Abstract
N6-methyladenine (6 mA) has emerged as a potential epigenetic marker in eukaryotic genomes, yet its precise distribution patterns and biological functions in plant genomes are still not fully understood. In this study, we investigated the occurrence, global levels, and distribution patterns of 6 mA in four rye species: Secale cereale, S. strictum, S. sylvestre, and S. vavilovii. Using multiple complementary approaches, i.e., ELISA, ultra-performance liquid chromatography tandem mass-spectrometry (UPLC-MS/MS), immunofluorescence, and high-throughput sequencing (PacBio and MeDIP-seq), 6 mA was detected in all the examined species. The level of 6 mA in the genomic DNA of the rye species, depending on the species and organ, ranged from approximately 1 to 10 per million nucleotides. Immunofluorescence revealed specific chromosomal distribution patterns of 6 mA. The 6 mA signal in the rye chromosomes was dispersed along all chromosome arms, while no 6 mA signals were detected in the centromeric and telomeric regions, indicating that large blocks of constitutive subtelomeric and pericentromeric heterochromatin along with functional telomeric and centromeric regions do not contain 6 mA residues. To precisely map the genomic distribution of 6 mA, a machine learning approach combining PacBio methylation calls with MeDIP-seq data was developed, which enabled the identification of 6 mA-enriched regions across the genomes of all the four rye species. The findings provide comprehensive insights into the presence and distribution of 6 mA in the rye genomes, contributing to the growing understanding of this modification’s potential role in the plant epigenetic regulation.
Similar content being viewed by others
Introduction
It had long been believed that 5-methylcytosine (5mC) was the only modified nitrogenous base in the DNA that played a role in the epigenetic regulation of the eukaryotic genome. However, there is increasing evidence that there are other base modifications in the DNA with a potential regulatory role1. One of the modifications that is considered important as an epigenetic marker is N6-methyladenine (6 mA)2. 6 mA was detected in the DNA of both plants and animals. The presence of 6 mA was reported in C. elegans3, D. melanogaster4, Tetrahymena5, zebrafish and pig6, mouse7, human8, Arabidopsis thaliana9, soybean10, sea buckthorn11, woodland strawberry12, rice13, lotus14 and fungi15. Thus, 6 mA appears to be widespread in eukaryotic genomes. There exists a clear pattern regarding the relative abundance of 5mC and 6 mA in different organisms16: genomes rich in 5mC contain relatively small amounts of 6 mA (e.g., plants and mammals), whereas organisms with low 5mC levels typically show a higher 6 mA content (e.g., unicellular eukaryotes). Although the level of 6 mA in the plant and mammalian DNA is very low (usually ranging from about 0.0001–0.0003% of adenines) in comparison with some bacteria species (3%) or the dinoflagellate Peridinium triquetrum (10%), this does not preclude 6 mA’s role as an important genome regulator16. However, despite so many studies, it was indicated, that the 6 mA presence in plants and animals is too questionable, and it should be considered as an epigenetic regulator only in basal fungi, ciliates and green algae17. The doubts still exist despite abundant evidence for the specific distribution and function of 6 mA in plant and animal genomes, demonstrating its potential functions, and the identification of some putative methyltransferases and demethylases of 6 mA along with proteins recognizing 6 mA in the DNA18. The specific distribution of the methylated motifs and the potential effects of such modifications in plant genomes remain poorly understood, hence further studies in other organisms are needed19.
In plants 6 mA is more abundant in the organellar than in the nuclear DNA10. It is highly probable that in mitochondria and chloroplasts of plants the adenine methylation patterns are enzymatically maintained20. Various reports regarding the location and function of 6 mA in the nuclear genome of plants were published. Numerous studies confirmed that there exists a genome-wide distribution of 6 mA. It was indicated that 6 mA enriches pericentromeric heterochromatin but is also present within gene-rich regions. Yet, it is not unequivocal whether the higher levels exist in genic or intergenic regions8,9,13,21. Even though the 6 mA amount was very low, its presence within promoters and gene bodies was positively or negatively correlated with expression8,11,12,13. In Arabidopsis thaliana 6 mA is distributed mainly in pericentromeric regions and can also be found in gene bodies rather than intergenic regions9. Liang et al.9 reported that 6 mA is an epigenetic mark of actively expressed genes in Arabidopsis. Moreover, it was found that the 6 mA frequency positively correlates with the gene expression level and the transition from vegetative to reproductive growth. In Ficus carica 6 mA was present in protein-coding genes and transposable elements22. Xie et al.12 reported that 6 mA sites are broadly distributed across the Fragaria vesca genome, and the modification sites were located within intergenic regions. It was shown that the 6 mA modification influenced the gene transcription and was positively associated with the gene expression. Similarly to Arabidopsis, or woodland strawberry, 6 mA methylated sites were widely distributed in the sea buckthorn chromosomes11. In this species, the influence of 6 mA on the gene expression was evident, indicating that this modification should be considered as an epigenetic marker. This indication also comes from rice studies13, where 6 mA was found to enrich about 20% of genes. In promoters, 6 mA marked silent genes, but in gene bodies it correlated with the gene activity. Based on the different research, as can be seen, no clear regularities can be identified regarding the epigenetic role of 6 mA, hence further research is needed to help unravel this issue.
The methodology used to analyze the 6 mA presence and distribution plays a crucial role in obtaining reliable results. Given the low abundance of 6 mA in plant genomes, both bacterial contamination and RNA contamination can significantly impact results, as 6 mA occurs in the eukaryotic RNA. The selection of appropriate methods balancing sensitivity and specificity, along with stringent experimental controls, is particularly important. Technological limitations and antibody non-specificity may cause problems in the reliable quantitation of 6 mA. Various methods were employed to detect 6 mA in plant genomes, including dot blot, high-performance liquid chromatography combined with mass spectrometry, methylated DNA immunoprecipitation sequencing, methylated DNA with restriction enzyme digestion followed by sequencing, and third-generation high-throughput sequencing23. Recent advances in single-molecule sequencing technologies have significantly improved the accuracy of the 6 mA detection24.
This study provides insight into both the occurrence of 6 mA in rye and interspecific variation in the level and distribution of this modified nitrogenous base in the DNA. A combination of different methods was used to analyze the global level and distribution of 6 mA in the genomic DNA of four rye species: Secale cereale, Secale strictum, Secale sylvestre, and Secale vavilovii. Initially, to assess whether 6 mA is present in the rye genome, the DNA isolated from the coleoptiles, roots, and caryopses of four rye species using an ELISA method was tested. The results indicated the presence of 6 mA at low levels in the tested plant material, prompting a thorough analysis of global 6 mA levels using sensitive ultra-performance liquid chromatography tandem mass-spectrometry (UPLC-MS/MS). In addition to coleoptiles, roots, and caryopses, leaves and stems were also included in this analysis. To assess the chromosomal distribution of 6 mA in the four rye species, the immunofluorescence technique was employed. The PacBio sequencing of S. cereale genomic DNA was used to precisely determine genomic distribution patterns, utilizing the available high-quality reference genome for this species. Our goal was to integrate these PacBio-derived methylation calls with MeDIP-seq data by building a Python-based machine learning. A convolutional neural network (CNN) was prepared to predict methylation at different genomic positions. The CNN took in the local sequence context from the reference genome, PacBio 6 mA labels, and coverage profiles from the MeDIP-seq data. Once optimized, the trained model to MeDIP-seq data from four species: Secale cereale, Secale strictum, Secale sylvestre, and Secale vavilovii was applied.
Results
The global level of N6-methyladenine in the rye genomic DNA
The ELISA test clearly showed that 6 mA is present at detectable levels, in an amount of approximately 1–10 per million nucleotides on average, in the DNA isolated from coleoptiles, roots and caryopses. Slightly higher levels were detected in roots than coleoptiles, and in most of the species the lowest amount of 6 mA was found in caryopses (Supplementary Table S1). The lowest level was detected in S. sylvestre caryopses (1.68 ± 7.2E−15 6 mA/106 nucleotides) and the highest in S. cereale roots (10.71 ± 2.21 6 mA/106 nucleotides).
Since the preliminary ELISA test showed the presence of 6 mA in each species, accurate determination of the 6 mA level was carried out using sensitive UPLC–MS/MS. The global level of 6 mA in the genomic DNA of S. cereale, S. strictum, S. sylvestre and S. vavilovii was determined in coleoptiles, roots, caryopses, leaves and stems. The 6 mA levels varied among species and organs tested (Fig. 1; Supplementary Table S2). The greatest interspecific variation occurred in the in coleoptiles, roots and caryopses, slightly lower in leaves and stems. In coleoptiles the lowest 6 mA level was found in S. cereale i.e. 1.8 ± 0.02 6 mA/106 nucleotides, whereas in three remaining species it was higher, reaching the levels of approximately 5–7 6 mA per million nucleotides (Fig. 1). Even a greater variation occurred in the 6 mA levels in roots. As in the case of coleoptiles, the lowest amount of 6 mA was present in the genomic DNA of S. cereale roots, only 0.5 ± 0.15 6 mA/106 nucleotides. However, unlike coleoptiles, the amount of 6 mA varied greatly between the other three species (Fig. 1), ranging up to 14.75 ± 3.58 6 mA/106 nucleotides in S. sylvestre. Also, it should be noted that the values between the biological replicates were different (high standard deviation value). In caryopses, 6 mA levels were more even among species, but still, the S. cereale had the lowest level of all the species (2.48 ± 0.11 6 mA/106 nucleotides). In S. strictum and S. sylvestre the level was higher, 3.19 ± 0.37 and 3.72 ± 0.66 6 mA/106 nucleotides respectively, only in S. vavilovii exceeded the value of 4 6 mA per million nucleotides. Both in leaves and stems the levels were low, in almost all the samples below 1 6 mA per million nucleotides, only in S. strictum stems amounted to 1.28 ± 0.02 6 mA/106 nucleotides.
Global DNA N6-methyladenine (6 mA) levels in coleoptiles, roots, leaves, stems and caryopses of Secale cereale, Secale strictum, Secale sylvestre and Secale vavilovii obtained by the mass spectrometry analysis. The mean 6 mA amounts per million nucleotides (with standard deviations) are given in the table under the chart.
Summarizing the obtained results, it can be indicated that there is a visible variation between species and organs. Overall, the least 6 mA level was found in the genomic DNA of S. cereale compared to other species. Pearson coefficient was calculated to verify, whether the 6 mA levels in the analysed species correlate, and it turned out that the correlation is strong between S. sylvestre and S. strictum (R2 = 0.6479), S. sylvestre and S. vavilovii (R2 = 0.9031), S. strictum and S. vavilovii (R2 = 0.7058). The moderate degree of correlation was found between S. strictum and S. cereale (R2 = 0.4453). It can also be noticed that in all the species significantly less 6 mA was detected in the genomic DNA of stems and leaves as compared to roots, coleoptiles and caryopses (Fig. 1). In general, 6 mA levels in leaves and roots did not differ significantly (probabilty value p = 0.2719), and also 6 mA levels between coleoptiles and roots did not show significant differences (p = 0.2064). There were statistically significant differences between the remaining organs.
The distribution of N6-methyladenine in rye chromosomes
There are seven pairs of submetacentric chromosomes in the Secale karyotype. Chromosomes are identified on the basis of their length and both distribution and sizes of constitutive heterochromatin blocks (C or DAPI bands). The comparative analysis of 6 mA signal distribution in four rye species revealed that 6 mA is distributed along all chromosome arms. However, regions with higher or lower 6 mA signals density were clearly visible. In all the species there were no 6 mA signals in the centromeric and telomeric (including subtelomeric heterochromatin) regions (Figs. 2, 3 and 4B and D). Thus, the more subtelomeric heterochromatin was present in chromosomes, the smaller the range of chromosome arms was covered by the 6 mA signals. 1R, 2R, 3R and 7R chromosomes are to the greatest extent enriched in subtelomeric constitutive heterochromatin (Figs. 2 and 3). The remaining three pairs of rye chromosomes, i.e. 4R, 5R and 6R showed the occurrence of the 6 mA signal in a larger range, due to the lack of subtelomeric heterochromatin blocks on long arms (Figs. 2, 3 and 4E-F). In the rye species, there was also a lack of 6 mA in the area of intercalary heterochromatin bands (Fig. 4D-F). The thorough analysis of numerous metaphase plates showed an identical pattern of distribution of 6 mA IF signals along the chromosomes for each of a given species. Only in S. sylvestre intraspecific polymorphism was observed in the 6 mA distribution at the secondary constriction (Figs. 2, 3C and 4A).
N6-methyladenine (6 mA) distribution patterns in metaphase chromosomes of Secale species. The DNA is stained with DAPI (blue color); Green color (Alexa Fluor™ 488) marks the location of 6 mA in chromosomes. Images for DAPI and Alexa Fluor™ 488 are shown separately as well as merged. Scale bars represent 5 μm.
N6-methyladenine (6 mA) distribution patterns in chromosomes 1R and 6R of Secale species; (A) Chromosome 1R of S. vavilovii, the arrows show the secondary constriction; (B) Chromosome 1R of S. cereale, the arrows show the secondary constriction and the centromere; (C) Chromosome 1R of S. strictum, the arrows show the secondary constriction; (D) Chromosome 6R of S. sylvestre, the arrows show the centromere and the intercalary band on the long arm of the chromosome; (E) Chromosome 6R of S. vavilovii, the arrows show the intarcalary bands on the long arms; (F) Chromosome 6R of S. cereale, the arrows show the intercalary bands on the long arm. Scale bars represent 1 μm.
The comparison of N6-methyladenine genomic distribution in different rye species
Analysis of the genomic distribution of 6 mA (MeDIP sequencing and PacBio sequencing) was performed on the DNA extracted from rye coleoptiles. First, contamination in each dataset was assessed, and sequencing reads were taxonomically classified using Kraken25 with a custom database comprising all sequences of bacteria, archaea, viruses, protozoa, plasmids, and fungi in RefSeq, plus the human genome (GRCh38). Bracken26 was used to estimate the number of misclassified reads that could be reassigned to the target organism. Approximately 0.5–1.5% of reads per sample were identified as non-target contaminants and subsequently excluded from downstream analyses, ensuring high-quality data specifically representative of Secale genomes. The doubtless conclusion was that the methylation analysis applies only to the Secale genome. Approximately 82% of the PacBio reads uniquely aligned to the Secale cereale genome, indicating strong overall alignment performance despite the high proportion of repetitive DNA sequences (~ 90%). In the four Secale species analyzed (S. cereale, S. strictum, S. sylvestre, and S. vavilovii), protein-coding genes consistently displayed the highest density of 6 mA modifications, followed by the transposable element (TE) genes and, finally, pseudogenes. Among these species, S. vavilovii showed the greatest 6 mA density in protein-coding regions, reaching approximately 0.70%, whereas S. sylvestre, S. strictum, and S. cereale exhibited slightly lower densities of about 0.68%, 0.65%, and 0.61%, respectively (Fig. 5). A similar pattern was observed for TE genes, where 6 mA density ranged from around 0.40% in S. vavilovii to 0.35% in S. cereale. Only in S. cereale genome there were 3 sequences found, which may indicate the methylation of genes belonging to Ty1-copia retrotransposons. The pseudogenes had the lowest levels of methylation across all the species, with 6 mA densities spanning from 0.28% in S. vavilovii to 0.25% in S. cereale. These findings suggest that 6 mA marks are not uniformly distributed but are most concentrated in protein-coding regions across all the four species. The data also reveal a clear gradient in 6 mA density, with S. vavilovii consistently showing the highest levels in each gene category and S. cereale displaying the lowest. This pattern could imply species-specific regulatory roles of 6 mA modifications.
Distribution (density) of N6-methyladenine (6 mA) modification in Secale cereale, Secale strictum, Secale, sylvestre, Secale vavilovii genomes. The 6 mA densities indicate the proportion of adenines identified as methylated (6 mA) relative to the total number of adenines within each specific genomic category (protein-coding genes, TE genes, and pseudogenes).
No sequences were found for matching the pSc250, pSc119.2, or pSc200 repeat families, which are mainly located in subtelomeric heterochromatin regions. This confirms the results of immunofluorescence, which did not show the presence of 6 mA in the area of subtelomeric heterochromatin blocks.
The genomic distribution patterns of 6 mA modifications described here were obtained from high-confidence predictions generated by our integrated CNN-based approach. Specifically, the CNN model utilized PacBio methylation calls validated with orthogonal MeDIP-seq data as training inputs. This integrative approach enhanced accuracy and allowed consistent comparative analysis across the four rye species, ensuring robust and reliable determination of methylation densities and localization patterns within different genomic regions.
Our PacBio HiFi sequencing approach provided approximately 4× genome coverage, which is below the optimal threshold (~ 20×) recently recommended for highly accurate 6 mA detection27. Despite this limitation, our integration of PacBio data with MeDIP-seq substantially reduced potential artifacts. Indeed, we observed approximately 86% overlap between the PacBio-identified methylation sites and MeDIP-seq peaks, indicating a robust cross-validation between independent methodologies. To quantify the agreement between the two independent detection platforms, we intersected the final set of MeDIP-seq peaks with the PacBio HiFi methylation calls using Bedtools (minimum one-base overlap). Across all four Secale genomes this analysis identified high-confidence MeDIP peaks, coincided with at least one PacBio-validated 6 mA position. The converse comparison yielded a similar figure: PacBio 6 mA sites (86%) lay within a MeDIP peak. Taken together, these concordance values indicate that fewer than 14% of the MeDIP peaks lack PacBio support and therefore represent either genuine methylation events falling below the ~ 4× PacBio detection threshold or a small fraction of immunoprecipitation noise. These data provide a quantitative upper bound for potential false-negative calls attributable to limited long-read depth and underscore the robustness of the integrated map. Nevertheless, future studies employing deeper sequencing coverage or alternative technologies, such as nanopore sequencing, will further enhance the reliability and resolution of methylation mapping in rye genomes.
In the next step, the 4-bp sequences upstream and downstream of the 6 mA sites were extracted and searched for consensus motifs. The figure displays sequence that illustrates conserved motifs linked to methylated adenine sites, highlighting nucleotide preferences at various positions relative to the methylation site (Fig. 6). The height of each letter reflects the information content, indicating how conserved each nucleotide is at a specific position, while the size of each letter corresponds to its frequency. In all Secale genomes, a highly conserved “GAGG” sequence followed by a less conserved “CA” suggests a strong preference for guanine and adenine at the central positions. The prominent presence of adenine at the second position hints that it may serve as a key target for methylation or play a crucial role in how DNA-binding proteins recognize methylation sites. Meanwhile, the lower information content at flanking positions implies that the core motif is essential, but some variability in the surrounding bases is permissible. Furthermore, a prominent “A” is featured at the first and fourth positions, while a conserved “CGA” sequence is displayed in the center. The high information content of adenine at the initial position emphasizes its potential importance in the methylation recognition or binding affinity. The conservation of “CGA” suggests a structured recognition pattern, possibly critical for methylation-specific binding proteins or for influencing the local DNA structure. In contrast, the lower information content at the flanking regions indicates these positions can vary more without affecting methylation or binding efficiency.
Together, these data suggest that methylation is targeted to specific motifs enriched in conserved adenine and guanine residues rather than being randomly distributed. The presence of multiple conserved motifs points to a functional diversity in the methylation patterns, potentially linked to roles in transcriptional regulation, chromatin accessibility, or interactions with DNA-binding proteins. The observed conservation patterns offer insights into how methylation sites are selectively recognized, suggesting that some motifs may act as preferred targets for methylation or as binding sites for proteins that are sensitive to methylation.
Across the four Secale genomes analyzed, the majority of 6 mA sites consistently cluster within gene bodies, with intergenic and promoter regions accounting for the remainder (Fig. 7A). In S. cereale, 55% of 6 mA sites occur in gene bodies, whereas 25% map to intergenic regions and 20% to promoters. S. strictum displays a similar distribution, with 52% in gene bodies, 27% in intergenic segments, and 21% in promoters. S. sylvestre shows a slightly higher proportion of 6 mA in intergenic regions (32%) and a corresponding decrease in the gene-body localization (50%), while promoters harbor 18% of the total sites. S. vavilovii largely parallels S. strictum, with 52% of sites occurring in the gene bodies, 30% in intergenic regions, and 18% in promoters. Taken together, these findings indicate that the enrichment of 6 mA in Secale generally favors gene bodies as the primary site of methylation, although modest differences in the proportion of intergenic versus promoter‐associated 6 mA sites distinguish the four species.
Distribution of N6-methyladenine (6 mA) in genomic features of Secale genomes. (A) Number of 6 mA modification sites distributed into gene bodies, promoters and intergenic regions and (B) within gene bodies divided into exons (excluding 5´UTR and 3´ UTR in protein-coding genes), introns, and 5´ and 3´ UTRs.
Within the gene-body compartment, introns consistently harbor the largest share of 6 mA sites, ranging from 51% in S. cereale to 56% in S. sylvestre, with the remaining gene‐body methylation mostly localized to exons and only small fractions in the 5′ and 3′ untranslated regions (Fig. 7B). In S. cereale, 51% of intragenic 6 mA occurs in introns, 39% in exons, and 6% and 4% in the 5′ and 3′ UTRs, respectively. S. strictum follows a comparable distribution (55% introns, 36% exons, 4% 5′UTR, 5% 3′UTR), whereas S. sylvestre has slightly more 6 mA in introns (56%) and correspondingly fewer exonic sites (38%). S. vavilovii is again very similar to S. strictum, with 55% of intragenic 6 mA located in introns, 38% in exons, 3% in the 5′UTR, and 4% in the 3′UTR. Overall, these results highlight a broadly conserved pattern in which gene bodies constitute the principal 6 mA reservoir, primarily within introns, yet modest differences among the four species indicate potential variation in how 6 mA is distributed between exonic versus regulatory regions of the gene body.
In all four Secale genomes examined (S. cereale, S. strictum, S. sylvestre, and S. vavilovii), the 6 mA methylation signal around the transcription start site (TSS) from − 2 kb to + 2 kb exhibits a pronounced trough precisely at the TSS and strong enrichment in the immediately flanking upstream and downstream regions (Fig. 8). For 6 mA-methylated genes, the log₂(IP_RPKM/input_RPKM) ratio typically rises to about 0.40–0.45 at ∼1 kb downstream of the TSS in each species, compared with approximately − 0.50 at the TSS itself. This local depletion at the TSS creates a clear discontinuity between the upstream (− 2 kb) and downstream (+ 2 kb) segments, which is seen both in protein‐coding and transposable element genes. In contrast, genes that were not identified as 6 mA‐methylated show only modest enrichment values (close to 0.0) in the flanking regions and dip to about − 0.30 to − 0.40 at the TSS, indicating that the dramatic trough is largely driven by 6 mA‐methylated loci.
Although the overall shape of the methylation profiles remains consistent across the four Secale species, subtle differences in the amplitude of enrichment can be observed. S. cereale and S. strictum each exhibit a steep drop from ∼0.40–0.45 in the upstream region to around − 0.50 at the TSS, whereas S. sylvestre and S. vavilovii show a similar but slightly less pronounced depletion (∼0.45). In all cases, the 6 mA-methylated genes display a regular periodicity extending up to 2 kb on both sides of the TSS, suggesting a conserved pattern of methylation organization across these genomes. Heatmaps of relative methylation levels (ranging from − 2.0 to + 2.0) reinforce the meta‐analysis, showing that the local 6 mA depletion at the TSS is consistently present in all the samples and is surrounded by periodic peaks of higher methylation density. When analyzed together, these results indicate that the discontinuous 6 mA distribution near the promoter region is a hallmark of Secale genomes, reflecting both the pronounced dip at the TSS and the characteristic oscillations of 6 mA in the proximal gene body and upstream regulatory regions.
The provided Fig. 9 illustrates the results of a GO enrichment analysis based on the methylation data (Fig. 9A), categorized into a molecular function (MF), biological process (BP), and cellular component (CC). The analysis revealed a significant enrichment of GO terms (Fig. 9B) related to molecular functions critical to nucleic acid interactions. Terms associated with signal receptor binding, long-chain fatty acid transporter activity, nucleotide transmembrane transporter, and catalytic activities, such as ABC-type transporter activity, showed low false discovery rate (FDR) values, indicating major statistical significance. For example, the prominent bubbles with large sizes for terms such as identical protein binding suggest that a substantial number of genes involved in fundamental protein interactions are subject to methylation. This result highlights the role of methylation in modulating the gene expression at the molecular level.
Gene ontology (GO) enrichment analysis. (A) Volcano plot for all 6 mA-methylated protein-coding genes. (B) Molecular function, (C) Biological process, (D) Cellular component GO enrichment categories of high-methylation-density protein-coding genes in Secale genomes. All protein-coding genes were classified by the parameter K = log2(FC) into genes with high 6 mA density (K > 1), and low 6 mA density (K < − 1); FC is the fold change between particular 6 mA density of a gene and the average value. The methylated protein-coding genes were subjected to GO analysis, and the results were used for GO mapping and classification. The GO categories are listed with the false discovery rate-adjusted p-value < 0.01.
In the biological process category (Fig. 9C), methylation was strongly associated with GO terms such as peroxisome organization, peroxisomal membrane transporter, and protein import into peroxisome matrix. These terms consistently exhibited low FDR values, suggesting that methylation significantly impacts subcellular organelle dynamics and metabolic pathways. Additionally, processes such as nitrogen compound transport, fatty acid beta-oxidation, and fatty acid metabolic process appeared among the enriched terms, indicating that methylation plays a crucial role in the regulation of peroxisomal function and broader metabolic adaptation. The larger bubble sizes for these terms reflect the involvement of a substantial number of methylated genes, further supporting their biological relevance. The notable enrichment of processes like protein import into peroxisome matrix, docking and regulation of peroxisome size underscores the potential impact of methylation on organellar biogenesis and maintenance, while nucleotide transport highlights the role of nucleic acid transactions in this regulatory network.
The cellular component category (Fig. 9D) highlighted enrichment in terms such as peroxisomal membrane, organelle membrane, and membrane, underscoring the localization of methylation marks in membrane-associated compartments. The significant FDR values for terms like cytoplasm and intracellular membrane-bounded organelle point to the broad distribution of methylation targets within the cell. Additionally, the association with peroxisomal importer complex and integral component of peroxisomal membrane suggests specific roles for methylation in facilitating transport and assembly processes in peroxisomes. The presence of glyoxysome and integral component of membrane and cytosol among the enriched terms indicates potential roles in specialized metabolic pathways and membrane-related regulatory mechanisms.
Across all three categories, the lowest FDR values (e.g., < 1e-9 in the MF and BP categories) were observed for terms associated with core functions such as transmembrane transport, peroxisome organization, and membrane localization. These findings indicate that methylation strongly targets regions critical for maintaining cellular integrity and regulating gene expression in response to metabolic demands. The smaller bubble sizes for certain terms, despite low FDR values, suggest that some processes may involve fewer genes but still exhibit high specificity for methylation. The consistent enrichment of terms across MF, BP, and CC categories highlights methylation as a key regulatory mechanism with broad impacts on cellular function. The variation in bubble size and FDR values suggests differential roles for methylation across various biological pathways and cellular locations. Collectively, these results underscore the importance of methylation in regulating metabolic functions, membrane dynamics, and peroxisomal processes, providing insights into its role in cellular adaptation, organelle maintenance, and gene silencing mechanisms. Further experimental validation is warranted to investigate specific genes and pathways identified in this analysis.
Discussion
In this study, the level and distribution patterns of 6 mA in the genome of four rye species were determined. To date, there are few reports on 6 mA occurrence, location, or potential function in the plant genome regulation. Additionally, due to the very small amounts of 6 mA in the plant genome, there are opinions indicating that bacterial contamination, RNA contamination, technological limitations, and antibody non-specificity may distort research results and thus, the evidence hitherto published is not sufficient to support the presence of 6 mA in the plant or animal nuclear DNA17,28,29, indicating that further studies are required. If certain repeatable patterns of 6 mA distribution are demonstrated, this cannot be a matter of chance. Hence, we did not focus on a single species, but made a preliminary comparative analysis on four, relatively genetically similar species belonging to one genus. Although the matter of Secale classification is still under debate, according to Germplasm Resource Information Network (GRIN), there are four species recognized in the Secale genus: S. cereale, S. strictum, S. vavilovii, S. sylvestre. Thus, all the rye species were included in this study. Phylogenetic analyses and studies of genetic diversity within the genus Secale indicate that S. cereale is most closely related to S. vavilovii, which is considered its putative ancestor30. During rye evolution, S. sylvestre is believed to have diverged first, preceding the separation of S. strictum, S. vavilovii, and S. cereale31. Genotyping-by-sequencing analysis revealed that S. cereale shares 95.2% of polymorphic sites with S. vavilovii, 65.8% with S. strictum, and 51.1% with S. sylvestre31. Similarly, DArT marker analyses clustered S. cereale and S. vavilovii together, S. strictum formed a separate cluster, while S. sylvestre appeared the most divergent32,33. This pattern was also supported by SSR (Simple Sequence Repeats) analyses and variation in nuclear EST (Expressed Sequence Tags) sequences, which confirmed low genetic differentiation between S. cereale and S. vavilovii, and higher divergence of S. sylvestre34. Despite these general patterns, regional genomic comparisons reveal more complex relationships. For example, a comparative analysis of chromosomes 1R and 4R in the S. cereale inbred line ‘Lo7’ revealed unexpected similarity with S. sylvestre in these regions35, highlighting that localized genomic divergence may not always align with whole-genome phylogenetic relationships. Crucially, the Secale genome comprises up to 90% repetitive sequences36,37. Their amplification, together with transposable element activity, have significantly shaped genome evolution38, hence, the genomes of Secale species are differentiated mainly in terms of the number and distribution of these types of sequences. Given that 6 mA is primarily enriched in regions associated with coding sequences, the global amount and distribution patterns of 6 mA may not directly correspond to overall genome divergence or phylogenetic distance. Instead, the observed similarity in 6 mA profiles among species may reflect a comparable number and organization of gene-rich regions, which are potentially subject to regulation by 6 mA.
Measurable levels of 6 mA in each rye species were detected. Comparing the level of 6 mA in the root, coleoptile, and caryopsis with the level in leaf and stem, one can see a lower level of the latter two as compared to the others, even though in this scale of quantity it does not constitute an enormous difference. It can be indicated that despite the greatest care in removing microbial contamination, even a minimal contamination quantity can affect the result. This was confirmed by the sequencing results, where sequences from microbiome (genomes of bacteria, archaea, and fungi) were found in the reads. In the LC-MS/MS studies of 6 mA levels in rice and Arabidopsis, bacterial contamination was excluded9,39. However, when analyzing the results conducted in Arabidopsis, a higher level of 6 mA is clearly visible in the root than, for example, in the stem, similar to rye. Since the presence of bacterial contamination in both rice and Arabidopsis was verified using PCR-based approaches using universal 16 S primers, it is possible that the samples contain the DNA of microbes, the presence of which cannot be confirmed using these primers. In this work, the presence of bacterial contamination in coleoptiles was confirmed, and the conclusion was that it is especially true for roots. Surface sterilization protocols primarily remove or reduce epiphytic microbes on the outer plant surface but do not fully remove endophytes or rhizosphere-associated microbes (those intimately associated with the root surface and apoplast). As a result, the rhizosphere harbors a diverse and abundant microbial community compared to aboveground plant parts (leaves, stems). Root exudates (carbohydrates, amino acids, secondary metabolites) attract and support a dense microbial population. It, therefore, seems that in all species and organs, this modification’s level may be similar and slightly above or below 0.0001% of DNA nucleotides, i.e. about 0.0004% of adenines. In the genome of 9-day-old Arabidopsis seedlings, the 6 mA level was 0.048% 6 mA/A9. However, there was quite a large variation between individual organs, with the highest amount detected in roots (0.138% 6 mA/A) and the lowest in stems (0.006% 6 mA/A). A very similar amount, approximately 0.04% of 6 mA, was found in the genome of wild and cultivated soybeans10. Also, it should be noted, that 6 mA densities in nuclear genome between wild and cultivated soybeans were similar, as observed at interspecific level in rye. Higher levels of 6 mA were detected in the rice genome, where levels were determined in 3-week-old seedlings. The content ranged from 0.15 to 0.55% and was similar to that observed in wheat, maize, sorghum, Setaria italica, and Medicago39. In the Herrania umbratica genome, the level of this modification was determined to be 0.048% 6 mA/A. Also, a similar amount of 6 mA was found in sea buckthorn, where the level in whole seedlings was 0.016% 6 mA/A, in early fruits 0.154% 6 mA/A, and in leaves 0.008% 6 mA/A11. The 6 mA content represented approximately 0.139% of the total adenines in the genomic DNA of Fragaria vesca12. Therefore, comparing the amount of 6 mA in the rye genome with the amount in the genomes of other plants, it is visible that it is lower. However, such a small amount does not exclude 6 mA as a potential epigenetic regulator. Additionally, it may indicate its presence in unique rather than repetitive sequences. It should be noted that species of the Secale genus have a vast genome of size ~ 7–8 Gb, and a high content of repetitive sequences of 85-90%37. Hence, if 6 mA occurs mainly in gene-rich areas, the amount of 6 mA in rye should be lower compared to species that have less repetitive sequences. Indeed, only 25 − 32% (depending on rye species) of 6 mA occurs in intergenic regions. In Oryza, slightly more, 32 − 42%39. Since 6 mA occurs mainly in genes, a much lower percentage of this modification in rye is expected as compared to rice, due to the fact that rice has a much smaller genome (~ 7–8 Gbp vs. ~380–430 Mb). However, there remains the issue of the methylation of transposable element genes, especially retrotransposons, which enrich the rye genome. In the rice genome 6 mA is found mainly in protein-coding genes, however, it is also present in TE genes39. In Arabidopsis, protein-coding genes and transposable elements genes were the two largest groups of 6 mA-methylated genes9. These results were also supported by 6 mA methylome study on the lotus genome, in which 6 mA enriched mostly genic regions, but also, in smaller quantities in transposable elements14. Also, the 6 mA distribution density in all the gene regions (0.049%) was higher than that in the intergenic region (0.025%) of Herrania umbratica genome40. Such a pattern of 6 mA distribution would be consistent with the presented distribution in rye chromosomes, along the course of which genes and mobile elements occur41. The first attempts using indirect immunofluorescence gave a positive result, but the signal was so weak that it was difficult to record images, as fluorescence faded quickly. Therefore, signal amplification was used, which allowed the visualization of the areas of chromosomes in which 6 mA occurs. In all the species, the distribution pattern is similar and, above all, the lack of 6 mA can be seen in most of the constitutive heterochromatin regions. Since rye chromosomes are large and contain large blocks of constitutive heterochromatin composed of tandemly repeated sequences, this is clearly visible. As in rye, in Arabidopsis or rice 6 mA occurs along all chromosomes, but unlike in rye it enriches the pericentomeric heterochromatin regions9,39. This may be related to the content of different types of sequences, e.g. TEs in which 6 mA occurs. In rye chromosomes, the density of transposable elements (TEs) is relatively low in distal chromosomal regions. Among the two most abundant retrotransposon families, gypsy-like elements are predominantly localized in pericentromeric regions, with their abundance decreasing toward the telomeres on chromosomes 1R–7R. In contrast, copia-like retrotransposons show an opposite distribution, being more abundant along chromosome arms37. A similar chromosomal distribution pattern has been observed for protein-coding genes, which are least abundant in centromeric regions and become progressively more frequent toward the distal ends of chromosome arms35,37. The observed immunolocalization pattern of 6 mA, with signals detected along chromosome arms and excluded from centromeric and telomeric regions, corresponds well with the distribution of genes and copia-like elements. Pseudogenes are diffusely distributed, and have been reported even within blocks of tandemly repeated sequences42. However, their low abundance in centromeric and subtelomeric heterochromatin, combined with the overall low levels of 6 mA, may explain the absence of detectable signal in these regions.
6 mA chromosomal pattern of distribution is very consistent with the location of 5mC enriched regions in rye chromosomes43, although the quantity of these modifications is incomparable. There are definitely more TEs along the length of the arms than protein-coding genes, and almost all transposable elements are fully methylated at 5mC in plants44. In contrast, as mentioned above, in rice genome only 14% of TE contained 6 mA, among which about 20–25% of retroelements were marked by 6 mA13. Yet, crosstalk among 5mC and 6 mA in coding genes cannot be excluded, and some research support this indication11.
The distribution of 6 mA modifications observed in the Secale genomes, mostly concentrated in protein-coding genes, followed by TEs and pseudogenes, as mentioned above, aligns closely with the previous studies on 6 mA patterns in other plant species9,39. In our analysis, protein-coding genes in Secale consistently exhibited the highest 6 mA density, peaking at about 0.70% in S. vavilovii and gradually decreasing through S. sylvestre (0.68%), S. strictum (0.65%), and S. cereale (0.61%). This pattern is similar to findings by Liang et al.9, who reported that protein-coding genes made up the majority of 6 mA-marked genes in the Arabidopsis thaliana Col genome, outnumbering transposable elements and other categories. Although the exact frequencies differ, the overall trend of protein-coding genes showing the most extensive 6 mA modifications remains consistent. The enrichment of 6 mA within gene bodies also reflects the results of Zhang et al.39, who found that most 6 mA-modified genes in rice were protein-coding and that 6 mA patterns were largely conserved across different rice cultivars. Interestingly, Zhang et al.39 also documented variation in 6 mA levels among related accessions, which parallels our findings at interspecific level in Secale, with S. vavilovii exhibiting the highest 6 mA density and S. cereale the lowest. This suggests that lineage-specific epigenetic regulation could explain the differences in 6 mA density observed among the Secale species. Additionally, the scarcity of 6 mA in pseudogenes observed here aligns with Liang et al.9, who reported that pseudogenes rarely contained 6 mA sites compared to coding sequences.
Although TEs displayed lower 6 mA densities than protein-coding genes, the detection of three Ty1-copia-derived sequences with 6 mA in S. cereale indicates that retrotransposons can acquire this modification in Secale. This finding is consistent with Liang et al.9, who observed substantial 6 mA enrichment in TEs, especially those located in heterochromatic regions. Similarly, Zhang et al.39 identified distinct 6 mA patterns near the start sites of various TE families in rice, suggesting that 6 mA might play a role in regulating transposon activity or silencing.
Since it turned out that most 6 mA residues occur in coding areas, to infer putative biological functions of 6 mA its density in promoters and gene bodies was analyzed. As the analysis showed, 6 mA occurs in the promoter regions, but a much larger amount of this modification occurs in the gene body. A precise analysis of the distribution within the gene body showed that more than 50% of the 6 mA contained in the gene body occurs in introns and almost 40% in exons. An almost identical distribution of 6 mA occurs in rice genes39. Oryza and Secale belong to the Poaceae family, explaining such a high similarity. Additionally, the same distribution patterns in species belonging to the same genus may support the potential of this modification as an epigenetic regulator. A slightly different pattern of distribution was observed in the Arabidopsis genome, where 6 mA was more abundant in intergenic regions than in gene bodies. Moreover, within gene bodies 6 mA occurred mainly in exons (70–80%), and much less was to be found in introns, 14–20%9. An even more diverged pattern of 6 mA distribution was found in soybean, where in the nuclear DNA only about 10% of 6 mA occurred in gene bodies, with the remainder in intergenic regions10. Also, intergenic regions were mostly enriched in 6 mA in Fragaria vesca genome12. Despite the different amounts, patterns and densities in individual regions of the different plant genomes, there is evidence of an influence of 6 mA on the gene expression in plants13,45.
The presence of multiple conserved 6 mA motifs in Secale genomes, including the characteristic GAGG sequence and its surrounding bases, closely resembles patterns observed in other species. In this study, a consensus motif, which is highly enriched in guanine and adenine residues was identified. One variant of this motif contains a central CGA sequence, with a strong preference for adenine at the initial position. This arrangement suggests a structured sequence context that could be critical for protein binding or for the recognition of methylation sites. Interestingly, the GAGG motif detected in Secale parallels similar sequences reported in C. elegans3 and later in rice39, implying that such motifs might be conserved across different taxa. Moreover, the observed preference for adenine residues in the flanking regions aligns with the findings of Zhang et al.39, who described motifs in Nip and 93 − 11 cultivars that typically retain two conserved adenines at both ends with a cytosine in the middle. This pattern is also consistent with motifs reported in Arabidopsis thaliana9, suggesting a common regulatory framework for 6 mA sites across plant species.
Additionally, our data reveal motifs containing A and G at key positions, which resemble the AGGCAGCG sequence reported by Hao et al.46. This recurring motif pattern suggests that adenine and guanine-rich sequences may act as structural signatures recognized by the methylation machinery. The conserved nature of these motifs implies that they might play a universal role in providing high-affinity binding sites for proteins associated with methylation or in influencing the local chromatin structure.
Interestingly, the partial variability observed in the flanking nucleotides indicates that while the core motifs are conserved, they can tolerate some sequence flexibility without losing their ability to direct 6 mA placement. This suggests a dual strategy where a conserved set of core sequences ensures reliable methylation targeting, while species-specific variations allow for the adaptable and nuanced gene regulation.
To determine whether 6 mA can affect the gene expression in rye, the distribution of 6 mA was determined within 2 kb around the transcription start site. The distinct trough of 6 mA observed at the TSS, followed by an enrichment of 6 mA immediately downstream, was evident in all the four Secale genomes. This pattern aligns closely with the previous studies suggesting that 6 mA is strategically localized in actively transcribed regions39. In this analysis, 6 mA-methylated genes showed a sharp decrease in methylation at the TSS (∼−0.50 in log₂[IP_RPKM/input_RPKM]), which transitioned into a pronounced enrichment about 1 kb downstream. This organized distribution hints at a regulatory role for 6 mA, possibly influencing how genes are turned on and off. The difference in methylation patterns between 6 mA-modified and unmodified genes suggests that the TSS acts as a critical control point for where 6 mA accumulates. This observation is consistent with Liang et al.9, who reported a similar depletion of 6 mA at or near the TSS, especially in transposable elements, followed by a rapid increase immediately downstream.
These oscillatory methylation patterns might be linked to active transcription. Zhang et al.39 found higher levels of 6 mA in more actively expressed genes, particularly when 6 mA was present in promoter and gene body regions. This suggests that the dip in methylation at the TSS might help regulate gene activation, while the flanking peaks could enhance transcription by affecting chromatin accessibility or by attracting regulatory proteins. Although Liang et al.9 reported less pronounced patterns for protein-coding genes in their study, our findings suggest that in Secale, both protein-coding and transposable element genes follow a similar pattern of 6 mA depletion at the TSS, surrounded by elevated methylation levels. This difference might reflect distinct evolutionary pressures on epigenetic regulation or differences in experimental methods and sequencing depth. Interestingly, S. cereale and S. strictum displayed a steeper drop in 6 mA at the TSS compared to S. sylvestre and S. vavilovii, indicating subtle differences in methylation dynamics among these closely related species. However, all four species shared a consistent pattern of 6 mA distribution, extending up to 2 kb upstream and downstream of the TSS. This regular spacing suggests a conserved epigenetic framework in Secale, which could help coordinate the gene expression efficiently.
The periodic peaks of 6 mA around the TSS may support transcriptional regulation by creating a structured chromatin environment, similar to the role of 5mC39. Overall, the findings are consistent with previous research9,39, emphasizing the importance of TSS-centered 6 mA organization while also highlighting species-specific variations. These variations might reflect the evolutionary flexibility of 6 mA methylation in Secale.
GO enrichment analysis connecting 6 mA methylation to signaling receptor binding, peroxisome organization, and nucleotide transport in Secale is consistent with findings in other plant species. For example, in Arabidopsis thaliana, Liang et al.9 found that 6 mA-methylated genes were significantly enriched in categories related to protein binding. Similarly, Zhang et al.39 reported that genes with high levels of 6 mA in rice were associated with various molecular functions, particularly binding activities. These parallels suggest that 6 mA-modified genes across different plant genomes might share core roles in transmembrane transport and organelle organization.
In the present analysis, the enrichment of GO terms such as signaling receptor binding, identical protein binding, and ABC-type transporter activity supports the idea that 6 mA serves as an epigenetic marker influencing molecular interactions39. Additionally, the low FDR values observed for processes like peroxisome organization and protein import into peroxisome matrix suggest that 6 mA might significantly impact organellar dynamics and transcriptional control. Interestingly, the association of 6 mA with pathways related to fatty acid beta-oxidation and fatty acid metabolic process in Secale mirrors findings in Paspalum vaginatum reported by Hao et al.46. In that study, genes with high and moderate levels of 6 mA were linked to biosynthesis, metabolism, and stress response pathways. This similarity suggests that the enrichment of 6 mA in genes related to fatty acid metabolic process in Secale might play a role in adaptation to stress, similar to the patterns seen in P. vaginatum.
The peroxisomal localization of many enriched GO terms, such as those linked to the peroxisomal membrane and intracellular membrane-bounded organelle, further underscores the role of 6 mA in gene regulation within these compartments. This is consistent with evidence that 6 mA can influence transmembrane transporter activity. At the same time, the association of 6 mA with the cytosol suggests that it might also play a role in additional metabolic processes.
To sum up, the amount and pattern of 6 mA genomic distribution is relatively similar in S. cereale, S. strictum, S. sylvestre and S. vavilovii. The global level of 6 mA is low, about slightly above or below 0.0001% of DNA nucleotides. However, it is dispersed in all chromosome arms, but not in blocks of constitutive heterochromatin. The presence of 6 mA mostly in gene bodies and promoters was confirmed by 6 mA methylated DNA-immunoprecipitation-sequencing and SMRT-PacBio-seq: single molecule-PacBio sequencing technology. The concentration of methylated adenine surrounding the TSS genes indicates its potential role in the gene expression regulation. To assess the potential role of 6 mA as an epigenetic mark, its transgenerational inheritance must be evaluated. In Caenorhabditis elegans, heritable 6 mA patterns have been observed47,48. However, no direct evidence currently supports the inheritance of 6 mA in plants. A major challenge is the low abundance of this modification in plant genomes, which complicates its detection and quantitative analysis at a scale suitable for inheritance studies. In plants, epigenetic memory is primarily associated with cytosine methylation, histone modifications, and RNA-mediated mechanisms49. Whether 6 mA also contributes to this epigenetic regulatory network remains unclear and will require further investigation.
The analysis presented here is a preliminary examination of the presence and location of 6 mA in rye genome and strongly indicates the need for further research. The obtained results indicate the need for a more in-depth analysis, planned to correlate the distribution patterns of 6 mA, but also 5mC and 5-hydroxymethylcytosine with the gene expression. The research shall provide much broader insight into the epigenetic potential of these modifications and their crosstalk in rye, shedding some light on the rye epigenome.
Materials and methods
Plant material
The study encompassed four rye species: Secale cereale L. cv. Imperial, Secale strictum C.Presl (accession no. 789/95), Secale sylvestre Host (accession no. 839/96), and Secale vavilovii Grossh (accession no. K1773). Caryopses were sourced from the Botanical Garden of the Polish Academy of Science (Warsaw, Poland) and the National Plant Germplasm Inspection Station (Laurel, MD, USA).
Methods
DNA isolation
Caryopses germinated under laboratory conditions at room temperature. Coleoptiles were harvested from seven-day-old seedlings grown in darkness at room temperature. For root, stem, and leaf collection, three-day-old seedlings were transferred to soil-filled pots. Further cultivation was conducted in the experimental field of the West Pomeranian University of Technology in Szczecin.
Prior to DNA isolation, all plant organs underwent stringent surface sterilization to minimize microbial contamination. The sterilization protocol consisted of sequential treatments: 1 min immersion in Incidin™ Liquid (Ecolab, Germany), followed by 1 min treatment with 70% (v/v) ethanol, and 5-min incubation in 0.1% (v/v) sodium hypochlorite solution supplemented with Tween-20. All the samples were thoroughly rinsed with deionized water after each step.
For ELISA and mass spectrometry analyses, genomic DNA was extracted using the Plant & Fungi DNA Purification Kit (Eurx®, Poland). Tissue homogenization was performed using a FastPrep™ homogenizer (MP™ Biomedical, USA) with FastPrep™ Lysing Matrix A (MP™ Biomedical, USA), following the manufacturer’s protocol. The DNA was eluted with sterile, DNase-free water (Sigma-Aldrich, Germany). Additional RNase treatment was performed in the post-isolation stage: the samples were incubated with RNase A (1 µl of 1 mg/ml; Sigma-Aldrich, Germany) at 37 °C for 1 h, which was followed by purification using the GeneMATRIX PCR/DNA Clean-Up Purification Kit (Eurx®, Poland).
For 6 mA-enriched fragment immunoprecipitation and PacBio sequencing, the genomic DNA was isolated using the CTAB method. Fresh tissue (2 g of coleoptiles) was ground to a fine powder in liquid nitrogen using a pre-chilled mortar and pestle. The powder was transferred to the extraction buffer (2× CTAB: 100 mM Tris pH 8.0, 1.4 M NaCl, 20 mM EDTA, 2% w/v CTAB, 0.1% w/v PVP, 0.2% v/v β-mercaptoethanol) and incubated at 65 °C for 2 h. The lysate was mixed with an equal volume of chloroform: isoamyl alcohol (24:1, v/v) for 30 min. Subsequent to centrifugation (3,000 rpm, 10 min), the aqueous phase was collected, and the DNA was precipitated with 2-propanol. The precipitate was recovered, washed twice with 70% (v/v) ethanol, air-dried, and resuspended in DNase-free water. The RNA contamination was removed by RNase A treatment (10 mg/ml) at 37 °C for 2 h. The DNA was further purified by two sequential extractions with phenol: chloroform: isoamyl alcohol (25:24:1, v/v/v) (Sigma-Aldrich, Germany) followed by two chloroform: isoamyl alcohol (24:1, v/v) extractions. The DNA was precipitated by adding 0.1 volume of 3 M sodium acetate (pH 8.0) and absolute ethanol, followed by centrifugation (14,000 rpm, 30 min, 4 °C). The final DNA pellet was washed with 70% (v/v) ethanol, air-dried, and dissolved in the TE buffer. The DNA quality and quantity were assessed using 0.8% (w/v) agarose (Serva, Germany) gel electrophoresis in 1×TBE buffer and the spectrophotometric analysis (NanoDrop™ 2000c; Thermo Fisher Scientific™, USA).
ELISA
MethylFlash™ m6A DNA Methylation ELISA Kit (Epigentek, USA) was used to assess the global level of 6 mA in the rye genomic DNA. The manufacturer’s protocol was followed with 200 ng of the DNA input. Three biological replicates of each sample were applied to the 48-well plate. Also, two replicates of negative control and two replicates each of positive control (0.002 ng/µl, 0.005 ng/µl, 0.01 ng/µl, 0.02 ng/µl, 0.1 ng/µl) were applied to the plate. The absorbance was read on a BioTek (USA) EL800 microplate reader at 450 nm. The BioTek (USA) Gen5 software was used to capture the data from the microplate reader.
DNA hydrolysis and mass spectrometry
The DNA samples were completely dried in a SpeedVac system. Then the pellet was dissolved in 50 µL of MilliQ-grade deionized water and mixed with 50 µL of NP1 buffer (200 mM ammonium acetate (Sigma-Aldrich, Germany Cat. Nr 73594), 0.2 mM ZnCl2 (POCH, Poland Cat. Nr 264170113); pH 4.6). Nuclease P1 (100 U, New England Biolabs, USA Cat. Nr M0660S) and tetrahydrouridine (10 mg/ml, Merck Germany Cat. Nr 584222) were added to the mixture and incubated at 37 °C for 3.5 h. Next, 13 µL of 10% (v/v) NH4OH (J.T.Baker Cat. Nr 4807-05) and 12 µL of Shrimp Alkaline Phosphatase (rSAP, New England Biolabs, USA Cat. Nr M0371L) were added to the samples following two hours of incubation at 37 °C. All the hydrolysates were ultrafiltered prior to injection and concentrated in a SpeedVac to a final volume of 10 µL. The DNA hydrolysates were spiked with the internal standard in the volumetric ratio 4:1, to a concentration of 5 fmol/µL of [D3]-N6-mdA, and analyzed using UPLC-MS/MS.
Chromatographic separation was achieved using a method described by Starczak et al.50 based on a UPLC system with a photo-diode array detector (used for the quantification of canonical deoxynucleosides) and a tandem quadrupole mass spectrometer (Xevo TQ-XS, Waters) using the following columns: Waters Cortecs T3 column (150 mm×3 mm, 1.6 μm) with a precolumn. A flow rate of 0.5 mL/min was used, with an injection volume of 2 µL and gradient elution for 10 min using a mobile phase of 0.05% acetate (A) and acetonitrile (B) (0.7-5% B for 5 min, followed by the column washing with 30% acetonitrile and re-equilibration with 99% A for 3.6 min).
All the samples were analyzed in three to five technical replicates and the technical mean was used for further calculation. The quantities of canonical deoxynucleosides were determined by UV detection at 260 nm for 2’-deoxythymidine (dT), and at 280 nm for 2’-deoxyguanosine (dG). The total deoxynucleosides amount (dN) calculated as doubled sum of dT and dG was used as a reference for the quantitative expression of modified ones.
Immunofluorescence
Slide preparation was carried out as followed by the previously described methodology (Kalinka and Achrem, 2020). 6 mA chromosomal distribution was determined with the N6-Methyladenosine (m6A) Recombinant Rabbit Monoclonal Antibody (Catalog no MA5-33030, Invitrogen, USA) and Alexa Fluor™ 488 Tyramide SuperBoost™ Kit Goat anti-Rabbit IgG (Invitrogen, USA). Three types of controls for the IF study were employed: no primary antibody control, no secondary antibody control, and isotype control. Additionally, detection of 5-methylcytosine and 5-hydroxymethylcytosine was performed as a control43,51. As the first step of IF, 100 µl of 3% (v/v) hydrogen peroxide solution was applied to each slide covered with glass coverslips and incubated for 1 h at room temperature. The slides were washed three times for 5 min with 1× PBS at room temperature. 100 µl of blocking buffer (Component A from SuperBoost™ Kit) was applied to each slide, covered with a glass coverslip, and incubated for 1 h at room temperature. The cover glass was removed and directly 50 µL of primary antibody solution (2 µg/µL in 1% w/v BSA; 1×PBS, pH 7.4) was applied to the slide, covered with a coverslip and incubated overnight at 4 °C in a humidity chamber. The lides were washed three times for 10 min with 1× PBS at room temperature. 100 µl of secondary antibody (Component B from SuperBoost™ Kit) was dropped onto the slide. The preparation was covered with a coverslip, placed in a humid chamber and incubated for 1 h at room temperature. The slide was washed three times with 1× PBS, for 10 min each wash. The tyramide working solution and reaction stop reagent were prepared according to the manufacturer’s instruction. 100 µl of the tyramide working solution was applied to the cells and incubated for 2.5 min at room temperature. The reaction was stopped with 100 µl of stop reagent. The slides were washed three times for 5 min with 1× PBS at room temperature. A small drop of Fluoroshield™ with DAPI (Sigma-Aldrich, Germany) was used as a mounting solution.
Immunoprecipitation
5 µg of the genomic DNA isolated from coleoptiles diluted in 450 µl of TE buffer was sonicated with the Sonoplus HD 3100 (Bandelin, Germany) device into 100–600 bps-long fragments. The conditions of DNA shearing were optimized, and the following parameters were used: 30% power output during 4 pulses of sonication, 15 s each, followed by forty-seconds rest on ice between each pulse. The DNA fragment sizes were checked with 2% (w/v) agarose gel electrophoresis. The DNA was incubated at 100 °C for 10 min and snap-cooled on ice for 5 min. 50 µl of 10× IP buffer (100 mM Na-phosphate pH 7.0, 1.4 M NaCl, 0.5% v/v TritonX-100) was added to each sample. Subsequently, 1 µl of antibody (N6-Methyladenosine (m6A) Recombinant Rabbit Monoclonal Antibody, Catalog no MA5-33030, Invitrogen, USA) was mixed with the DNA solution. Normal, purified rabbit IgG was used as a negative control antibody in the parallel experiment. Samples were incubated for 3 h on an orbital rotator at 4 °C. The pre-wash of 40 µl of magnetic Dynabeads™ M-280 sheep anti-rabbit IgG (Invitrogen, USA) was carried out with 1 ml of PBS buffer with the addition of 0,1% (w/v) BSA for 5 min. Afterwards, the beads were collected with a magnetic separation rack. The pre-wash step was repeated and the Dynabeads were suspended in 40 µl of 1× IP buffer. Dynabeads in the IP solution were mixed with the DNA sample on a rotator at 4 °C for 16 h. The dynabeads were collected with magnetic separation rack and the solution was removed. The dynabeads were washed with 1 ml of 1× IP buffer at room temperature for 10 min. The washing step was repeated two more times. 250 µl of digestion buffer (5mM Tris pH 8.0, 1 mM EDTA pH 8.0, 0.05% w/v SDS) and 5 µl of 20 mg/ml proteinase K (Eurx®, Poland) were added to Dynabeads. The samples were incubated at 50 °C for 3 h on a rotator. The dynabeads were collected with a magnetic separation rack and the DNA solution was transferred to the new tube. For the DNA extraction the phenol-chloroform method was applied. The DNA was precipitated with 1/10 vol of 3 M NaOAc (pH 8.0), 1 ml of absolute ethanol and 1 µl of glycogen (20 mg/ml). The sample was carefully mixed and centrifuged (14,000 rpm, 30 min, 4 °C). The precipitate was washed twice with 70% (v/v) ethyl alcohol, air-dried, and dissolved in 100 µl of TE buffer.
Library preparation and sequencing
All the quality controls of the DNA samples and sequencing procedures, including sample preparation, library construction, and data generation were performed by BGI Genomics (China). For every Secale species, genomic DNA was isolated from a pool of ≥ 30 seedlings harvested at the same developmental stage. The pooled DNA served as input for a single PacBio HiFi library and a single MeDIP-seq library; no additional sequencing replicates were generated. For the Secale cereale genomic DNA (isolated from coleoptiles) analysis, sequencing was conducted using the PacBio Revio platform, generating 28.97 GB of the clean data output. The immunoprecipitated DNA samples enriched with 6 mA from all four Secale species were sequenced using the DNBseq platform with paired-end 100 bp reads strategy (Table 1), yielding ~ 4 GB of clean data per sample.
Data analysis
ELISA and mass spectrometry
To quantify the amount of 6 mA, a standard curve was generated in the Microsoft Excel software. OD values (negative control subtracted) versus the amount of positive control at each concentration point were plotted and the slope (OD/ng) using linear regression was determined. The percentage of 6 mA was calculated with the formulas given by the manufacturer of the ELISA kit.
The ELISA and mass spectrometry data were processed in Microsoft Excel software. Means and SDs were calculated from 3 biological replicates. The Shapiro-Wilk test was used to determine if ELISA and MS data follow a normal distribution, and the F-test was used to determine the significance level. In all the cases, p < 0.05 was considered statistically significant. MS data were used to calculate Pearson’s correlation coefficients, describing interrelationships between species.
Immunofluorescence
For the image acquisition and analysis an Axio Imager Z2 epifluorescence microscope (Carl Zeiss, Germany) and GenASIs software (Applied Spectral Imaging, USA) were used. From each species, 30 preparations were analysed (six preparations per cultivar/population of each species). In each preparation, high quality metaphase spreads were captured, and out of these three were analysed in detail. Based on the images, idiograms showing the distribution of 6 mA in each species were created.
PacBio HiFi sequencing and data preprocessing
The genomic DNA was sequenced using the PacBio Revio platform, generating circular consensus sequencing (CCS) reads with high accuracy. The sequencing produced an average of 1,915,236 reads per sample, totaling 28.97 Gbp of data. The mean read length was 15,127 bp with an N50 of 15,239 bp, and an average Phred quality score of 34. Raw BAM files obtained from the sequencing platform were converted to FASTQ format using the bam2fastq utility from SMRT Link (Pacific Biosciences). Reads shorter than 500 bp were filtered out using seqtk (v1.3) to improve the downstream analysis accuracy. Contamination assessment and filtering were performed using Kraken with a custom-built database containing RefSeq sequences from bacteria, archaea, viruses, protozoa, plasmids, fungi, and the human genome (GRCh38). Following initial classification by Kraken, Bracken was employed to estimate and quantify potential misclassified reads. The Secale cereale reference genome (GCA_902687465.1_Rye_Lo7_2018_v1p1p1_genomic.fna.gz) was downloaded. The reference genome was indexed using pbmm2 (v1.4.0) to facilitate efficient alignment of long reads. Filtered HiFi reads were aligned to the reference genome using pbmm2 with the CCS preset to leverage the high accuracy of the reads. Read group information was added for sample identification. Alignment quality was assessed using alignment statistics to ensure high mapping rates and accuracy.
6 mA methylation sites were detected using the ipdSummary tool from SMRT Analysis (v10.1). This tool computes the inter-pulse duration (IPD) ratios, which indicated nucleotide modifications based on polymerase kinetics. The --methylFraction option calculated the fraction of molecules methylated at each site, and the --identify m6A parameter specified the detection of 6 mA modifications. The resulting GFF file was filtered to retain only 6 mA sites. To identify regions with significant methylation enrichment, the 6 mA sites were converted to BED format for compatibility with peak-calling tools. Peaks were called using MACS2 (v2.2.7.1) with the broad peak option suitable for detecting diffuse methylation regions. The estimated genome size for Secale cereale (~ 7.9 × 10^8 bp) was used.
We generated a set of validated 6 mA sites and their corresponding methylation fraction estimates, which served as our high-confidence training dataset (see MeDIP-seq).
Sequences flanking the methylated adenines were extracted to identify consensus motifs associated with 6 mA sites. A window of ± 4 bp around each methylated site was considered. Bedtools (v2.30.0) were used for sequence extraction. Motif discovery was performed using MEME-ChIP from the MEME Suite (v5.4.1). Parameters were set to identify motifs between 6 and 12 bp in length. The analysis provided consensus motifs and their statistical significance.
Methylation sites were annotated with genomic features using bedtools and the Secale cereale gene annotation file (GTF format). Custom Python scripts were used to categorize methylation sites into genomic regions: promoters (within 2 kb upstream of transcription start sites), exons, introns, 5′ and 3′ untranslated regions (UTRs), and intergenic regions. Statistical analyses were performed in R (v4.1.0) to determine the distribution of 6 mA sites. Chi-square tests were conducted to assess whether the observed distribution deviated from expected proportions based on the genome annotation.
Gene IDs associated with methylated regions were extracted for the Gene Ontology (GO) analysis. The topGO package was used to identify enriched GO terms in biological processes, molecular functions, and cellular components.
All the statistical analyses were performed using R (v4.1.0) and Python (v3.8) with appropriate libraries. P-values were adjusted for multiple comparisons using the Benjamini-Hochberg procedure where applicable. Significance thresholds were set at p < 0.05.
The following software and tools were used in this study: SMRT Link (v10.1; Pacific Biosciences), Seqtk (v1.3), Pbmm2 (v1.4.0; Pacific Biosciences), SMRT Analysis (v10.1), Bedtools (v2.30.0), MEME Suite (v5.4.1), R (v4.1.0) (with packages: topGO, ggplot2, VennDiagram, ggseqlogo, dplyr), Python (v3.8) (with packages: matplotlib, pandas, numpy, seaborn), MACS2 (v2.2.7.1), DeepTools (v3.5.1), HISAT2 (v2.2.1), StringTie (v2.1.4), FastQC (v0.11.9), Trimmomatic (v0.39), Samtools (v1.11).
Sequencing of N6-methyadenosine immunoprecipitated DNA (MeDIP-seq)
Reads preprocessing was executed using BBTools (BBMap and BBDuk, version 38.96, available at https://sourceforge.net/projects/bbmap/), following the Reads QC Workflow. This preprocessing involved quality trimming, adapter trimming, and low quality ends trimming (if necessary). Duplicated reads were removed either by SAMtools markdup52 or UMI-tools dedup53. The MeDIP-seq data were then aligned to a reference sequence genome (Secale cereale reference genome Rye_Lo7_2018_v1p1p1, GenBank: GCA_902687465.1) using BWA54. The alignments were saved in SAM format, converted to BAM format and sorted with SAMtools52. Methylated mapped region was extracted as BED format by SAMtools and BEDtools55. These BED files were supplied to MEDIPS56, package for analyzing data derived from MeDIP experiments followed by sequencing (MeDIP-seq). For the characterization of the methylated regions, CEAS57 was used to calculate the fraction of regions present in different gene regions (introns, coding exons, 5´UTRs, 3′UTRs, and distal intergenic regions), promoters, bidirectional promoters, and regions downstream of gene bodies. 6 mA distribution was calculated as log2(IPRPKM/inputRPKM + 1), RPKM value from the regions before TSS 500 bp.
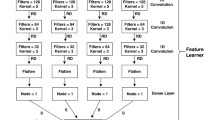
IPD signals indicative of 6 mA methylation, obtained using the ipdSummary tool from SMRT Analysis (v10.1), constituted a set of validated 6 mA sites and their corresponding methylation fraction estimates that served as our high-confidence training data set. Next, a Python-based machine learning workflow was employed to leverage these PacBio-derived methylation calls for guiding the annotation of MeDIP-seq data. The workflow was implemented in Jupyter Notebooks and relied on several core libraries: NumPy (v1.21.2) for numerical computations, pandas (v1.3.3) for data structuring, Biopython (v1.79) for sequence handling, and PySAM (v0.17.0) for BAM/CRAM file parsing. The machine learning model was designed to high-accuracy PacBio methylation calls and MeDIP-seq coverage data. Specifically, we constructed a deep convolutional neural network (CNN) implemented using TensorFlow (v2.7.0). The CNN model was trained to predict the probability of 6 mA methylation at each genomic position based on a combination of sequence context, PacBio-derived high-confidence methylation labels, and normalized MeDIP-seq coverage signals. Before training, coverage signals to account for variations in immunoprecipitation efficiency across samples were normalized. In developing the classifier we first assembled a high-confidence training set from the Secale cereale PacBio HiFi run. Adenines called as 6 mA by ipdSummary with a methylFraction ≥ 0.8 were treated as positives, while adenines with methylFraction ≤ 0.05 and lying ≥ 500 bp from any positive were sampled in equal number as negatives. To prevent information leakage, chromosomes 1–5 were used for training, chromosome 6 for validation during hyper-parameter tuning, and chromosome 7 for a final blind test. For every candidate adenine we built a feature tensor that combines a one-hot-encoded 101-bp reference sequence window centred on the site, PacBio kinetic metrics (mean IPD ratio and pulse-width, per strand), local MeDIP-seq coverage vectors (30 bp bins in a ± 250 bp window for both IP and input tracks, normalised for GC bias), and contextual covariates such as mappability and GC content. This tensor is fed into a three-block 1-D convolutional neural network (filter sizes 64 → 128 → 256, kernel 5, ReLU activation) followed by two fully connected layers with dropout 0.3. Class imbalance is mitigated with focal loss, and model selection is guided by Bayesian optimisation on the validation set.
Limitation
A significant limitation was encountered in that no orthogonal validation data, such as targeted methylation assays, was available to confirm the reliability of the predicted methylation sites. This absence of external validation datasets restricted the ability to quantify the false-positive or false-negative rates accurately, leaving the model’s performance reliant solely on internal cross-validation metrics and the consistency of the PacBio-derived signals. Nevertheless, intersecting our predictions with an independent MeDIP-seq dataset revealed that 86% peaks co-localised with PacBio-validated 6 mA sites, so the residual 14% provides a quantitative upper bound on potential false negatives attributable to the ~ 4 × HiFi depth and underscores the overall completeness of the integrated map. Although the machine learning framework demonstrated promising predictions in silico, the lack of independent data prevented a definitive assessment of the approach’s robustness in broader contexts or under varying experimental conditions.
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository and accession number can be found at: https://www.ncbi.nlm.nih.gov/, accession number PRJNA1249262.
References
Kumar, S., Chinnusamy, V. & Mohapatra, T. Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front. Genet. 9, 640. https://doi.org/10.3389/fgene.2018.00640 (2018).
Breiling, A. & Lyko, F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenet Chromatin. 8, 24. https://doi.org/10.1186/s13072-015-0016-6 (2015).
Greer, E. L. et al. DNA methylation on N6-adenine in C. elegans. Cell 161, 868–878. https://doi.org/10.1016/j.cell.2015.04.005 (2015).
Zhang, G. et al. N6-methyladenine DNA modification in Drosophila. Cell 161, 893–906. https://doi.org/10.1016/j.cell.2015.04.018 (2015).
Wang, Y., Chen, X., Sheng, Y., Liu, Y. & Gao, S. N6-adenine DNA methylation is associated with the linker DNA of H2A.Z-containing well-positioned nucleosomes in pol II-transcribed genes in Tetrahymena. Nucleic Acids Res. 45, 11594–11606. https://doi.org/10.1093/nar/gkx883 (2017).
Liu, J. et al. Abundant DNA 6 ma methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 7, 13052. https://doi.org/10.1038/ncomms13052 (2016).
Yao, B. et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. https://doi.org/10.1038/s41467-017-01195-y (2017).
Xiao, C. L. et al. N6-Methyladenine DNA modification in the human genome. Mol. Cell. 71, 306–318e7. https://doi.org/10.1016/j.molcel.2018.06.015 (2018).
Liang, Z. et al. DNA N6-adenine methylation in Arabidopsis thaliana. Dev. Cell. 45 (3), 406–416e. https://doi.org/10.1016/j.devcel.2018.03.012 (2018).
Yuan, D. H. et al. DNA N6-methyladenine modification in wild and cultivated soybeans reveals different patterns in nucleus and cytoplasm. Front. Genet. 11, 736. https://doi.org/10.3389/fgene.2020.00736 (2020).
Zhang, G., Diao, S., Song, Y., He, C. & Zhang, J. Genome-wide DNA N6-adenine methylation in sea Buckthorn (Hippophae rhamnoides L.) fruit development. Tree Physiol. 42, 1286–1295. https://doi.org/10.1093/treephys/tpab177 (2022).
Xie, S. Q. et al. N6-methyladenine DNA modification in the woodland strawberry (Fragaria vesca) genome reveals a positive relationship with gene transcription. Front. Genet. 10, 1288. https://doi.org/10.3389/fgene.2019.01288 (2020).
Zhou, C. et al. Identification and analysis of adenine N6-methylation sites in the rice genome. Nat. Plants. 4, 554–563. https://doi.org/10.1038/s41477-018-0214-x (2018).
Zhang, Y. et al. 6 mA DNA methylation on genes in plants is associated with gene complexity, expression and duplication. Plants 12, 1949–1949. https://doi.org/10.3390/plants12101949 (2023).
Mondo, S. J. et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 49, 964–968. https://doi.org/10.1038/ng.3859 (2017).
O’Brown & Greer, E. L. N6-methyladenine: a conserved and dynamic DNA mark. Adv. Exp. Med. Biol. 945, 213–246. https://doi.org/10.1007/978-3-319-43624-1_10 (2016).
Bochtler, M. & Fernandes, H. DNA adenine methylation in eukaryotes: enzymatic mark or a form of DNA damage? BioEssays 43, 2000243. https://doi.org/10.1002/bies.202000243 (2021).
Li, H. et al. DNA N6-methyladenine modification in eukaryotic genome. Front. Genet. 13, 914404. https://doi.org/10.3389/fgene.2022.914404 (2022).
Jiménez-Ramírez, I. A., Pijeira-Fernández, G. & Moreno-Cálix, D. M. De-la-Peña, C. Same modification, different location: the mythical role of N6-adenine methylation in plant genomes. Planta 256, 9. https://doi.org/10.1007/s00425-022-03926-y (2022).
Fedoreyeva, L. I. & Vanyushin, B. F. N6-Adenine DNA‐methyltransferase in wheat seedlings. FEBS Lett. 514, 305–308. https://doi.org/10.1016/s0014-5793(02)02384-0 (2002).
Zhu, S. et al. Mapping and characterizing N6-methyladenine in eukaryotic genomes using single-molecule real-time sequencing. Genome Res. 28, 1067–1078. https://doi.org/10.1101/gr.231068.117 (2018).
Usai, G. et al. DNA modification patterns within the transposable elements of the Fig (Ficus carica L.) genome. Plants 10, 451–451. https://doi.org/10.3390/plants10030451 (2021).
Chachar, S. et al. Harnessing current knowledge of DNA N6-methyladenosine from model plants for non-model crops. Front. Genet. 12, 668317. https://doi.org/10.3389/fgene.2021.668317 (2021).
Zhao, L. Y., Song, J., Liu, Y., Song, C. X. & Yi, C. Mapping the epigenetic modifications of DNA and RNA. Protein Cell. 11, 792–808. https://doi.org/10.1007/s13238-020-00733-7 (2020).
Wood, D. E., Salzberg, S. L. & Kraken,. Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. https://doi.org/10.1186/gb-2014-15-3-r46 (2014).
Lu, J., Breitwieser, F. P., Thielen, P. & Salzberg, S. L. Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 3, e104. https://doi.org/10.7717/peerj-cs.104 (2017).
Li, H., Niu, J., Sheng, Y., Liu, Y. & Gao, S. SMAC: identifying DNA N6-methyladenine (6 mA) at the single-molecule level using SMRT CCS data. Brief. Bioinform. 26, bbaf153. https://doi.org/10.1093/bib/bbaf153 (2025).
O’Brown, Z. K. et al. Sources of artifact in measurements of 6 ma and 4mC abundance in eukaryotic genomic DNA. BMC Genom. 20, 445. https://doi.org/10.1186/s12864-019-5754-6 (2019).
Douvlataniotis, K., Bensberg, M., Lentini, A., Gylemo, B. & Nestor, C. E. No evidence for DNA N6-methyladenine in mammals. Sci. Adv. 6, eaay3335. https://doi.org/10.1126/sciadv.aay3335 (2020).
Sun, Y. et al. Population genomic analysis reveals domestication of cultivated Rye from weedy Rye. Mol. Plant. 15, 552–561. https://doi.org/10.1016/j.molp.2021.12.015 (2022).
Schreiber, M., Himmelbach, A., Börner, A. & Mascher, M. Genetic diversity and relationship between domesticated Rye and its wild relatives as revealed through genotyping-by‐sequencing. Evol. Appl. 12, 66–77. https://doi.org/10.1111/eva.12624 (2018).
Bolibok-Brągoszewska, H., Targońska, M., Bolibok, L., Kilian, A. & Rakoczy-Trojanowska, M. Genome-wide characterization of genetic diversity and population structure in Secale. BMC Plant. Biol. 14, 184. https://doi.org/10.1186/1471-2229-14-184 (2014).
Hawliczek, A. et al. Selective sweeps identification in distinct groups of cultivated Rye (Secale cereale L.) germplasm provides potential candidate genes for crop improvement. BMC Plant. Biol. 23, 323. https://doi.org/10.1186/s12870-023-04337-1 (2023).
Maraci, Ö., Özkan, H. & Bilgin, R. Phylogeny and genetic structure in the genus Secale. PLoS ONE. 13, e0200825. https://doi.org/10.1371/journal.pone.0200825 (2018).
Rabanus-Wallace, M. T. et al. Chromosome-scale genome assembly provides insights into Rye biology, evolution and agronomic potential. Nat. Genet. 53, 564–573. https://doi.org/10.1038/s41588-021-00807-0 (2021).
Bauer, E. et al. Towards a whole-genome sequence for Rye (Secale cereale L). Plant. J. 89, 853–869. https://doi.org/10.1111/tpj.2017.89.issue-5 (2016).
Li, G. et al. A high-quality genome assembly highlights Rye genomic characteristics and agronomically important genes. Nat. Genet. 53, 574–584. https://doi.org/10.1038/s41588-021-00808-z (2021).
Evtushenko, E. V. et al. The expansion of heterochromatin blocks in Rye reflects the co-amplification of tandem repeats and adjacent transposable elements. BMC Genom. 17, 337. https://doi.org/10.1186/s12864-016-2667-5 (2016).
Zhang, Q. et al. N6-methyladenine DNA methylation in Japonica and indica rice genomes and its association with gene expression, plant development, and stress responses. Mol. Plant. 11, 1492–1508. https://doi.org/10.1016/j.molp.2018.11.005 (2018).
Luan, M. W. et al. DNA N6-methyladenosine modification role in transmitted variations from genomic DNA to RNA in Herrania umbratica. BMC Genom. 20, 508. https://doi.org/10.1186/s12864-019-5776-0 (2019).
Kalinka, A. & Achrem, M. Reorganization of wheat and Rye genomes in octoploid triticale (× Triticosecale). Planta 247, 807–829. https://doi.org/10.1007/s00425-017-2827-0 (2017).
Achrem, M. & Kalinka, A. Tracking of intercalary DNA sequences integrated into tandem repeat arrays in Rye Secale vavilovii. Acta Soc. Bot. Pol. 86, 3548. https://doi.org/10.5586/asbp.3548 (2017).
Kalinka, A. & Achrem, M. The distribution pattern of 5-methylcytosine in Rye (Secale L.) chromosomes. PLoS ONE. 15, e0240869. https://doi.org/10.1371/journal.pone.0240869 (2020).
Feng, S. et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U S A. 107, 8689–8694. https://doi.org/10.1073/pnas.1002720107 (2010).
Karanthamalai, J., Chodon, A., Chauhan, S. & Pandi, G. DNA N6-methyladenine modification in plant genomes - a glimpse into emerging epigenetic code. Plants 9, 247. https://doi.org/10.3390/plants9020247 (2020).
Hao, J. S. et al. Distribution pattern of N6-methyladenine DNA modification in the seashore paspalum (Paspalum vaginatum) genome. Front. Plant. Sci. 13, 922152. https://doi.org/10.3389/fpls.2022.922152 (2022).
Ma, C. et al. N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell. Biol. 21, 319–327. https://doi.org/10.1038/s41556-018-0238-5 (2019).
Wan, Q. L. et al. N6-methyldeoxyadenine and histone methylation mediate transgenerational survival advantages induced by hormetic heat stress. Sci. Adv. 7, eabc3026. https://doi.org/10.1126/sciadv.abc3026 (2021).
Cao, S. & Chen, Z. J. Transgenerational epigenetic inheritance during plant evolution and breeding. Trends Plant. Sci. 29, 1203–1223. https://doi.org/10.1016/j.tplants.2024.04.007 (2024).
Starczak, M., Gawronski, M., Olinski, R. & Gackowski, D. Quantification of DNA modifications using two-dimensional ultraperformance liquid chromatography tandem mass spectrometry (2D-UPLC-MS/MS). Methods Mol. Biol. 2198, 91–108. https://doi.org/10.1007/978-1-0716-0876-0_8 (2021).
Kalinka, A. et al. DNA 5-hydroxymethylcytosine level and its chromosomal distribution in four Rye species. J. Exp. Bot. 74, 3488–3502. https://doi.org/10.1093/jxb/erad102 (2023).
Li, H. et al. The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079. https://doi.org/10.1093/bioinformatics/btp352 (2009).
Smith, T., Heger, A. & Sudbery, I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 27, 491–499. https://doi.org/10.1101/gr.209601.116 (2017).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. https://doi.org/10.1093/bioinformatics/btp324 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. https://doi.org/10.1093/bioinformatics/btq033 (2010).
Lienhard, M., Grimm, C., Morkel, M., Herwig, R. & Chavez, L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 30, 284–286. https://doi.org/10.1093/bioinformatics/btt650 (2014).
Shin, H., Liu, T., Manrai, A. K. & Liu, X. S. CEAS: cis-regulatory element annotation system. Bioinformatics 25, 2605–2606. https://doi.org/10.1093/bioinformatics/btp479 (2009).
Acknowledgements
We would like to thank Magdalena Achrem for her help with cytogenetic analyses.
Funding
Co-financed by the Minister of Science under the “Regional Excellence Initiative” Program for 2024–2027 (RID/SP/0045/2024/01).
Author information
Authors and Affiliations
Contributions
AK: conceptualization and supervision; AK, MS and DG: investigation; AK, DC-L, MS and DG: methodology; AK and DC-L: formal analysis, visualization; AK, DC-L, MS and DG: original draft preparation, review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kalinka, A., Cembrowska-Lech, D., Starczak, M. et al. Distribution patterns of N6-methyladenine in the rye genome. Sci Rep 15, 26066 (2025). https://doi.org/10.1038/s41598-025-11699-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-11699-z