a complex with 1.94 Å bond distance. 9e also exhibited π-σ bond with Leu1198, π-Sulphur interaction with Trp1209, π-π interactions with Phe1131 and π-alkyl interactions with Val1121, Val1135 amino acid residues in the active pocket as presented in Fig. 8.
Putative binding interaction of 9e in the active pocket of CA-II (PDB ID# 3V7X). A free open access software [ChemDraw v20.1 available at. https://disk.yandex.com/d/sY_jZQuwkLDLIg] was used.
The x-ray crystallographic structure of CA-IX is homo-tetrameric, hence, for docking analysis, chain A was selected which contained a co-crystalized ligand (5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl) thiophene-2-sulfonamide). This co-crystalized ligand was chosen as a reference for docking and re-docked for validation of docking protocol and an RMSD value of 1.23 Å was found (Fig. S12). In molecular docking studies, 9d exhibited a binding energy of −6.2629 Kcal/mole within the active pocket of CA-IX protein with PDB ID: 5FL4. Here, 9d exhibited two hydrogen bondings with Thr200, involving SO2 and NH2 of sulphonamide moiety. An important interaction was a coordination between the nitrogen of the Sulfonamide moiety and co-factor Zn++ with bond length of 1.80 Å. In addition, two carbon-hydrogen linkages were developed between the oxygen of the sulfonyl group and His94, Leu199 amino acid residues in the active pocket of the target protein. A π-σ bond and π-Sulphur interaction were also observed with amino acid residues of Leu199 and Trp210, respectively. π-alkyl linkages were also developed with Val121, Val130, and Leu134 amino acid residues as presented in Fig. 9.
Putative binding interaction of 9d in the active pocket of CA-IX (PDB ID# 5FL4). A free open access software [ChemDraw v20.1 available at. https://disk.yandex.com/d/sY_jZQuwkLDLIg] was used.
CA-XII crystallographic structure (PDB ID: 4HT2) also contained homo-tetramer chains. In this case, chain A was chosen for docking analysis, which contains a co-crystalized ligand (4-[(4,6-dimethylpyrimidin-2-yl) thio]−2,3,5,6-tetrafluorobenzenesulfonamide). This co-crystalized ligand was redocked and obtained an RMSD value of 0.83 Å (Fig. S13). In molecular docking studies, 9b exhibited a high binding energy of ΔG= −7.1974 Kcal/mole which is comparable to that of the co-crystallized ligand with ΔG= −7.2245 Kcal/mole, as presented in Table 5. The selected docked pose of 9b reveals that there were three hydrogen bondings with Ala129, Thr198, and Thr199 involving oxygen atoms of sulfonyl and carbonyl moieties, as presented in Fig. 7. In addition to this, a π-σ bond with Leu197 and π-alkyl interactions with amino acids residues Val119, and Ala129 were also observed. The carbonyl’s moiety oxygen atom (C = O) linked with the zinc metal and made a bond distance of 1.99 Å.
In Silico ADME and Pharmacokinetic parameters
SwissADME is a free online tool used to analyze the pharmacokinetic characteristics, including ADME (absorption, distribution, metabolism, and excretion). This tool is also used to assess the drug-ability of low molecular weight derivatives44. This software predicts the ADME values of recently synthesized sulfonamide derivatives using certain algorithms, and the results are shown in Table 6. To better represent GI (gastrointestinal) permeability, these results were exhibited as a boiled egg plot in Fig. 10. Among them, most of the derivatives show better GIT absorption and do not have the ability to cross the BBB (blood-brain barrier) which may be advantageous in reducing unwanted CNS effects. Here, the drug-likeness of synthesized compounds was measured based on certain recognized drug-ability rules, like Lipinski’s rule of five and PAINS criteria. Lipinski’s rule states that for a compound to be a likely successful lead candidate, orally absorbed, it must obey at least 3 of the following: <5 hydrogen bond donor groups, < 10 hydrogen bond acceptor groups, a molecular mass of < 500 Da, calculated lipophilicity (MLOGP) of < 5, and a number of rotatable bonds < 10. In addition, for a potential drug candidate, the TPSA should be in a range of 0-140Å for better absorption of a drug candidate. The results indicate that most of our synthesized compounds fulfil the drug-ability criteria and may prove to be better batter drug candidates. Log[S] value indicates the water solubility of compounds, among the synthesized derivatives, 9d was found to be a water-soluble compound and the rest of the molecules were moderately soluble in water. These synthesized derivatives serve as promising drug candidates because the produced compounds comply with drug-ability criteria.
Materials and methods
General description
All the required chemicals and reagents in our research have been purchased from Alfa Aesar and Sigma Aldrich (USA). Solvents have been purified by distillation technique and rotary apparatus before use. Chromatographic techniques including thin-layer chromatography (TLC) and column chromatography have been employed to separate, identify and purify the desired products. Silica gel of various pore sizes (70–230 mesh) as well as (230–400 mesh) has been used as a stationary phase in column chromatography. Reaction monitoring was accompanied by TLC cards (PF254-coated) and UV lamps having characteristic wavelengths of 254 to 365 nm to visualize spots. Verification of product structures has been achieved through an NMR instrument of 600 MHz (Bruker spectrophotometer) using deuterated DMSO as a solvent to obtain1H NMR spectra. Coupling constant and chemical shifts were calculated and recorded in units such as Delta ppm and Hertz (Hz) respectively. The LECO 630–200–200 TruSpec CHNS micro-analyzer was applied for elemental analysis.
General procedure to synthesize 4’-amino-[1,1’-biphenyl]−4-sulfonamide derivatives (9a-e)
Chan Lam cross-coupling reaction protocol was followed to synthesize new compounds (9a-e)41,45,46. A 200 mL round bottom flask was rinsed with ethyl acetate before use. 4’-amino-[1,1’-biphenyl]−4-sulfonamide (0.8 mmol, 1.0 eq) was taken in the flask in the presence of 10 mL methanol. In the next step, 2 mL distilled water was added in the flask. Subsequently, aryl boronic acid (1.6 mmol, 2.0 eq), and TMEDA (1.6 mmol, 2.0 eq) were added to the reaction mixture. After the addition of Cu(OAc)2 (0.8 mmol, 1.0 eq) the blend was agitated overnight in the presence of ambient oxygen. After intervals, reaction progress was examined by TLC cards under a UV lamp. Filtration of the reaction mixture was carried out on reaction completion followed by the evaporation of solvent to get concentrated product. Separation of desired products was carried out through column chromatography. Combination of solvents (n-hexane/ethyl acetate) was used in multiple gradient ratios as a mobile phase in column chromatography. Prepared and purified compounds (9a-e) were analyzed by NMR spectroscopy40.
Structure Elucidation of 4’-amino-[1,1’-biphenyl]−4-sulfonamide derivatives
4’-((4-chlorophenyl)amino)-[1,1’-biphenyl]−4-sulfonamide (9a)
Dark reddish Sticky solid, MP = 190–192°C1. H-NMR (600 MHz, DMSO-d6) δ 9.48 (s, 1 H), 8.15 (s, 2 H), 7.33 (d, J = 8.4 Hz, 2 H), 7.20 (d, 2 H), 6.97–6.92 (m, 4 H), 6.53 (d, J = 8.4 Hz, 2 H), 5.93 (s, 2 H)13. C-NMR (150 MHz, DMSO-d6) δ 153.1, 143.4, 139.6, 131.6, 129.4, 129.2, 125.2, 123.0, 122.7, 118.7, 117.7, 113.0. Analytic calculations for C18H15ClN2O2S (358.84): C, 60.25; H, 4.21; N, 7.81; S, 8.93. Experimental: C, 60.29; H, 4.25; N, 7.88; S, 8.87.
Methyl 4-((4’-sulfamoyl-[1,1’-biphenyl]−4-yl)amino)benzoate (9b)
Orange yellowish crystalline solid, MP = 138–141°C1. H NMR (600 MHz, DMSO-d6) δ 8.68 (s, 1 H), 7.81 (d, J = 8.6 Hz, 2 H), 7.54–7.40 (m, 3 H), 7.12–6.93 (m, 5 H), 6.61 (d, 2 H), 5.93 (d, 2 H), 3.82 (s, 3 H)13. C NMR (150 MHz, DMSO-d6) δ 166.5, 149.4, 146.9, 137.7, 133.1, 131.5, 129.1, 127.9, 122.3, 121.1, 119.2, 114.0, 113.0, 60.7. Analytic calculations for C20H18N2O4S (382.43): C, 62.81; H, 4.74; N, 7.33; S, 8.38. Experimental: C, 62.85; H, 4.79; N, 7.21; S, 8.32.
4’-((3,5-dimethyl phenyl)amino)-[1,1’-biphenyl]−4-sulfonamide (9c)
Blackish solid, MP = 129–133°C1. H NMR (600 MHz, DMSO-d6) δ 9.07 (s, 1 H), 8.08 (d, 2 H), 7.77–7.33 (m, 6 H), 6.89 (s,2 H), 6.64–6.50 (m, 3 H)0.2.32(s, 6 H)13. C-NMR (150 MHz, DMSO-d6) δ 153.1, 143.4, 139.6, 131.6, 129.4, 129.2, 125.2, 123.0, 122.7, 118.7, 117.7, 113.0, 22.5. Analytic calculations for C20H20N2O2S (352.45):C, 68.16; H, 5.72; N, 7.95; S, 9.10. Experimental: C, 68.22; H, 5.79; N, 7.86; S, 9.09.
4’-(pyridin-4-ylamino)-[1,1’-biphenyl]−4-sulfonamide (9d)
Brown solid, MP = 201–209°C1. HNMR (600 MHz, DMSO-d6) δ 10.06 (s, 1 H), 8.24–8.05 (m, 2 H), 7.80–7.61 (m, 2 H), 7.40 (ddd, 3 H), 7.07–6.84 (m, 2 H), 6.65–6.45 (m, 3 H), 5.88 (d, 2 H)13. C-NMR (150 MHz, DMSO-d6) δ 159.3, 152.7, 140.2, 133.9, 131.7, 128.6, 127.4, 120.9, 119.8, 112.5, 112.4. Analytic calculations for C17H15N3O2S (325.39): C, 62.75; H, 4.65; N, 12.91; S, 9.85. Experimental: C, 62.79; H, 4.72; N, 12.86; S, 9.78.
4’-((3-chloro-4-fluorophenyl)amino)-[1,1’-biphenyl]−4-sulfonamide (9e)
Lumps of blackish purple color, MP = 222–224°C1H NMR (600 MHz, DMSO-d6) δ 9.52 (s, 1 H), 8.14 (d, J = 7.9 Hz, 2 H), 7.34 (d, 2 H), 7.21 (t, J = 9.0 Hz, 2 H), 6.98–6.94 (m, 3 H), 6.93–6.90 (m, 4 H)13. C-NMR (150 MHz, DMSO-d6) δ 153.1, 150.3, 142.0, 139.4, 131.9, 129.1, 125.1, 122.9, 118.9, 117.8, 117.6, 117.0, 116.2, 113.0. Analytic calculations for C18H14ClFN2O2S (376.83): C, 57.37; H, 3.74; N, 7.43; S, 8.51. Experimental: C,57.59; H,3.77; N,7.31; S, 8.47.
Carbonic anhydrase Inhibition assay
A slightly modified previously designed protocol43 was used to carry out carbonic anhydrase inhibition activities of newly synthesized molecules (9a-e). The present method is based on hCA hydrolyzing the p-nitrophenyl acetate to p-nitrophenol, followed by spectrophotometrical determination. The blend consisted of 10 µL (0.5 mM) of sample compound dissolved in 1% DMSO, 60 µL (50 mM) of Tris-sulfate buffer solution with pH 7.6 having 0.1 mM ZnCl2, and carbonic anhydrase enzyme of about 10 µL. Blending and preincubation of added ingredients were carried out for 10 min at 25°C. The plate was read at 348 nm by a 96-well plate reader. Freshly prepared p-nitrophenyl acetate was used and 6 mM stock (using < 5% acetonitrile in buffer) was taken for the preparation. To get more clarity, 0.6 mM concentration was attained by the addition of a reaction mixture of about 20 µL. The total volume of the reaction mixture was kept at 100 µL. The incubation was carried out for 30 min at 37°C and followed by blending of all the ingredients and afterward noted a reading at 348 nm. Acetazolamide and 1% DMSO were used as positive and negative controls respectively. The results are calculated using the following formula and reported as percent inhibitions. The given results are the mean of three separate experiments(± SEM).
Inhibition (%) = 100 (Abs of test comp/Abs of control) ×100.
A few compounds were selected, which exhibited inhibition activities greater than 50% at 1.0 mM concentration. For the measurement of the IC50 value of selected compounds, further evaluation was done through dilution serial.
In Silico analysis
Ligand Preparation
Structures of all synthesized compounds were drawn in the widely used chemical structure drawing software [ChemDraw v20.1. https://disk.yandex.com/d/sY_jZQuwkLDLIg]. The geometry of these drawn structures was optimized by using the MMFF94 utility of Chem3D software and structures were saved in sdf format. Optimized structures of compounds were loaded in AutoDock tools and saved in pdbqt format for further molecular docking studies.
Protein Preparation and molecular Docking studies
X-rays crystallographic structures of the target enzymes were taken from PDB (Protein Data Bank) with PDB IDs 3V7X (CA II), 5FL4 (CA IX), and 4HT2 (CA XII). These X-ray crystallographic structures were inserted into Visualizer (Discovery Studio) and water molecules, unwanted ligands as well as hydrogen atoms were removed. Co-crystallised ligand of each target protein was chosen to define the active pockets and attributes of active pockets were analyzed. These co-crystallized inhibitors were also selected as reference ligands in molecular docking studies. These proteins were taken as pdb format for protein preparation and loaded in AutoDock47. Kollman charges were removed and these protein structures in pdbqt format. Grid dimensions were set manually as noted from Discovery Studio Visualizer for co-crystallized ligand. Further, docking studies were continued with AutoDock and binding interactions of each pose were analyzed in Visualizer (Discovery Studio)48.
Conclusion
A novel series of compounds (9a-e) was synthesized through Chan Lam cross-coupling reactions between aryl/heteroaryl boronic acids and commercially available 4’-amino-[1,1’-biphenyl]−4-sulfonamide in the presence of Cu(OAc)2 catalyst to afford excellent yields. The synthesized compounds were spectroscopically characterized and followed by the testing techniques to check the selectivity of synthesized compounds (9a-e) as compared to standard drug (acetazolamide) against three hCA isoforms. Overexpression of hCA isozymes (hCA-IX, hCA-XII) results in various cancerous diseases. Almost all the analogs were found to be active against hCA isoforms and this data was also confirmed by molecular docking studies. It was noticed that 9e exhibited selective hCA-II inhibition activity with IC50 = 0.38 µM. In addition, molecular docking studies confirmed the high binding tendency of 9e with active sites of hCA-II. Furthermore, 9d expressed remarkable inhibition activity against hCA-IX with IC50 = 0.21 µM. Additionally, 9b was also found to reduce the enzymatic activity of hCA-XII having IC50 = 0.69 µM. The lead compounds showed maximum binding affinities with the target proteins. Cell viability studies of all the newly prepared analogs have been performed as compared to the cisplatin drug. Most of the synthesized compounds were found to be non-toxic to normal cell lines. To conclude, the conducted study suggests that 9e, 9d, and 9b fulfilled the drug-ability criteria and may be considered lead structures to develop more potential hCA inhibitors to treat cancerous disorders.
Data availability
Data is provided within the manuscript or supplementary information files.
Change history
15 August 2025
The original online version of this Article was revised: The Acknowledgements, as well as the Funding section in the original version of this Article contained an error, where the grant number was erroneously given as “(RGP2/376/45)”. The correct grant number is “(RGP2/329/46)” for the Acknowledgements and for the Funding section, respectively. The original article has been corrected.
References
Supuran, C. T., Scozzafava, A. & Casini, A. Carbonic anhydrase inhibitors. Med. Res. Rev. 23, 146–189. https://doi.org/110.1002/med.10025 (2003).
Supuran, C. T. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nature reviews Drug discovery 7, 168–181. https://doi.org/110.1038/nrd2467 (2008).
Neri, D. & Supuran, C. T. Interfering with pH regulation in tumours as a therapeutic strategy. Nature reviews Drug discovery 10, 767–777. https://doi.org/710.1038/nrd3554 (2011).
Carta, F. & Supuran, C. T. Diuretics with carbonic anhydrase inhibitory action: a patent and literature review (2005–2013). Expert opinion on therapeutic patents 23, 681–691. https://doi.org/610.1517/13543776.13542013.13780598 (2013).
Kumar, A., Siwach, K., Supuran, C. T. & Sharma, P. K. A decade of tail-approach based design of selective as well as potent tumor associated carbonic anhydrase inhibitors. Bioorg. Chem. 126, 105920. https://doi.org/105910.101016/j.bioorg.102022.105920 (2022).
Supuran, T. C. Structure and function of carbonic anhydrases. Biochem. J. 473, 2023–2032. https://doi.org/2010.1042/BCJ20160115 (2016).
Zaib, S. et al. New aminobenzenesulfonamide–thiourea conjugates: synthesis and carbonic anhydrase Inhibition and Docking studies. Eur. J. Med. Chem. 78, 140–150. https://doi.org/110.1016/j.ejmech.2014.1003.1023 (2014).
Aggarwal, M. & McKenna, R. Update on carbonic anhydrase inhibitors: a patent review (2008–2011). Expert opinion on therapeutic patents 22, 903–915. https://doi.org/910.1517/13543776.13542012.13707646 (2012).
De Simone, G., Alterio, V. & Supuran, C. T. Exploiting the hydrophobic and hydrophilic binding sites for designing carbonic anhydrase inhibitors. Expert opinion on drug discovery 8, 793–810. https://doi.org/710.1517/17460441.17462013.17795145 (2013).
Supuran, C. T. Carbonic anhydrases as drug targets—general presentation. Drug Des. Zinc-Enzyme Inhibitors: Funct. Struct. Disease Applications, 15–38 (2009).
Supuran, C. T. & Scozzafava, A. Carbonic anhydrase inhibitors and their therapeutic potential. Expert Opinion on Therapeutic Patents 10, 575–600. https://doi.org/510.1517/13543776.13543710.13543775.13543575 (2000).
Supuran, C. T. Structure-based drug discovery of carbonic anhydrase inhibitors. Journal of enzyme inhibition and medicinal chemistry 27, 759–772. https://doi.org/710.3109/14756366.14752012.14672983 (2012).
Liao, S. et al. Expression of cell surface transmembrane carbonic anhydrase genes CA9 and CA12 in the human eye: overexpression of CA12 (CAXII) in glaucoma. J. Med. Genet. 40, 257–261. https://doi.org/210.1136/jmg.1140.1134.1257 (2003).
Saied, S. et al. Discovery of indolinone-bearing benzenesulfonamides as new dual carbonic anhydrase and VEGFR-2 inhibitors possessing anticancer and pro-apoptotic properties. Eur. J. Med. Chem. 259, 115707. https://doi.org/115710.111016/j.ejmech.112023.115707 (2023).
Pastorek, J. & Pastorekova, S. in Seminars in cancer biology. 52–64. https://doi.org/10.1016/j.semcancer.2014.1008.1002 (Elsevier).
SupuranC. T. Carbonic anhydrase inhibitors as emerging agents for the treatment and imaging of hypoxic tumors. Expert Opin. Investig. Drugs. 27, 963–970. https://doi.org/910.1080/13543784.13542018.11548608 (2018).
Supuran, T. C Experimental carbonic anhydrase inhibitors for the treatment of hypoxic tumors. J. Experimental Pharmacol. 603–617. https://doi.org/610.2147/JEP.S265620 (2020).
Supuran, C. T. Carbonic anhydrase inhibitors. Bioorganic & medicinal chemistry letters 20, 3467–3474. https://doi.org/3410.1016/j.bmcl.3405.3009 (2010). (2010).
Schulze Wischeler, J. et al. Bidentate Zinc Chelators for α-Carbonic Anhydrases that Produce a Trigonal Bipyramidal Coordination Geometry. ChemMedChem 5, 1609–1615. https://doi.org/1610.1002/cmdc.201000200 (2010).
Khan, K. M. et al. Evaluation of bisindole as potent β-glucuronidase inhibitors: synthesis and in Silico based studies. Bioorg. Med. Chem. Lett. 24, 1825–1829. https://doi.org/1810.1016/j.bmcl.2014.1802.1015 (2014).
Supuran, C. T. & Scozzafava, A. Carbonic anhydrase inhibitors. Curr. Med. Chemistry-Immunology Endocr. Metabolic Agents. 1, 61–97. https://doi.org/10.2174/1568013013359131 (2001).
Silver, L. H. & Group, T. B. P. T. S. Clinical efficacy and safety of brinzolamide (Azopt™), a new topical carbonic anhydrase inhibitor for primary open-angle glaucoma and ocular hypertension. American journal of ophthalmology 126, 400–408. https://doi.org/410.1016/S0002-9394(1098)00095-00096 (1998).
Pinard, M. A., Boone, C. D., Rife, B. D., Supuran, C. T. & McKenna, R. Structural study of interaction between Brinzolamide and dorzolamide Inhibition of human carbonic anhydrases. Bioorg. Med. Chem. 21, 7210–7215. https://doi.org/7210.1016/j.bmc.2013.7208.7033 (2013).
Nakano, T. et al. Effects of brinzolamide, a topical carbonic anhydrase inhibitor, on corneal endothelial cells. Advances in therapy 33, 1452–1459. https://doi.org/1410.1007/s12325-12016-10373-y (2016).
Iester, M. Brinzolamide. Expert Opin. Pharmacother. 9, 653–662. https://doi.org/610.1517/14656566.14656569.14656564.14656653 (2008).
Stams, T. et al. Structures of murine carbonic anhydrase IV and human carbonic anhydrase II complexed with brinzolamide: molecular basis of isozyme-drug discrimination. Protein Sci. 7, 556–563. https://doi.org/510.1002/pro.5560070303 (1998).
Yuan, L. et al. Carbonic anhydrase 1-mediated calcification is associated with atherosclerosis, and methazolamide alleviates its pathogenesis. Frontiers in Pharmacology 10, 766. https://doi.org/710.3389/fphar.00766 (2019). (2019).
Fossati, S. et al. The carbonic anhydrase inhibitor methazolamide prevents amyloid beta-induced mitochondrial dysfunction and caspase activation protecting neuronal and glial cells in vitro and in the mouse brain. Neurobiol. Dis. 86, 29–40. https://doi.org/10.1016/j.nbd.2015.1011.1006 (2016).
Teppema, L. J., Bijl, H., Gourabi, B. M. & Dahan, A. The carbonic anhydrase inhibitors methazolamide and acetazolamide have different effects on the hypoxic ventilatory response in the anaesthetized cat. The Journal of Physiology 574, 565–572. https://doi.org/510.1113/jphysiol.110528 (2006). (2006).
Supuran, C. T. Indisulam: an anticancer sulfonamide in clinical development. Expert opinion on investigational drugs 12, 283–287. https://doi.org/210.1517/13543784.13543712.13543782.13543283 (2003).
Monção, C. C. et al. Indisulam reduces viability and regulates apoptotic gene expression in pediatric high-grade glioma cells. Biomedicines 11, 68–83. https://doi.org/10.3390/biomedicines11010068 (2022).
de Oliveira, K. N. et al. Synthesis and antidepressant-like activity evaluation of sulphonamides and sulphonyl-hydrazones. Bioorganic & medicinal chemistry 19, 4295–4306. https://doi.org/4210.1016/j.bmc.4205.4056 (2011). (2011).
Kurt, B. Z. et al. Synthesis, antioxidant and carbonic anhydrase I and II inhibitory activities of novel sulphonamide-substituted coumarylthiazole derivatives. Journal of enzyme inhibition and medicinal chemistry 31, 991–998. https://doi.org/910.3109/14756366.14752015.11077823 (2016).
Onoabedje, E. A., Ibezim, A., Okoro, U. C. & Batra, S. New sulphonamide pyrolidine Carboxamide derivatives: synthesis, molecular docking, antiplasmodial and antioxidant activities. PLos One. 16, 1–19. https://doi.org/10.1371/journal.pone.0243305 (2021).
Güngör, S. A. et al. Biological evaluation and Docking study of Mono-and Di‐Sulfonamide derivatives as antioxidant agents and acetylcholinesterase inhibitors. Chem. Biodivers. 19, 1–9. https://doi.org/10.1002/cbdv.202200325 (2022).
Verma, S. K. et al. Antibacterial activities of sulfonyl or sulfonamide containing heterocyclic derivatives and its structure-activity relationships (SAR) studies: A critical review. Bioorg. Chem. 105, 1–14. https://doi.org/10.1016/j.bioorg.2020.104400 (2020).
Angeli, A. et al. Pyrazolo [4, 3-c] pyridine sulfonamides as carbonic anhydrase inhibitors: synthesis, biological and. Silico Stud. Pharmaceuticals. 15, 1–23. https://doi.org/10.3390/ph15030316 (2022).
Chen, J. Q., Li, J. H. & Dong, Z. B. A review on the latest progress of Chan-Lam coupling reaction. Advanced Synthesis & Catalysis 362, 3311–3331. https://doi.org/3310.1002/adsc.202000495 (2020).
Raghuvanshi, D. S., Gupta, A. K. & Singh, K. N. Nickel-mediated N-arylation with arylboronic acids: an avenue to Chan–Lam coupling. Org. Lett. 14, 4326–4329. https://doi.org/4310.1021/ol3021836 (2012).
Rizwan, K. et al. Copper-mediated N-arylation of methyl 2-aminothiophene-3-carboxylate with organoboron reagents. Tetrahedron letters 56, 6839–6842. https://doi.org/6810.1016/j.tetlet.6810.6080 (2015). (2015).
Farahat, A. A. & Boykin, D. W. Copper (I) 3-methylsalicylate mediates the Chan–Lam N-arylation of heterocycles. Synth. Commun. 45, 245–252. https://doi.org/210.1080/00397911.00392014.00961196 (2015).
Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 65, 55–63. https://doi.org/10.1016/0022-1759(1083)90303-90304 (1983).
Hussain, Z. et al. Synthesis and evaluation of amide and thiourea derivatives as carbonic anhydrase (CA) inhibitors. ACS Omega. 7, 47251–47264. https://doi.org/47210.41021/acsomega.47252c06513 (2022).
Daina, A., Michielin, O. & Zoete, V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717. https://doi.org/42710.41038/srep42717 (2017).
Wexler, R. P., Nuhant, P., Senter, T. J. & Gale-Day, Z. J. Electrochemically enabled chan–lam couplings of Aryl boronic acids and anilines. Org. Lett. 21, 4540–4543. https://doi.org/4510.1021/acs.orglett.4549b01434 (2019).
Battula, S., Subbareddy, G. & Chakravarthy, I. A mild and efficient copper-catalyzed N-arylation of unprotected sulfonimidamides using boronic acids. Tetrahedron Lett. 55, 517–520. https://doi.org/510.1016/j.tetlet.2013.1011.1084 (2014).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of computational chemistry 30, 2785–2791. https://doi.org/2710.1002/jcc.21256 (2009).
Biovia, D. S. Discovery studio visualizer. San Diego CA USA 936, 240–249 (2017).
Acknowledgements
The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through the large group Research Project under grant number (RGP2/329/46). The present data are the part of M. Phil thesis of Moniba Sharif. The authors strongly acknowledge the material synthesis lab of Government College University, Faisalabad.
Funding
The authors strongly acknowledge the material synthesis lab of Government College University, Faisalabad. The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through the large group Research Project under grant number (RGP2/329/46).
Author information
Authors and Affiliations
Contributions
MS, AM did analysis, experiments and Manuscript write-up, AK, MA, MUQ, TAS did visualization, review and editing, NR did Supervision, revised, AM did Software editing, TAS, AAMAH and MHAM, IIS provided resources and funding to the lab.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sharif, M., Mahmood, A., Kanwal, A. et al. Facile synthesis of aminobiphenyl sulfonamides via Chan–Lam coupling and their biological evaluation as potent carbonic anhydrase inhibitors. Sci Rep 15, 25661 (2025). https://doi.org/10.1038/s41598-025-10048-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10048-4