Abstract
Humanized transgenic mice carrying human genes are useful for research on gene function and disease. Bacterial artificial chromosomes (BACs) that carry human genomic sequences with regulatory elements enable the expression of transgenes at physiological levels in vivo. To study complex biological phenomena involving multiple genes, techniques for co-introducing transgenes into mice have been developed; however, the introduction of multiple BACs remains laborious. The simultaneous integration of multiple gene loading vectors (SIM) system was developed to incorporate three or more gene-loading vectors (GLVs) using a mouse artificial chromosome (MAC) vector. This system allows for simultaneous site-specific incorporation of three GLVs into a single MAC with only one screening. However, the capacity for large constructs, such as BACs, has yet to be evaluated. This study is the first to demonstrate the development of multi-BAC transchromosomic (Tc) mice targeting the human leukocyte antigen (HLA) class I gene cluster (HLA-A, HLA-B, HLA-C) and beta-2-microglobulin (B2M) using the SIM system. By constructing a MAC using three BACs containing these genomic regions, we successfully generated HLA class I Tc mice. The technology to generate multi-BAC Tc mice will accelerate the analysis of complex life mechanisms involving multiple factors.
Similar content being viewed by others
Introduction
Transgenic (Tg) animals are indispensable for analyzing the function of target genes in vivo. Humanized Tg mice engineered to carry human genes are particularly valuable, recapitulating numerous human phenotypes and diseases.
Common vector systems, such as viruses, plasmids, and bacterial artificial chromosomes (BACs), have been employed to generate Tg mice. BACs harboring human genomic sequences (< 300 kb) have the potential to enhance the reproducibility of humanized animal models because they can include gene regulatory regions, such as promoters and enhancers. Tg mice carrying BACs have been widely developed, demonstrating stable expression and, in some cases, physiological regulation of human transgene expression in vivo1.
Researchers are increasingly generating multi-gene Tg mice to study complex biological phenomena involving interactions and cooperation between multiple factors. For example, multifactorial interactions play crucial roles in the immune and nervous systems, prompting the development of humanized models that integrate these factors2,3. Instead of conventional Tg mouse generation through random integration, site-specific insertion of large DNA segments, including BACs, has become a common approach for controlled transgene expression. Furthermore, methods for the site-specific introduction of multiple large (> 10 kb) gene-loading vectors (GLVs) have been explored4,5 but require screening for each GLV insertion and long production periods.
The simultaneous integration of multiple GLVs (SIM) system, which has been developed to accommodate three or more GLVs, employs a chromosomal gene-delivery vector platform known as a mouse artificial chromosome (MAC)6. Briefly, a MAC was constructed by truncating a natural mouse chromosome at a site adjacent to the centromeric region via telomere seeding, resulting in the deletion of whole genomic regions while retaining the mouse centromere, followed by the incorporation of acceptor sites, such as loxP7. This MAC was maintained in cells and mice without integration into the host genome, and has been used to generate humanized mice8,9,10. Mice carrying MACs or other foreign chromosomes, known as transchromosomic (Tc) mice, are generated by loading a target gene into a MAC and transferring it into mouse embryonic stem (ES) cells. The SIM system allows for the simultaneous site-specific insertion of GLVs into a single MAC using Cre recombinase and Bxb1/PhiC31 integrase. It requires a minimum of one selection marker, and only one screening is needed for every three GLVs inserted under copy number control. Previously demonstrated only with plasmids, application of the SIM system to BACs could enable the simultaneous integration of multiple BAC-scale GLVs into a MAC. This advancement is expected to accelerate the generation of Tc mice by reducing the number of required screenings, while enabling copy number control and physiological transgene expression.
In this study, we have demonstrated the application of the SIM system in the development of multi-BAC Tc mice by targeting the human leukocyte antigen (HLA) gene cluster, which plays a crucial role in antigen presentation and T-cell-mediated immunity in humans. Classical HLA class I molecules, including HLA-A, B, and C, are encoded by genes located across a 1.8-Mb region on human chromosome 6. They form complexes with human beta-2-microglobulin (hB2M), encoded by human chromosome 15, and are expressed on the surface of nucleated cells. The generation of mouse models carrying HLA is particularly significant given the species differences in the peptide repertoires presented by H-2 in mice and HLA in humans, despite their homology. For applications such as peptide vaccine development, several HLA knock-in mice have been generated11,12,13; however, generating mice with multiple HLAs is complex, and includes multiple crossbreeding steps14.
Specifically, we used three BAC vectors (59–180 kb each) containing the genomic and regulatory regions of HLA-A, HLA-B, and HLA-C, as well as hB2M, for simultaneous assembly of the HLA class I haplotype on the MAC. This study has achieved two firsts: the co-transduction of three BACs and the co-transduction of HLA-A, HLA-B, HLA-C, and hB2M in mice using the SIM system. It also serves as proof of concept for generating HLA class I Tc mice.
Results
Construction of a MAC carrying HLA class I gene regions using the SIM system
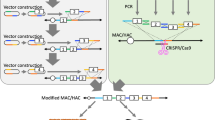
First, HLA class I-containing BACs were modified to carry the following recombination cassettes (hereafter referred to as SIM cassettes) for MAC loading: loxP and Bxb1 attP on the first cassette (C1), Bxb1 attB and PhiC31 attP on the second cassette (C2), and PhiC31 attB and the hypoxanthine–guanine phosphoribosyl transferase (HPRT) gene exon 3–9 (3′HPRT) on the third cassette (C3). Each SIM cassette also contains homology arms (SACB and F1) of 500 bp each that match regions on the BACs (Supplementary Fig. S1), enabling insertion of the SIM cassettes via homologous recombination in Escherichia coli (E. coli). SIM cassettes C1, C2, and C3 were incorporated into HLA-A BAC (CH501-309N1, GenBank: AL671277.4, A*03:01:01:01), HLA-B/C BAC (CH501-248L24, GenBank: AL671883.3, B*07:02:01:01, C*07:02:01:03), and hB2M BAC (RP11-292P13, GenBank: AC018901.8), respectively (Supplementary Figs. S2 and S3). PCR amplification of the backbone and cassette junction using primers designed against the outside and inside of each arm (SACB arm junction: SACB F/R; F1 arm junction: F1 F/R) confirmed the insertion of the respective SIM cassettes in the modified BACs (Supplementary Tables S1 and S2). The three SIM cassette-carrying BACs were then co-transfected with Cre, PhiC31, and Bxb1 expression vectors into CHO cells with stable maintenance of a MAC (Fig. 1a) pre-engineered to carry CAG promoter-driven enhanced green fluorescent protein (CAG-EGFP), the HPRT gene exon1-2 (5′HPRT), loxP, phosphoglycerate kinase (PGK) promoter-driven expression of hygromycin (PGK-HygR), and puromycin (PGK-PuroR)15. Loading of all three BACs via the SIM system reconstitutes the HPRT gene (5′HPRT on the MAC and 3′HPRT on the third cassette), creating hypoxanthine-aminopterin-thymidine (HAT) resistance. To obtain CHO cells carrying the MAC loaded with three recombinant BACs, we used both HAT-based drug selection and MAC-derived GFP fluorescence, yielding 33 GFP-positive and drug-resistant clones.
Construction of HLAcI-MAC using the SIM system in CHO cells. (a) Schematic representation of the SIM system used to load three HLA class I bacterial artificial chromosomes (BACs) onto a mouse artificial chromosome (MAC). (b) Genomic PCR analysis of the retention of the HLA and hB2M genes on HLAcI-MAC in CHO cells using the following PCR primer sets: HLA-A, HLA-A F/R; HLA-C, HLA-C F/R; HLA-B, HLA-B F/R; and hB2M, hB2M F1/R1) (see Supplementary Table S1 for primer sequences). The electrophoresis data are shown in Supplementary Fig. S4. (c) Representative fluorescence in situ hybridization images showing the retention of HLAcI-MAC in metaphase CHO cells using two probes: mouse Cot-1 DNA (red) strains MAC and HLA-B/C BAC (green). Yellow arrowheads indicate the HLAcI-MAC, enlarged in the insets. Scale bars: 10 μm.
Next, we performed genomic PCR on the candidate clones. First, we performed PCR to detect the HPRT junction and the HLA-A gene (Supplementary Figs. S5 and S6). In the first screening, 29 out of 33 clones (87.9%) showed reconstitution of the HPRT locus, and 16 out of 33 clones (48.5%) carried the HLA-A gene on the first cassette. Clones that were positive for both PCR were then subjected to additional PCR analyses targeting HLA-B/C, hB2M, and broader regions of the HLA loci using HLA-long PCR primers. The results of these follow-up PCRs are provided in Supplementary Figs. S7 and S8. Based on these analyses, 6 out of 33 clones (18.2%) appeared to carry all three GLVs—HLA-A BAC, HLA-B/C BAC, and hB2M BAC—as expected. Representative PCR results of clones #18 − 1 and #25 − 6, which were used in subsequent experiments, are shown in Fig. 1b, Supplementary Figs. S4 and S9a. Next-generation sequencing (NGS) analysis revealed that clones #18 − 1 and #25 − 6 carried three regions of the human genome: locus 29,922,541 − 30,093,940 of NC_000006 containing HLA-A, locus 31,264,805 − 31,436,351 of NC_000006 containing HLA-B/C, and locus 44,676,664 − 44,724,505 of NC_000015 containing hB2M. These findings demonstrated that full-length, transduced HLA-A, HLA-B/C, and hB2M gene regions had been retained, without deletions, in the host CHO cells.
Fluorescent in situ hybridization (FISH) analysis confirmed the insertion of the HLA class I gene regions in the MAC (hereafter referred to as HLAcI-MAC) and demonstrated that there were no unexpected insertions into genomic regions of the host CHO cells (Fig. 1c, Supplementary Fig. S9b, Supplementary Table S3).
HLA class I molecules are membrane-bound glycoproteins composed of a heterodimer of an HLA-encoded α-chain and hB2M. The hB2M protein does not bind to the cell membrane directly but rather forms a complex with the α-chain expressed on the membrane surface. Therefore, the expression of HLA class I molecules in CHO HLAcI-MAC clones was analyzed using an anti-HLA-ABC antibody that recognizes the shared regions of HLA-A, HLA-B, HLA-C, and hB2M16 (Supplementary Table S4). Retention of the MAC was confirmed by detecting GFP fluorescence signals. The parental strain CHO MAC, which does not carry any HLA genes, was employed as an HLA class I−/GFP+ control, while the human fibrosarcoma tumor cell line HT1080 was employed as a positive control for HLA detection (Supplementary Figs. S10 and S11). Flow cytometry (FCM) analyses showed HLA class I expression in the two CHO HLAcI-MAC clones, albeit at a lower level than that in HT1080. GFP was expressed at similar levels in parental CHO MAC and the CHO HLAcI-MAC clones. These results indicated that the HLAs carried by HLAcI-MAC were properly expressed in CHO cells.
Transfer of HLAcI-MAC into mouse ES cells
HLAcI-MAC was transferred into mouse ES cells using the microcell-mediated chromosome transfer (MMCT) method17. Briefly, donor microcells from the two CHO HLAcI-MAC clones were fused with mouse ES cells of the XO ES9 line18 (Fig. 2a).
Establishment of mouse ES cells carrying HLAcI-MAC. (a) Schematic representation of the MMCT method used to transfer HLAcI-MAC from CHO cells to mouse ES cells. (b) Bright-field (BF) and fluorescence microscopy images of mouse ES cells following HLAcI-MAC transduction, showing GFP expression from the MAC. Scale bars: 100 μm. (c) Representative FISH images showing the retention of HLAcI-MAC in metaphase mouse ES cells using two probes: human Cot-1 (red) and mouse minor satellite DNA (green). The arrowhead indicates the HLAcI-MAC. Scale bar: 10 μm. (d) Flow cytometry analysis of cell-surface expression of HLA-ABC, HLA-A3, HLA-B7, and HLA-C under treatment with interferon (IFN)-γ in a representative ES HLAcI-MAC clone. Gating strategies are described in Supplementary Fig. S13. Percentages of HLA class I (HLA-A3, B7, or C) and fluorescein isothiocyanate-conjugated GFP double-positive cell populations are indicated in the upper right quadrants.
After drug selection, we obtained 16 and two ES clones containing HLAcI-MAC derived from the donors #18 − 1 and #25 − 6, respectively. Genomic PCR using primers designed against internal regions of the HLA and hB2M genes confirmed the retention of these regions in 13 out of 16 #18-1-derived clones and one of the two #25-6-derived clones (Supplementary Fig. S12). Examination of these clones under a fluorescence microscope confirmed that the MAC-derived GFP was retained (Fig. 2b). FISH analysis was then performed to confirm the stable and independent retention of HLAcI-MAC in ES clones. Chromosomes were identified using two probes: human Cot-1 DNA, which labels human genomic sequences in BACs, and mouse minor satellite DNA, which strongly labels mouse centromeres. FISH analysis confirmed the normal karyotype and the independent retention of HLAcI-MAC in the ES clone (ES HLAcI-MAC #3 and #7) derived from #18 − 1 (Fig. 2c, Supplementary Table S5).
FCM analysis was performed to determine whether the transferred HLAs were expressed as proteins at the ES cell membrane surface. HLA class I expression at the cell surface was analyzed using anti-HLA-ABC, anti-HLA-A3, anti-HLA-B7, and anti-HLA-C antibodies specific for the transfected BACs containing HLA. Retention of HLAcI-MAC was confirmed by detecting GFP fluorescence signals. The parental strain XO ES9 was used as a negative control, while Jurkat#2H4, an immortalized line of human T lymphocytes with the same HLA type as the transduced HLA genes, was used as a positive control. Populations with double-positive expression of HLA-A3+/HLA-ABC+, HLA-B7+/HLA-ABC+, and HLA-C+/HLA-ABC+ were identified in a representative ES HLAcI-MAC clone (Fig. 2d, Supplementary Fig. S13). Thus, the FCM analysis confirmed that the transduced HLA genes were expressed in ES HLAcI-MAC clones. Two clones (#3 and #7) with a target karyotype of > 70% and confirmed HLA expression were used in the following experiments.
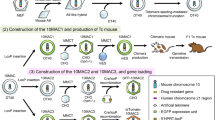
Maintenance of HLAcI-MAC in Tc mice
Chimeric mice were generated by injecting the ES HLAcI-MAC clones #3 and #7 into embryos (8-cell stage) of wild-type mice and transplanting them into the uteri of putative mothers (Supplementary Table S6). The chimeric mice with high chimerism (95–100%), as judged by coat color, were crossed with wild-type mice to test for germline transmission in the offspring, referred to as HLAcI Tc mice. Tc mice derived from clone #7 were used in subsequent experiments. To detect the presence of HLAcI-MAC in these mice, we performed FISH analysis of peripheral blood mononuclear cells (PBMCs), which confirmed the retention of HLAcI-MAC in addition to the endogenous mouse chromosomes (Fig. 3a). Furthermore, the retention of HLAcI-MAC in multiple tissues (brain; thymus; heart; liver; lung; spleen; kidney; skeletal muscle; small intestine; and testis) was confirmed by FISH analysis and GFP fluorescence (Fig. 3b, c, Supplementary Table S7). These findings indicated that the MAC was retained in each cell across tissues throughout the mouse body. Additionally, reverse transcription (RT)-PCR analysis confirmed the gene expression of HLA-A, HLA-B, HLA-C, and hB2M in all analyzed tissues (Fig. 3d, Supplementary Fig. S14 and S15). Notably, the GFP-positive rate of PBMCs from HLAcI Tc mice of generations F2 to F9 was stable, at < 87% (Fig. 3e). Collectively, these findings indicated that the mice had stably retained HLAcI-MAC throughout the body and were expressing its HLA genes.
Generation and analysis of HLAcI Tc mice. (a) Representative image of FISH analysis of metaphase peripheral blood mononuclear cells (PBMCs) from HLAcI Tc mice (F3, male, 6 w) showing the retention of HLAcI-MAC using two probes: human Cot-1 (red) and HLA-A BAC (green). The arrowhead indicates the HLAcI-MAC, enlarged in the inset. Scale bar: 10 μm. (b) Retention rate of the HLAcI-MAC in various Tc mouse tissues analyzed by interphase FISH, obtained by calculating the percentage of HLA-A probe-labeled MAC-retaining cells among the total cells (range: n = 66–114 per tissue) (F5, male, 9 w). (c) Bright-field (BF) and fluorescence microscopy images of different tissues from HLAcI Tc mice (same as Fig. 3b) carrying HLAcI-MAC, as indicated by GFP expression. Scale bars: 5,000 μm. (d) RT-PCR analysis of HLA expression in the following tissues derived from Tc (F5, male, 9 w) and wild-type (WT) mice (male, 17 w): brain (Br), thymus (Thy), heart (He), liver (Li), lung (Lu), spleen (Sp), kidney (Ki), skeletal muscle (Sk), small intestine (In), and testis (Te). The PCR primer sets (HLA-A: cDNA HLA-A F/R; HLA-C: cDNA HLA-C F/R; HLA-B: cDNA HLA-B F/R; and cDNA hB2M: cDNA hB2M F/R) are shown in Supplementary Table S1. The electrophoresis data are shown in Supplementary Fig. S14. Internal control data are shown in Supplementary Fig. S15. (e) Percentage of GFP-positive PBMCs from three different HLAcI Tc mice per indicated generation (F2–F9). Mice were 5–9 weeks old with mixed sexes. Bars represent mean ± SD.
Expression of HLA class I molecules in HLAcI Tc mice
FCM analysis confirmed that the HLA-A, HLA-B, HLA-C, and hB2M proteins were expressed on the surface of PBMCs from HLAcI Tc mice (Fig. 4a, Supplementary Fig. S16). HLA-A3 and HLA-B7 were found to be strongly expressed, while HLA-C was only slightly expressed. The GFP-positive ratio of lymphocytes from HLAcI Tc mice averaged 78.0% (n = 8, F1) (Supplementary Table S8). As expected, there was no expression of HLA-A, HLA-B, or HLA-C in wild-type negative control mice.
(a) Representative results of flow cytometry (FCM) analysis of HLA class I expression the surface of PBMCs derived from HLAcI Tc mice (F1, male, 5 w) and control wild-type (WT) mice (male, 9 w). Gating strategies are described in Supplementary Fig. S16. Percentages of HLA-ABC and fluorescein isothiocyanate-conjugated GFP double-positive cell populations are shown in the upper right quadrants. (b) Mean fluorescence intensity (MFI) obtained by FCM, indicating the expression levels of HLA-A3, B7, and C in IFN-γ treated (+) vs. untreated (-) PBMCs of HLAcI Tc mice (F9, male, 15 w). The data represent mean values from three mice. Statistical analysis was performed using a paired t-test.
Although HLA class I is constitutively expressed, it is well established that Interferon (IFN)-γ plays a key role in upregulating HLA expression during immune responses, particularly under conditions such as infection and inflammation19. This upregulation is mediated through IFN-γ-responsive elements in the promoter regions of HLA genes. In addition, differences in the sequences of promoter regions containing IFN-stimulated response elements upstream of HLA genes have been reported to affect the extent of the response to IFN signaling20. Specifically, HLA-B is more strongly upregulated by IFN-γ than HLA-A and HLA-C. Therefore, we evaluated whether the physiological upregulation of HLA expression in response to IFN-γ is recapitulated in our HLAcI Tc mice, which carry a BAC containing the HLA gene along with its native regulatory elements. PBMCs derived from HLAcI Tc mice were cultured for 48 h, with and without IFN-γ treatment. Expression levels of HLA-A and HLA-B, but not HLA-C, were significantly enhanced under treatment with IFN-γ (Fig. 4b), with HLA-B showing a greater increase than HLA-A. Therefore, this result indicated that physiological regulation of expression by IFN-γ is recapitulated in HLAcI Tc mice with BAC-scale genomic sequences, as well as differential regulation of HLA class I molecules.
Discussion
In this study, the adaptive range of the SIM system was extended to accommodate simultaneous loading of three BACs. Furthermore, BAC-SIM-loaded HLA class I and hB2M genes (including their native regulatory regions) exhibited physiological expression patterns in mice, thereby validating the utility of BAC-based model animals. This study represents the first simultaneous transduction of three BACs, and the first simultaneous transduction of HLA-A, HLA-B, HLA-C, and hB2M in mice.
The SIM system was previously demonstrated using plasmids only, but has now been shown to be feasible with three BACs. Conventional humanized animals carrying multiple long (> 150 kb) DNA fragments have been generated through multistep mating of genetically modified mice or by sequential linkage of gene fragments21,22. These processes demand significant labor and time. Alternatively, mice have been generated using two BACs sequentially loaded onto human artificial chromosomes23. In this method, additional BAC loading is limited by the availability of selection markers and recombination sites beyond those used for the first two BACs. The BAC-SIM method has overcome this bottleneck, enabling the successful loading of three BACs using one selection marker. Furthermore, the SIM system has the potential for loading even more BACs, with a previous study reporting that an additional three plasmids can be loaded after the initial three plasmids, for a total of six sequential GLVs6.
Regarding the impact of each site-specific recombination event on clone recovery, since the acquisition of drug resistance relies on the successful execution of all three site-specific recombination events (Cre, PhiC31, and Bxb1), the efficiency and coordination of these recombination systems are likely to significantly affect the overall efficiency of clone recovery. Although it has been reported that the recombination efficiencies of PhiC31 and Bxb1 are comparable to that of Cre24 detailed evaluation and further optimization may be necessary to achieve maximal efficiency in the system.
In this study, we conducted extensive PCR analysis to evaluate the integration of the introduced GLVs. Among the 33 candidate clones, 29 (87.8%) were positive for the reconstruction of the HPRT-junction, and among these 29 clones, 16 (55.2%) were also positive for the HLA-A gene located on the first BAC-SIM cassette. However, only 6 of the 33 clones (18.2%) were found to have incorporated all three GLVs (HLA-A BAC, HLA-B/C BAC, and hB2M BAC) as intended, based on broader genomic PCR analysis of the HLA region. These results suggest that, while partial integration is relatively common, complete and intended incorporation of all elements occurs at a lower rate. By contrast, a previous study of three plasmids loaded via the SIM system reported 100% PCR-positive rates of both the HPRT-junction and HPRT gene on the loaded plasmids (5/5 randomly chosen clones). This difference may be attributable to transfection efficiency or structural fragility, given that BACs are significantly larger than plasmids. Notably, we observed more frequent deletions in the second BAC (HLA-B/C BAC), whereas fewer deletions were detected in the hB2M BAC (Supplementary Fig. S7). Given that HLA-A BAC (180 kb) and HLA-B/C BAC (129 kb) are substantially larger than hB2M BAC (59 kb), this suggests that BAC size may influence structural stability and contribute to the differences in positive clone rates. These findings support verifying construct integrity through PCR at multiple sites, followed by NGS analysis, when using the BAC-SIM system.
The surface expression of HLA molecules was low in ES HLAcI-MAC cells and HLAcI Tc mice (Figs. 2d and 4a). A possible explanation for this is that the binding of endogenous mouse B2m to HLA reduces the rate of complex formation between hB2M and HLA. Previous studies have compared the expression levels of HLA in mouse cells co-transfected with hB2M or mouse B2m and found that HLA was highly expressed when co-transfected with hB2M25,26. While mouse B2m was also reported to form a complex with HLA, albeit to a lesser extent than hB2M. Therefore, knockout of mouse B2m may improve HLA expression in this system.
In the present analysis, HLA expression was induced by IFN-γ in mouse cells (Fig. 4b). MHC class I molecules are regulated through both a homeostatic expression mechanism and an IFN-γ-mediated mechanism. Compared with untreated cells, IFN-γ-treated cell populations showed a tendency of increased HLA-A and HLA-B expression. This suggested that the HLA genes and surrounding regulatory regions introduced via BAC were functional. Considering that HLA and B2M reportedly have distinct regulatory regions27, Tg mice with knock-in of a chimeric HLA-A (α1/α2 domains of HLA-A and α3 domain of mouse H-2) within the mouse B2m region12 might exhibit abnormal expression patterns. In addition, the regulatory regions vary across each HLA allele e.g., HLA-A and HLA-B), suggesting the possibility that each HLA allele possesses distinct expression control mechanisms and produces different peptides28,29. HLAcI-MAC concatenating BACs that including native regulatory regions are expected to more accurately replicate physiological regulatory expression patterns in vitro and in vivo.
MACs serve as the platforms for the SIM system and have been transferred into multiple cell types, including pluripotent stem cells from humans, mice, rats, pigs, cows, and other animals30. Therefore, it is possible to concatenate regions scattered on different chromosomes into MACs using multiple BACs, which can then be transferred into various cell types. We anticipate that the BAC-SIM method will facilitate the generation of multifactorial cell/animal Tc models, accelerating investigations of complex life mechanisms and disease mechanisms, and the development of therapeutic interventions.
Methods
Ethics declarations
All animal experiments were approved by the Tottori University Animal Experimentation Committee (permit numbers: 20-Y-13, 21-Y-26, 22-Y-36). All experiments were performed in accordance with the ARRIVE guidelines and relevant local guidelines and regulations.
Construction of plasmids and BAC vectors
The SIM cassette (C1, C2, and C3) plasmids were constructed by modifying previously reported 1C, 2A, and 3A vectors6 used to insert GLVs into BACs via homologous recombination. The homology arms, comprising a SACB arm with linked multicloning site (SACB/MCS) plasmid and an F1 arm fragment, were synthesized by Eurofins (Huntsville, AL, USA), with the corresponding sequences of each shown in Supplementary Fig. S1. Next, the F1 arm fragment and the ampicillin resistance gene were ligated into the SACB/MCS plasmid, resulting in the construction of the F1/SACB/MCS plasmid. SIM cassette fragments from the 1C, 2A, 3A plasmids were ligated into the multiple cloning site of the F1/SACB/MCS plasmid, resulting in construction of the C1, C2, and C3 plasmids. The modification in BAC-containing E. coli was performed using the linearized C1, C2, and C3 plasmids, and the SIM cassettes were incorporated into BAC by homologous recombination via the SACB and F1 arms. The plasmids and BACs were extracted from E. coli using NucleoBond Xtra Midi and BAC kits (Macherey-Nagel, Nordrhein-Westfalen, Germany), respectively.
Cell culture
Hprt-deficient (Hprt−/−) CHO cells (JCRB0218) were obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). The MAC (MAC4) used here, which was generated in a previous study, contains a mouse centromere, GFP flanked by HS4 insulators, 5′HPRT, loxP, PGK-HygR, PGK-Puro, and telomeres7,15 (Fig. 1a). Hprt−/− CHO cells containing the MAC (CHO MAC) were cultured at 37 °C and 5% CO2 in Ham’s F12 medium (Fujifilm-Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA), 1% penicillin/streptomycin (PS; Fujifilm-Wako), and 500 µg/mL hygromycin B (Fujifilm-Wako). CHO MAC cells containing a reconstructed HPRT gene with the desired target genes were selected in Ham’s F12 medium supplemented with 10% FBS, 1% PS, and 2% HAT (Sigma-Aldrich).
Mouse embryonic fibroblasts (MEFs) were isolated from 13.5-day wild-type C57BL/6J embryos (CLEA, Tokyo, Japan). The MEFs were maintained at 37 °C and 5% CO2 in Dulbecco’s modified Eagle medium (DMEM; Fujifilm-Wako) supplemented with 10% FBS, and were treated with 10 µg/mL mitomycin C (Kyowa Kirin, Tokyo, Japan). The day before seeding mouse ES cells, feeder layers were created by seeding mitomycin C-treated MEFs at 2.0 × 105 cells per 6-cm dish coated with 0.1% gelatin and culturing in DMEM supplemented with 10% FBS.
The mouse ES cell line XO ES918, a subclone of TT2F (karyotype 39, XO), was cultured at 37 °C and 5% CO2 on the feeder layers of hygromycin-resistant MEFs in DMEM supplemented with 20% FBS, 1 mM sodium pyruvate (Gibco/Thermo Fisher Scientific; Waltham, MA, USA), 0.1 mM MEM non-essential amino acids solution (Gibco/Thermo Fisher Scientific), 0.1 mM 2-mercaptoethanol (Thermo Fisher Scientific), 2 mM L-glutamine (Gibco/Thermo Fisher Scientific), and 1,000 units/mL ESGRO Recombinant Mouse LIF Protein (Sigma-Aldrich).
Human sarcoma-derived HT1080 cells were obtained with permission from the American Type Culture Collection (ATCC, CCL-121) and were cultured at 37 °C and 5% CO2 in DMEM supplemented with 10% FBS and 1% PS.
Human T-cell leukemia-derived Jurkat#2H4 cells were cultured at 37 °C and 5% CO2 in RPMI-1640 with L-Glutamine and Phenol Red (Fujifilm-Wako) supplemented with 10% FBS and 1% PS.
Peripheral blood mononuclear cells (PBMCs) were cultured at 37 °C and 5% CO₂ in RPMI-1640 with L-glutamine and Phenol Red supplemented with 20% FBS, 2.75 mM 2-mercaptoethanol, 3 µg/mL PHA-M (Sigma-Aldrich), 3 µg/mL concanavalin A (Sigma-Aldrich), and 10 µg/mL LPS (Sigma-Aldrich).
In vitro IFN-γ stimulation
Mouse PBMCs in 100 µL of mouse peripheral blood and 1.0 × 106 Jurkat#2H4 cells were stimulated by adding recombinant mouse IFN-γ (BioLegend, San Diego, CA, USA) or recombinant human IFN-γ (BioLegend) to a final concentration of 1,000 units/mL or 50 ng/mL in 2 mL of culture medium in a 35-mm dish, respectively, for 48 h31. After culture, mouse peripheral blood was lysed for 15 min at room temperature using a 1× dilution of 10× RBC Lysis Buffer (Sony Biotechnology, San Jose, CA, USA) and mouse PBMCs were purified. FCM analysis was then performed as described below.
Mice
Chimeric mice were produced by injecting ES cells containing HLAcI-MAC into 8-cell stage embryos derived from Jcl: ICR mice (CLEA) which were then transferred into pseudopregnant Jcl: ICR females. Chimeric mice with 95–100% coat color chimerism were used for germline transmission.
Chimeric mice were mated to C57BL/6 N (B6N; CLEA) and the pups were confirmed to retain MAC by the GFP positivity. Subsequently, F2 and subsequent generations of mice were produced and maintained by mating GFP-positive pups of the same generation. The animal facility was specific-pathogen free, and experimental and control animals were kept in a controlled ambient temperature environment with a 12 h light/dark cycle. Mice were anesthetized with isoflurane for the collection of blood.
Loading of GLVs using the SIM system
Simultaneous integration of GLVs followed a previously reported method6 in which GLVs 1–3 were co-transfected with expression vectors encoding Cre recombinase, Bxb1 integrase, and PhiC31 integrase into CHO MAC cells using Lipofectamine LTX (Invitrogen/Thermo Fisher Scientific), in accordance with the manufacturer’s protocol. Briefly, GLVs 1, 2, and 3 (5 µg each) and the three recombinase/integrase expression vectors (500 ng each), were transfected into 70–80% confluent target cells in 10-cm dishes seeded in 10 mL PS-free medium on the day before transfection. Two mixtures were prepared and incubated at room temperature for 10 min: 3 mL Opti-MEM (Thermo Fisher Scientific) and 54 µL Lipofectamine LTX (lipo-premix); and 3 mL Opti-MEM and 16.5 µg DNA (DNA-premix). DNA-premix was then added dropwise to lipo-premix and incubated for 20 min before being added to cells. Prewashed cells were incubated with 4 mL of fresh Opti-MEM, followed by the addition of 6 mL of the DNA–lipid complex and incubated for up to 24 h at 37 °C and 5% CO2. Transfected cells were reseeded the next day, and 2% HAT and 500 µg/mL hygromycin B were added to the medium at 2 days post transfection. Fourteen days later, HAT-resistant colonies were picked and expanded for further analyses. To confirm the reconstitution of the HPRT gene, genomic PCR was performed using the primer sets shown in Supplementary Table S1 and the cycling conditions shown in Supplementary Table S2.
MMCT
HLAcI-MAC in CHO cells was transferred into mouse ES cells via MMCT, using a previously described method17. Briefly, the ES cells fused with CHO MAC-derived microcells were selected in medium containing 50 µg/mL hygromycin B for approximately 10 days to obtain hygromycin B-resistant and GFP-positive clones. The clones were identified by genomic PCR and FISH analyses.
Genomic PCR
Genomic DNA of cells and mouse tissues were extracted using the standard isopropanol precipitation method. Primer sets for the detection of recombination junctions and the targeted HLA and hB2M gene sites are shown in Supplementary Table S1. PCR was performed using KOD FX (TOYOBO, Osaka, Japan) or TaKaRa LA Taq (Takara Bio, Shiga, Japan). PCR cycling conditions are shown in Supplementary Table S2. PCR products were resolved by electrophoresis on agarose gels followed by staining with ethidium bromide. The GeneRuler 1 kb Plus DNA Ladder (Thermo Fisher Scientific) was used as a band size marker.
RT-PCR
Total RNA was isolated from cells grown to 80% confluency in 6-well plates using an RNeasy Plus Mini Kit (QIAGEN, Venlo, The Netherlands) following the manufacturer’s protocol. First-strand complementary DNA (cDNA) synthesis was performed using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific) following the manufacturer’s protocol. Primer sets for the detection of HLA and human B2M transcription32 are shown in Supplementary Table S1. PCR was performed using KOD One (TOYOBO). PCR cycling conditions are shown in Supplementary Table S2. Ribosomal protein L27 (Rpl27) was used as an internal control for human and mouse genes33.
FISH analysis
For preparation of chromosome specimens, cultured cells were treated with one of the following agents: 1.0 µg/mL colcemid, a cell cycle arrest reagent, for 2 h in CHO cells; 1 µg/mL metaphase arresting solution (Genial Helix, Chester, UK) for 90 min in mouse ES cells; or 0.5 µg/mL metaphase arresting solution for 2 h in mouse lymphocytes. Next, the cells were treated with 0.075 M KCl for 15 min and fixed with Carnoy’s solution (3:1 ratio of methanol: acetic acid by volume). Chromosome specimens were prepared by spreading fixed cells onto glass slides, followed by FISH staining using previously reported methods34. To prepare specimens for analyzing the stability of HLAcI-MAC in various mouse tissues, tissue samples were sectioned, stamped onto the glass slides, and fixed with Carnoy’s solution, and subsequently subjected to FISH staining. The following target DNA: probe combinations were utilized as previously described9: mouse DNA: mouse Cot-1 (Invitrogen/Thermo Fisher Scientific), human DNA: human Cot-1 (Invitrogen/Thermo Fisher Scientific), HLA-A:CH501-309N1, HLA-B/C: CH501-248L24, hB2M:RP11-292P13, and mouse minor satellite: mouse minor satellite DNA. Each DNA probe was labeled with biotin or digoxigenin. Background hybridization by the human BAC sequence probe was suppressed by adding a 5 times concentration of human Cot-1 DNA to the probe (described as “5× suppression” in the Supplementary Table legend). Whole chromosomes were indicated using staining with 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). Metaphase images were captured using an Axio Imager Z2 (Carl Zeiss, Jena, Germany) and Metafer software (Meta Systems, Altlussheim, Germany), and analyzed using ISIS software (Meta Systems). Interphase images were captured using an Axio Imager Z2 and analyzed using ISIS software.
FCM analysis
To evaluate the phenotype of HLAcI Tc mice, they were compared with roughly age-matched wild-type mice of a similar genetic background. Mouse PBMCs were isolated from adult male and female mice using aseptic procedures, and lysed for 15 min at room temperature using a 1× dilution of 10× RBC Lysis Buffer (Sony Biotechnology). CHO and mouse ES cells, without or without carriage of HLAcI-MAC, were also collected. To analyze the expression of cell-surface proteins, 5.0 × 105 cells per sample were washed and then resuspended in 100 µL staining buffer (phosphate-buffered saline with 5% FBS), stained with the antibodies shown in Supplementary Table S4 for 30 min on ice, and washed. Viability was assessed using 0.5 µg/mL DAPI, and cells were analyzed using a CytoFLEX S (Beckman Coulter, Brea, CA, USA).
Next-generation sequencing (NGS)
Whole-genome sequencing was performed in Hokkaido System Science Co., Ltd. (Sapporo, Hokkaido, Japan). DNA-seq libraries were prepared using a PCR-free method and sequenced on an Illumina NovaSeq 6000 platform with 150 bp paired-end reads, yielding approximately 90 GB of data per sample. Sequence analysis was conducted using the CLC Genomics Workbench software (QIAGEN).
Statistical analysis
Statistical significance was determined by the paired t-test. P-values of < 0.05 were considered significant.
Data availability
All data generated or analyzed during this study are included in this article and its supplementary information files and all whole-genome sequencing raw read data were deposited at the DDBJ Sequence Read Archive under the accession no. PRJDB20537.
References
Li, F. et al. Generation and expression analysis of BAC humanized mice carrying HLA-DP401 haplotype. Anim. Models Exp. Med. 4, 116–128 (2021).
Scheikl, T., Pignolet, B., Mars, L. T. & Liblau, R. S. Transgenic mouse models of multiple sclerosis. Cell. Mol. Life Sci. 67, 4011–4034 (2010).
Tian, S., Ye, T. & Cheng, X. The behavioral, pathological and therapeutic features of the triple Transgenic alzheimer’s disease (3 × Tg-AD) mouse model strain. Exp. Neurol. 368, 114505 (2023).
Brosh, R. et al. A versatile platform for locus-scale genome rewriting and verification. Proc. Natl. Acad. Sci. 118, e2023952118 (2021).
Blanch-Asensio, A. et al. STRAIGHT-IN enables high-throughput targeting of large DNA payloads in human pluripotent stem cells. Cell. Rep. Methods. 2, 100300 (2022).
Suzuki, T., Kazuki, Y., Oshimura, M. & Hara, T. A novel system for simultaneous or sequential integration of multiple Gene-Loading vectors into a defined site of a human artificial chromosome. PLOS ONE. 9, e110404 (2014).
Takiguchi, M. et al. A novel and stable mouse artificial chromosome vector. ACS Synth. Biol. 3, 903–914 (2014).
Shimoya, K. et al. Mice carrying the full-length human Immunoglobulin loci produce antigen-specific human antibodies with the lambda light chain. iScience 27, 111258 (2024).
Kazuki, Y. et al. A non-mosaic transchromosomic mouse model of Down syndrome carrying the long arm of human chromosome 21. eLife. 9, e56223 (2020).
Satofuka, H. et al. Efficient human-like antibody repertoire and Hybridoma production in trans-chromosomic mice carrying megabase-sized human Immunoglobulin loci. Nat. Commun. 13, 1841 (2022).
Romero-Castillo, L. et al. Human MHC class II and invariant chain Knock‐in mice mimic rheumatoid arthritis with allele restriction in immune response and arthritis association. Adv. Sci. 11, 2401513 (2024).
Harada, N. et al. Generation of a novel HLA class I Transgenic mouse model carrying a Knock-in mutation at the β2-Microglobulin locus. J. Immunol. 198, 516–527 (2017).
Dewan, A. E., Koentgen, F., Johannesen, M. K., Du Pre, M. F. & Sollid, L. M. Generation of an HLA-DQ2.5 Knock-In mouse. ImmunoHorizons F. 5, 25–32 (2021).
Akram, A. & Inman, R. D. Co-expression of HLA-B7 and HLA-B27 alleles is associated with B7-restricted immunodominant responses following influenza infection. Eur. J. Immunol. 43, 3254–3267 (2013).
Uno, N. et al. Development of a safeguard system using an episomal mammalian artificial chromosome for gene and cell therapy. Mol. Ther. - Nucleic Acids. 4, e272 (2015).
Kievits, F. & Ivanyi, P. Monomorphic Anti-HLA monoclonai antibody (W6/32) recognizes polymorphic H- 2 Heavy-Chain determinants exposed by association with bovine or human but not murine B2-Microglobulin. Hum. Immunol. 20, 115–126 (1987).
Suzuki, T., Kazuki, Y., Oshimura, M. & Hara, T. Highly efficient transfer of chromosomes to a broad range of target cells using Chinese hamster ovary cells expressing murine leukemia Virus-Derived envelope proteins. PLOS ONE. 11, e0157187 (2016).
Tomizuka, K. et al. Functional expression and germline transmission of a human chromosome fragment in chimaeric mice. Nat. Genet. 16, 133–143 (1997).
Keskinen, P., Ronni, T., Matikainen, S., Lehtonen, A. & Julkunen, I. Regulation of HLA class I and II expression by interferons and influenza A virus in human peripheral blood mononuclear cells. Immunology 91, 421–429 (1997).
Sznarkowska, A., Mikac, S. & Pilch, M. MHC class I regulation: the origin perspective. Cancers 12, 1155 (2020).
Macdonald, L. E. et al. Precise and in situ genetic humanization of 6 Mb of mouse immunoglobulin genes. Proc. Natl. Acad. Sci. 111, 5147–5152 (2014).
Lee, E. C. et al. Complete humanization of the mouse Immunoglobulin loci enables efficient therapeutic antibody discovery. Nat. Biotechnol. 32, 356–363 (2014).
Hasegawa, Y. et al. Generating a Transgenic mouse line stably expressing human MHC surface antigen from a HAC carrying multiple genomic BACs. Chromosoma 124, 107–118 (2015).
Yamaguchi, S. et al. A method for producing Transgenic cells using a Multi-Integrase system on a human artificial chromosome vector. PLoS ONE. 6, e17267 (2011).
Barkal, A. A. et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat. Immunol. 19, 76–84 (2018).
Perarnau, B. M., Gillet, A. C., Hakem, R., Barad, M. & Lemonnier, F. A. Human beta 2-microglobulin specifically enhances cell-surface expression of HLA class I molecules in transfected murine cells. J. Immunol. 141, 1383–1389 (1988).
Wang, F. et al. Downregulating human leucocyte antigens on mesenchymal stromal cells by epigenetically repressing a β2-microglobulin super-enhancer. Nat. Biomed. Eng. 8, 1682–1699 (2024).
van den Elsen, P. J. Expression regulation of major histocompatibility complex class I and class II encoding genes. Front. Immunol. 2(48), 1–9 (2011).
Di, D., Nunes, J. M., Jiang, W. & Sanchez-Mazas, A. Like wings of a bird: functional divergence and complementarity between HLA-A and HLA-B molecules. Mol. Biol. Evol. 38, 1580–1594 (2021).
Moriwaki, T., Abe, S., Oshimura, M. & Kazuki, Y. Transchromosomic technology for genomically humanized animals. Exp. Cell. Res. 390, 111914 (2020).
Jung, V. et al. Human chromosomes 6 and 21 are required for sensitivity to human interferon gamma. Proc Natl. Acad. Sci. USA. 84, 4151−4155 (1987).
Wang, Y. et al. The Plasticity of Mesenchymal Stem Cells in Regulating Surface HLA-I. iScience. 15, 66–78 (2019).
Campbell, A. E. et al. NuRD and CAF-1-mediated Silencing of the D4Z4 array is modulated by DUX4-induced MBD3L proteins. eLife. 7, e31023 (2018).
Miyamoto, H. et al. Rapid human genomic DNA cloning into mouse artificial chromosome via direct chromosome transfer from human iPSC and CRISPR/Cas9-mediated translocation. Nucleic Acids Res. 52, 1498–1511 (2024).
Acknowledgements
We thank M. Morimura and M. Hirose of Tottori University for assistance with analyzing obtained clones; Y. Sumida, E. Kaneda, K. Yoshida, M. Fukino, and A. Ashiba for assistance with generating and maintaining HLAcI Tc mice; and Dr. Y. Nakayama of Tottori University for assistance with FCM experiments. We are also grateful to Dr. H. Kugoh and Dr. M. Hiratsuka of Tottori University for critical discussions. Finally, we thank Michelle Kahmeyer-Gabbe, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.This study was performed in part at the Tottori Bio Frontier managed by Tottori Prefecture, and was partially supported by the Japan Agency for Medical Research and Development (AMED) under Grant Numbers JP25ama121046 (Y.K.), JP25gm1610006 (Y.K., K.T.), JP25bm1123038 (Y.K., N.U.), JP25gm0010010 (Y.K.), and; the Exploratory Research Center on Life and Living Systems (ExCELLS) program under Grant Number 21-101 (Y.K.); and JST CREST under Grant Number JPMJCR18S4 (Y.K.).
Author information
Authors and Affiliations
Contributions
N.K., T.M., N.U., S.A., T.S., K.T., and Y.K. planned the study; N.K., K.Y., M.S, Y.N., and T.E. performed vector construction; N.K., M.S., Y.M., W.N., R.Y., and Y.M. performed cell culture and mouse production experiments; N.K. and K.K. performed FISH analysis; N.K., T.M., W.N., and R.Y. performed FCM analysis; N.K., T.M., N.U., T.S, S.A, and Y.K. contributed to the analysis and discussion of the data. S.M. performed NGS analysis. N.K. wrote the manuscript, with contributions from each author, and T.M., N.U., S.A., T.S., K.Y., K.T., and Y.K. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kishima, N., Moriwaki, T., Uno, N. et al. Generation of transchromosomic mice harboring HLA-A/B/C and human B2M via mouse artificial chromosome and triple BAC integration. Sci Rep 15, 27852 (2025). https://doi.org/10.1038/s41598-025-13138-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-13138-5