Abstract
This multicenter imaging study aimed to establish and validate automated radiomics models predicting therapy response (TR) and minimal residual response (MRD) in newly diagnosed multiple myeloma (MM) from baseline MRI. Retrospectively, 118 MM patients from the GMMG-HD7 trial (EudraCT: 2017-004768-37) with data on TR and/or MRD after induction therapy and baseline MRI were included. Data were split by center into a training set (center 1–2; n = 79) and a test set (center 3–10; n = 39). TR was classified as very good partial response or better versus other. An in-house developed nnU-Net was used to automatically segment pelvic bone marrow for the subsequent extraction of 245 radiomics features and piriformis muscle for normalization. Random forest classifiers were trained using radiomics features only (I), radiomics features with additional confounders (II) or myeloma-relevant clinical features (III), or only clinical features (IV) to predict TR or MRD status. The area under the receiver operating characteristic curve (AUROC) was calculated to assess prediction performance. The prediction model using only radiomics features (I) showed the highest predictive performance for TR on the test set with an AUROC of 0.70. AUROC values for radiomics-based prediction of the MRD status (I-III) ranged from 0.54 to 0.52. In conclusion, our study demonstrated the potential of automated radiomics models from baseline MRI to non-invasively predict TR in MM on an independent, multicentric test set.
Similar content being viewed by others

Introduction
The clinical presentation of multiple myeloma (MM) can vary widely in terms of tumor burden, symptoms, tumor genetics, and outcome1. Newly diagnosed multiple myeloma (NDMM), as defined by the International Myeloma Working Group (IMWG), typically warrants therapy2. Eligible NDMM patients are often recommended intensive, high-dose treatment regimens followed by autologous transplantation3. Therapy response (TR), and occasionally the possible presence of minimal residual disease (MRD), is assessed after the administration of induction therapy4 and serve as early predictors for long-term outcome in MM5,6,7,8. TR is usually categorized based on serum or urine monoclonal protein (M-protein) or the serum free light chain ratio (SFLCR), and plasma cell infiltration (PCI) of the bone marrow (BM) according to the IMWG criteria. Next-generation sequencing (NGS) and next-generation flow (NGF) are used to determine MRD status and provide an estimate of the number of residual myeloma cells with a required minimum sensitivity of 1 in 105 nucleated cells4. Providing an accurate prognostic assessment for NDMM patients remains challenging1despite recent advances in treatment and overall patient outcome9,10. Non-invasive prediction of TR and MRD at baseline could potentially improve clinical decision making.
Whole-body MRI (WBMRI) provides comprehensive information on the BM status in MM11capturing the spatial heterogeneity of tumor manifestation12,13. Therefore, WBMRI allows the detection of diffuse infiltration pattern or focal lesions (FL)11. These features are of prognostic value, as shown for the presence of diffuse infiltration14,15 and FLs16,17 in NDMM, as well as for the detection of FLs during treatment assessment in MM18,19. The IMWG recommends WBMRI for baseline imaging20and both TR and MRD assessment can be complemented by WBMRI, which allows tracking of new and residual FLs21. Radiomic features encode high-dimensional image characteristics based on shape, signal intensity, or texture of the volume of interest22. Radiomics features, either on their own or in combination with clinical data, can be utilized to predict outcomes or histologic results and provide additional information about tumor characteristics23,24.
Previous radiomics studies in MM have provided valuable insights into predicting TR25,26 and MRD27 from baseline MRI. However, many of these important contributions have been based on single-center cohorts and have not yet included external validation, which may limit their generalizability and hinder clinical applicability. In addition, the manual or semi-automatic segmentation method of most existing pipelines introduces inter-observer variability and further constrains scalability, particularly in resource-limited settings. A fully automated, generalizable radiomics pipeline may overcome these limitations by improving reproducibility, reducing manual workload, and supporting large-scale implementation in clinical settings.
The aim of this study was to establish a fully automated machine learning model capable of predicting TR and MRD from baseline MRI, complemented by baseline clinical features, and to test the developed models on multicenter imaging data.
Materials and methods
For this retrospective multicenter study, appropriate ethical approval from the institutional review board was obtained (S-537/2020, clinical trial number: not applicable) with informed consent being waived. This study was performed in accordance with the Declaration of Helsinki and adhered to the relevant guidelines and regulations of our institution.
Study cohort
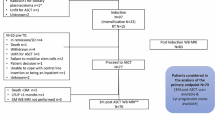
Unenhanced WBMRIs and clinical data were acquired within the phase 3 GMMG-HD7 trial (EudraCT: 2017-004768-37) of the German Speaking Myeloma Multicenter Group (GMMG) between 2018 and 2020 as well as 2021, respectively. Detailed inclusion criteria have been reported by Goldschmidt et al. in 202228. In addition, inclusion criteria in this study were a complete pelvis MRI performed before treatment start and availability of data on either or both MRD and TR status after induction. The GMMG-HD7 cohort with unenhanced baseline pelvis MRIs and corresponding clinical data was split by the location of the imaging centers. Data from centers 1 and 2 were used to train the machine learning classification models. Data from the centers 3–10 were included in the independent, external test set, which provided very heterogeneous testing conditions due to the heterogenous image acquisition settings of the included imaging data included. The respective flow chart is shown in Fig. 1. The imaging data has been used partly within different study cohorts in other studies from our institution28,29,30,31.
Flow chart. Baseline T1-weighted turbo spin echo sequence (T1w tse) MRIs with corresponding information on therapy response and/or minimal residual disease status after induction therapy from center 1 and 2 of the GMMG-HD7 trial were used in the training set. The independent, external, multicentric test set included baseline MRIs from center 3–10 with MRI scanners from various vendors and with different protocols.
Imaging
In this study, unenhanced coronal T1w turbo spin echo pelvis MRIs were used, which were acquired within the baseline WBMRIs up to 6 weeks prior to the start of therapy. Imaging was performed at different MRI scanners with diverse sequences from several vendors at multiple imaging centers. The MRI acquisition parameters can be found in Supplementary Table S1.
Clinical data collection
Clinical data used in this study was collected within the multicenter, randomized, active-controlled, phase 3 GMMG-HD7 trial with MRD status as the primary endpoint of the first part of the study, which has been reported elsewhere28. Baseline clinical data and MRIs were collected at study entry before therapy. Patients were assigned randomly to undergo three cycles of induction therapy, either with isatuximab in addition to lenalidomide, bortezomib, and dexamethasone or with lenalidomide, bortezomib, and dexamethasone alone. Treatment was assessed within 7 days after completion of the induction therapy using the IMWG criteria for TR and MRD status. TR included complete response (CR), very good partial response (VGPR), partial response, minimal response, stable disease, progressive disease4 and near-complete response (nCR), which was established as an additional class. MRD was evaluated using multiparametric NGF at a sensitivity cutoff of 1 tumor cell × 10⁵ nucleated cells28. Relevant clinical parameter collected before treatment included patient’s age, sex, body mass index (BMI), M-protein, SFLC-ratio, beta2-microglobulin, calcium levels, creatinine, lactate dehydrogenase (LDH), serum albumin, serum total protein, hemoglobulin, PCI, major histocompatibility complex (MHC) type, del(17p), gain(1q), t(4;14), and the treatment arm2,28,32,33. PCI was defined as the higher value of the histological or cytological derived PCI percentage in alignment with IMWG recommendations2. Cytogenetic aberrations were marked as clinical parameter by analogy to the R2-ISS criteria33. All clinical parameters were used as individual clinical features. Age, BMI and the treatment arm were marked as clinical confounders, since age and BMI are known to influence the BM signal34,35. Within the treatment regime of the GMMG-HD7 study, Isatuximab was associated with an elevated proportion of MRD-negative patients28.
Algorithmic architecture
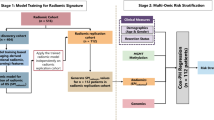
Figure 2 displays the algorithmic architecture of the study. A previously in-house-developed and externally validated nnU-Net segmentation algorithm was used to automatically segment the left and right pelvic BM, excluding the cortical bone, and the medial portion of the piriformis muscle on T1w images31.
Algorithmic concept of the study. In step I, the bone marrow of the pelvis and the piriformis muscle are automatically being segmented in the coronal T1-weighted turbo spin echo sequence by a previously trained nnU-Net and the bone marrow of each hip bone is individually labeled. Images are normalized to the mean signal intensity of the piriformis muscle and geometrically resampled. In step II, radiomics intensity and texture features are calculated from the pelvic bone marrow. In step III, radiomics and clinical features are utilized in machine learning models to predict therapy response and minimal residual disease status.
Prior to feature extraction, all MR images were resampled to a uniform voxel spacing that corresponded to the acquisition protocol of center 1, which served as the reference center due to its high image quality, representative acquisition parameters, and the largest group of patients having received a scan at center 1. This resampling step was applied to both training and external test datasets to reduce variability in spatial resolution across scanners and institutions. Following resampling, all images in both the test and training cohort underwent subject-specific intensity normalization using the mean signal intensity of the bilateral piriformis muscle tissue in each scan, following prior work31. This biologically grounded normalization approach accounts for inter-scanner variability by scaling each patient’s bone marrow signal to the internal reference tissue. Muscle-based normalization has been shown to improve inter-scanner comparability36 and enhances the repeatability and reproducibility of radiomics features across varying acquisition parameters37. No additional batch effect correction or statistical harmonization was applied to avoid potential information leakage between the training and independent test sets.
Radiomics first-order and texture features were calculated with the publicly available software MITK Phenotyping38. Radiomics features were computed individually for each hip bone, and the mean value was used for further analysis. Radiomics features with zero variance were excluded. Clinical features were also included in this study in alignment with the Radiomics Quality Score39. Missing clinical values were encoded as −1. Categorial variables were transformed using one-hot encoding40. The primary objective in this study was to predict TR, which was binarily classified as CR, nCR or VGPR versus worse response categories of therapy assessment, in line with the categorization within the GMMG-HD7 trial28. MRD negativity was the second target parameter to be predicted independently from TR. Four different random forest classifiers (RFC) were trained to predict each of the two target parameters MRD status and TR based on radiomics features only (I), radiomics features and clinical confounders (II), radiomics and clinical features (III), and clinical features only (IV). While prior work has identified subsets of radiomics features with high reproducibility across scanners37 all radiomics features were used in alignment with recent findings showing that RFCs trained on all features achieved superior performance for MRI-based prediction of clinical parameters in MM41. The RFC was chosen for its robustness in handling high-dimensional, heterogeneous data and its intrinsic ability to reduce overfitting by combining multiple trees in an ensemble, training each tree on a random subset of the data, and considering only a random subset of features for splitting at each node, which limits the ability of any single tree to overfit the data. This approach not only enhances generalizability but also provides built-in feature importance metrics, improving model interpretability. For the RFCs, default parameters were used with n_estimators = 10,000 and random_state = 0. All machine learning modeling was performed with Python 3.11.6 (Python Software Foundation, Wilmington, Delaware, USA), module scikit-learn 1.1.340. The prediction models were tested on the external, multicentric test set. The METhodological RadiomICs Score (METRICS)42 and the CheckList for EvaluAtion of Radiomics research (CLEAR)43 results are reported in Supplementary Tables S2 and S3.
Statistical analysis
The area under the receiver operating characteristic (AUROC) with a 95% confidence interval (95%-CI) was calculated to assess the performance of each prediction model. 95%-CIs were calculated following DeLong et al.44. The Youden index was calculated to define the optimal cutoff to calculate sensitivity, specificity, and F1 score. P <.05 were considered statistically significant. The Gini feature importance was used to report the relative influence of a radiomics feature on the prediction model. The statistical analysis was performed with Python version 3.11.6, modules scikit-learn40 matplotlib version 3.8.345 and seaborn version 0.13.246.
Results
Study cohort
Patient characteristics and MM-related parameters are shown for the training set and test set in Table 1. One hundred eighteen baseline MRIs of 118 patients from 10 imaging centers enrolled in the GMMG-HD7 trial were included. The training set comprised 79 MRIs of 79 patients from 2 centers. 39 MRIs of 39 patients from 8 different imaging centers were included in the test set (Fig. 1). One hundred seventeen MRIs had corresponding information on TR (training set: 78, test set: 39) and 114 MRIs had corresponding information on MRD status (training set: 75, test set: 39). Ninety-five first order and 150 texture radiomics features were used as an input for the RFCs in the prediction models (Supplementary Table S4). For some MRIs, corresponding clinical information was not available (no information: m-protein: 40/118, del17p: 4/118 and t(4;14): 4/118). There were no significant differences in age, BMI, and treatment arm between the training set and the test sets (p ≥.38).
Prediction of therapy response
Four models were trained for the prediction of binary classified TR. The performance metrics and respective ROC curves are shown in Table 2; Fig. 3. The model based on radiomics features only (I) achieved the best prediction performance for TR on the test set with an AUROC of 0.70. With radiomics models that also included confounders (II) or clinical features (III), the prediction performance was no better than the radiomics features only (I) model (both models with an AUROC of 0.69). For all models that included radiomics features (I-III), the prediction performance in AUROC value for TR was better than for the model using clinical features exclusively (IV; AUROC of 0.63). However, this tendency was not statistically significant (p ≥.68).
Predictive performance of the different machine learning models. (a) ROC values are displayed for the prediction of therapy response of the four different models (I–IV). (b) ROC values are shown for the prediction of minimal residual disease status of the four separate machine learning models (I–IV).
The heatmap presented in Fig. 4 color-encodes the clinical features and the 15 most important radiomics features of the pelvic BM for the radiomics features only model (I). No striking trend can be observed comparing radiomic signatures based on TR binarily classified. Radiomics features contributing most to the respective RFC for TR prediction included features that encode intensity range, maximum and minimum intensity: “first order numeric: maximum”, “first order numeric: range”, “first order histogram: maximum value” and “first order histogram: range value”.
Feature heatmap for the training and test set. Radiomic and clinical signatures of study subjects (given in columns) ordered by TR status in the training set (a) and test set (b). The 15 most important radiomics features are listed from top to bottom for the radiomics only model (I). The clinical features sex, therapy arm, cytogenetic aberration, and MHC complex are not included due to their categorial configuration. Clinical features with no information have been encoded white. (c) The 15 most important individual radiomics features are reported for the radiomics only model (I) by Gini feature importance calculations. (d) The color-coded z-score normalization of the clinical and radiomics features are given with standard deviations between − 3 and + 3.
Prediction of MRD status
For the prediction task MRD negativity, the performance for all radiomics based models (I–III) ranged from 0.52 to 0.54 for the test set. However, models I to III performed worse in absolute terms than the models to predict TR (Table 2; Fig. 3). The prediction performance of the model using clinical features only (IV) resulted in an AUROC of 0.35.
Discussion
In this study, we developed multiple machine learning models to predict both MRD and TR status based on radiomics features derived from baseline MRI and baseline clinical features. We subsequently investigated their performance on an external, multicentric data set. The algorithmic architecture leveraged a recently presented, nnU-Net-based automated segmentation tool capable of accurate, fully automated pelvic BM segmentation in T1w MRI31 allowing a completely automated workflow for radiomics-based prediction. Our results, based on the data from 10 different imaging centers, highlight the predictive value of comprehensive radiomics-encoded information from baseline MRI on TR in MM. The models showed robust performance on external test data, with AUROC values ranging from 0.69 to 0.70 across three radiomics-based model configurations for TR prediction (I-III). These data suggest that predictive radiomics models on baseline MRI in NDMM may provide additional information for clinical decision-making. However, no relevant predictive power could be shown for any of the machine learning models on MRD status.
In a previous study investigating radiomic models for the prediction of TR from baseline MRI, six different ML models were trained with a reported AUROC between 0.80 and 0.89 on an internal test set25. Wu and colleagues reported on a radiomics nomogram incorporating the ISS as an independent predictor for the prediction of TR from baseline MRI in a cohort of 123 MM patients, which resulted in an AUROC value of 0.87 compared to a radiomics-only model with an AUROC of 0.86 in their internal test set26. Importantly, these prior models were validated internally only, leaving it unclear whether and how the models would generalize in a multicentric clinical application24,47. In contrast, our study evaluated prediction performance on a multivendor, multiscanner test set from eight independent centers, providing a realistic estimate of generalizability for potential large-scale clinical deployment.
Also, all earlier models require time-intensive manual segmentation of the lumbar vertebrae before the radiomics features can be extracted, which further undermines future clinical application of said models. In contrast, our algorithmic concept incorporated a previously established nnU-net with a pelvic BM segmentation accuracy equal to that achieved by radiologists31. Hence, the all-automated algorithmic concept would allow to implement the presented prediction models in a clinical workflow and imaging platforms to enable scalable, routine use. In the future, patients predicted to have a poor treatment response may benefit from early, personalized cell therapies tailored to their specific disease profile. Reliable baseline prediction of treatment response may therefore contribute to more individualized therapy planning and ultimately improve patient prognosis.
In contrast to TR, our models failed to demonstrate predictive performance for MRD status. This finding differs from study results by Xiong et al., which included 83 MM patients and reported a strong prediction performance with AUROC values of up to 0.84 for their internal test set27. However, as with other previous studies, their model lacked external validation and relied on labor-intensive manual segmentation. Moreover, our multicentric design introduces real-world imaging heterogeneity, possibly contributing to more conservative—but clinically relevant—performance estimates.
The difference in predictive performance between TR and MRD may stem from the distinct clinical definitions of the prediction targets. IMWG standard criteria for TR are primarily based on clinical and serological biomarkers, such as m-protein levels and PCI4. Radiomics models have previously demonstrated their ability to capture MRI-derived information predictive for these biomarkers, particularly PCI31 which could make TR a more accessible and imaging-responsive prediction target. In contrast, MRD assessment allows to detect residual clonal plasma cells with high sensitivity, often revealing disease persistence in patients classified with CR status4. MRD negativity thus might represent deeper biological response that may no longer present with imaging features at baseline that prediction models are able to harness automatically, particularly when localized tumor burden is minimal or falls below the threshold of radiological detection. Our findings may therefore underscore the complexity of achieving MRD negativity and the need to include functional imaging modalities in future studies, such as Positron Emission Tomography or diffusion-weighted imaging.
With a median follow up time of 18 weeks from start to end of induction therapy in the underlying GMMG-HD7 study, our study subsequently focused on the prediction of short-term treatment assessment. Baseline clinical parameters associated with MRD and TR after induction therapy as primary/secondary endpoints are limited and have been reported to be only tumor genetics for MRD within the data of the phase 3 GMMG-HD7 trial used in the presented study28 and TR48.
Our findings underscore the limited performance of prediction models for TR or MRD after induction, if they solely incorporate clinical features. This finding emphasizes the potential of baseline prediction models that are based on, or are complemented by MRI information: Wu et al. reported a significantly worse predictive ability of a purely clinical model for TR, compared to a model that includes radiomics features26. For the MRD prediction task, Xiong et al. also found that only the PCI of the BM was associated with MRD status in a univariate analysis, and that a model combining this clinical feature with radiomics features from baseline MRI would show significantly better AUROC results for the prediction of the MRD status than the PCI alone27. Following expert recommendation39, we included 3 confounders (II) or 16 clinical features (III) in addition to the radiomics features in the prediction models, which lead to a similar prediction performance like the models that relied on radiomics features only (I). These results align with the presented recent studies, indicating that the inclusion of baseline clinical features into radiomics models did not substantially improve the predictive performance for MRD or TR status26,27,31.
Our study had several limitations, including its retrospective design. Even though the multicentric data were collected within a randomized controlled phase 3 trial, only a part of the participating patients had received a baseline MRI, resulting in a limited sample size. Radiomics features were extracted from T1w images only, and including information of additional MRI sequences may improve future prediction models. Another limitation is that the volume of interest was limited to the pelvic BM, which would not allow capturing the heterogeneity of MM tumor load distributed over the complete BM and as well as lesions. Recent efforts have explored automated segmentation of the whole-body skeleton29,49,50 or diffusion-weighted imaging30 which may provide additional information for prediction models through radiomics analysis.
In conclusion, our study showed that automated machine learning models based on radiomics features from baseline MRI have relevant predictive value regarding TR in MM. An independent, multicentric test set comprising data from 8 different centers was used to assess prediction performance. However, the performance of models predicting MRD status was negligible. These results suggest that non-invasive prediction of TR before therapy start could be implementable in the clinical workflow and might improve clinical decision-making. Further studies are warranted to prospectively confirm presented results and to explore whether prediction models can guide therapeutic decisions.
Data availability
The data supporting this study’s findings are derived from the ongoing GMMG-HD7 phase-3 trial containing protected patient health information and is subject to EU data protection regulations. Therefore, they are not publicly available. However, data from published portions of the trial may be available upon reasonable request through the corresponding author (FB; fabian.bauer@dkfz-heidelberg.de) to the principal investigator and the board of directors of the GMMG study group, subject to an approved data-sharing agreement and compliance with ethical guidelines.
References
Kumar, S. K. et al. Multiple myeloma. Nat. Rev. Dis. Primers. 3, 17046. https://doi.org/10.1038/nrdp.2017.46 (2017).
Rajkumar, S. V. et al. International myeloma working group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 15, e538–548. https://doi.org/10.1016/S1470-2045(14)70442-5 (2014).
Dimopoulos, M. A. et al. Multiple myeloma: EHA-ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 32, 309–322. https://doi.org/10.1016/j.annonc.2020.11.014 (2021).
Kumar, S. et al. International myeloma working group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 17, e328–e346. https://doi.org/10.1016/S1470-2045(16)30206-6 (2016).
van de Velde, H. J. et al. Complete response correlates with long-term survival and progression-free survival in high-dose therapy in multiple myeloma. Haematologica 92, 1399–1406. https://doi.org/10.3324/haematol.11534 (2007).
Chanan-Khan, A. A. & Giralt, S. Importance of achieving a complete response in multiple myeloma, and the impact of novel agents. J. Clin. Oncol. 28, 2612–2624. https://doi.org/10.1200/jco.2009.25.4250 (2010).
Munshi, N. C. et al. Association of minimal residual disease with superior survival outcomes in patients with multiple myeloma: A Meta-analysis. JAMA Oncol. 3, 28–35. https://doi.org/10.1001/jamaoncol.2016.3160 (2017).
Rawstron, A. C. et al. Minimal residual disease assessed by multiparameter flow cytometry in multiple myeloma: impact on outcome in the medical research Council myeloma IX study. J. Clin. Oncol. 31, 2540–2547. https://doi.org/10.1200/jco.2012.46.2119 (2013).
Binder, M. et al. Mortality trends in multiple myeloma after the introduction of novel therapies in the united States. Leukemia 36, 801–808. https://doi.org/10.1038/s41375-021-01453-5 (2022).
McCarthy, P. L. et al. Lenalidomide maintenance after autologous Stem-Cell transplantation in newly diagnosed multiple myeloma: A Meta-Analysis. J. Clin. Oncol. 35, 3279–3289. https://doi.org/10.1200/JCO.2017.72.6679 (2017).
Dimopoulos, M. A. et al. Role of magnetic resonance imaging in the management of patients with multiple myeloma: a consensus statement. J. Clin. Oncol. 33, 657–664. https://doi.org/10.1200/JCO.2014.57.9961 (2015).
Latifoltojar, A., Boyd, K., Riddell, A., Kaiser, M. & Messiou, C. Characterising Spatial heterogeneity of multiple myeloma in high resolution by whole body magnetic resonance imaging: towards macro-phenotype driven patient management. Magn. Reson. Imaging. 75, 60–64. https://doi.org/10.1016/j.mri.2020.10.005 (2021).
Bauer, F., Sauer, S., Weinhold, N., Delorme, S. & Wennmann, M. Smoldering) multiple myeloma: mismatch between tumor load estimated from bone marrow biopsy at Iliac crest and tumor load shown by MRI. Skeletal Radiol. 52, 2513–2518. https://doi.org/10.1007/s00256-023-04383-8 (2023).
Moulopoulos, L. A. et al. Diffuse pattern of bone marrow involvement on magnetic resonance imaging is associated with high risk cytogenetics and poor outcome in newly diagnosed, symptomatic patients with multiple myeloma: a single center experience on 228 patients. Am. J. Hematol. 87, 861–864. https://doi.org/10.1002/ajh.23258 (2012).
Moulopoulos, L. A. et al. Prognostic significance of magnetic resonance imaging of bone marrow in previously untreated patients with multiple myeloma. Ann. Oncol. 16, 1824–1828. https://doi.org/10.1093/annonc/mdi362 (2005).
Mai, E. K. et al. A magnetic resonance imaging-based prognostic scoring system to predict outcome in transplant-eligible patients with multiple myeloma. Haematologica 100, 818–825. https://doi.org/10.3324/haematol.2015.124115 (2015).
Walker, R. et al. Magnetic resonance imaging in multiple myeloma: diagnostic and clinical implications. J. Clin. Oncol. 25, 1121–1128. https://doi.org/10.1200/JCO.2006.08.5803 (2007).
Hillengass, J. et al. Changes in magnetic resonance imaging before and after autologous stem cell transplantation correlate with response and survival in multiple myeloma. Haematologica 97, 1757–1760. https://doi.org/10.3324/haematol.2012.065359 (2012).
Moreau, P. et al. Prospective evaluation of magnetic resonance imaging and [(18)F]Fluorodeoxyglucose positron emission tomography-Computed tomography at diagnosis and before maintenance therapy in symptomatic patients with multiple myeloma included in the IFM/DFCI 2009 trial: results of the IMAJEM study. J. Clin. Oncol. 35, 2911–2918. https://doi.org/10.1200/JCO.2017.72.2975 (2017).
Hillengass, J. et al. International myeloma working group consensus recommendations on imaging in monoclonal plasma cell disorders. Lancet Oncol. 20, e302–e312. https://doi.org/10.1016/S1470-2045(19)30309-2 (2019).
Lecouvet, F. E. et al. Imaging of treatment response and minimal residual disease in multiple myeloma: state of the Art WB-MRI and PET/CT. Skeletal Radiol. 51, 59–80. https://doi.org/10.1007/s00256-021-03841-5 (2022).
Lambin, P. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 48, 441–446. https://doi.org/10.1016/j.ejca.2011.11.036 (2012).
Aerts, H. J. The potential of Radiomic-Based phenotyping in precision medicine: A review. JAMA Oncol. 2, 1636–1642. https://doi.org/10.1001/jamaoncol.2016.2631 (2016).
Huang, E. P. et al. Criteria for the translation of radiomics into clinically useful tests. Nat. Rev. Clin. Oncol. 20, 69–82. https://doi.org/10.1038/s41571-022-00707-0 (2023).
Li, Y. et al. Radiomics models based on magnetic resonance imaging for prediction of the response to bortezomib-based therapy in patients with multiple myeloma. Biomed. Res. Int. 691:1246 (2022). https://doi.org/10.1155/2022/6911246
Wu, Z. et al. Spinal MRI-Based radiomics analysis to predict treatment response in multiple myeloma. J. Comput. Assist. Tomogr. 46, 447–454. https://doi.org/10.1097/RCT.0000000000001298 (2022).
Xiong, X. et al. Discriminating minimal residual disease status in multiple myeloma based on MRI: utility of radiomics and comparison of machine-learning methods. Clin. Radiol. 78, e839–e846. https://doi.org/10.1016/j.crad.2023.07.011 (2023).
Goldschmidt, H. et al. Addition of isatuximab to lenalidomide, bortezomib, and dexamethasone as induction therapy for newly diagnosed, transplantation-eligible patients with multiple myeloma (GMMG-HD7): part 1 of an open-label, multicentre, randomised, active-controlled, phase 3 trial. Lancet Haematol. 9, e810–e821. https://doi.org/10.1016/S2352-3026(22)00263-0 (2022).
Wennmann, M. et al. Combining deep learning and radiomics for automated, objective, comprehensive bone marrow characterization from Whole-Body MRI: A multicentric feasibility study. Invest. Radiol. 57, 752–763. https://doi.org/10.1097/RLI.0000000000000891 (2022).
Wennmann, M. et al. Deep learning for automatic bone marrow apparent diffusion coefficient measurements from Whole-Body magnetic resonance imaging in patients with multiple myeloma: A retrospective multicenter study. Invest. Radiol. 58, 273–282. https://doi.org/10.1097/RLI.0000000000000932 (2023).
Wennmann, M. et al. Prediction of bone marrow biopsy results from MRI in multiple myeloma patients using deep learning and radiomics. Invest. Radiol. 58, 754–765. https://doi.org/10.1097/RLI.0000000000000986 (2023).
Palumbo, A. et al. Revised international staging system for multiple myeloma: A report from international myeloma working group. J. Clin. Oncol. 33, 2863–2869. https://doi.org/10.1200/JCO.2015.61.2267 (2015).
D’Agostino, M. et al. Second revision of the international staging system (R2-ISS) for overall survival in multiple myeloma: A European myeloma network (EMN) report within the HARMONY project. J. Clin. Oncol. 40, 3406–3418. https://doi.org/10.1200/JCO.21.02614 (2022).
Poulton, T. B., Murphy, W. D., Duerk, J. L., Chapek, C. C. & Feiglin, D. H. Bone marrow reconversion in adults who are smokers: MR imaging findings. AJR Am. J. Roentgenol. 161, 1217–1221. https://doi.org/10.2214/ajr.161.6.8249729 (1993).
Lavdas, I. et al. Apparent diffusion coefficient of normal abdominal organs and bone marrow from Whole-Body DWI at 1.5 T: the effect of sex and age. AJR Am. J. Roentgenol. 205, 242–250. https://doi.org/10.2214/AJR.14.13964 (2015).
Wennmann, M. et al. Repeatability and reproducibility of ADC measurements and MRI signal intensity measurements of bone marrow in monoclonal plasma cell disorders: A prospective Bi-institutional multiscanner, multiprotocol study. Invest. Radiol. 57, 272–281. https://doi.org/10.1097/RLI.0000000000000838 (2022).
Wennmann, M. et al. In vivo repeatability and multiscanner reproducibility of MRI radiomics features in patients with monoclonal plasma cell disorders: A prospective Bi-institutional study. Invest. Radiol. 58, 253–264. https://doi.org/10.1097/RLI.0000000000000927 (2023).
Gotz, M., Nolden, M., Maier-Hein, K. M. I. T. K. & Phenotyping An open-source toolchain for image-based personalized medicine with radiomics. Radiother Oncol. 131, 108–111. https://doi.org/10.1016/j.radonc.2018.11.021 (2019).
Lambin, P. et al. Radiomics: the Bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 14, 749–762. https://doi.org/10.1038/nrclinonc.2017.141 (2017).
Pedregosa, F. et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Wennmann, M. et al. Reproducible radiomics features from Multi-MRI‐Scanner Test–Retest‐Study: influence on performance and generalizability of models. J. Magn. Reson. Imaging. https://doi.org/10.1002/jmri.29442 (2024).
Kocak, B. et al. METhodological radiomics score (METRICS): a quality scoring tool for radiomics research endorsed by EuSoMII. Insights Imaging. 15, 8. https://doi.org/10.1186/s13244-023-01572-w (2024).
Kocak, B. et al. CheckList for evaluation of radiomics research (CLEAR): a step-by-step reporting guideline for authors and reviewers endorsed by ESR and EuSoMII. Insights Imaging. 14, 75. https://doi.org/10.1186/s13244-023-01415-8 (2023).
DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–845 (1988).
Hunter, J. D. & Matplotlib A 2D graphics environment. Comput. Sci. Eng. 9, 90–95. https://doi.org/10.1109/MCSE.2007.55 (2007).
Waskom, M. L. & Seaborn Statistical data visualization. J. Open. Source Softw. 6, 3021. https://doi.org/10.21105/joss.03021 (2021).
Park, J. E. et al. Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur. Radiol. 30, 523–536. https://doi.org/10.1007/s00330-019-06360-z (2020).
Goldschmidt, H. et al. Response-adapted Lenalidomide maintenance in newly diagnosed myeloma: results from the phase III GMMG-MM5 trial. Leukemia 34, 1853–1865. https://doi.org/10.1038/s41375-020-0724-1 (2020).
Bauer, F. et al. Advanced automated model for robust bone marrow segmentation in whole-body MRI. Acad. Radiol. https://doi.org/10.1016/j.acra.2024.12.060 (2025).
Akinci D’Antonoli, T. et al. TotalSegmentator MRI: Robust Sequence-independent Segmentation of Multiple Anatomic Structures in MRI. Radiology 314, e241613. https://doi.org/10.1148/radiol.241613 (2025).
Acknowledgements
The authors thank the German Speaking Myeloma Multicenter Group (GMMG) for providing data from the GMMG-HD7 trial (EudraCT: 2017-004768-37).
Funding
Open Access funding enabled and organized by Projekt DEAL. The authors state that this work has not received any funding. MW has been funded by a fellowship of the DKFZ Clinician Scientist Program, supported by the Dieter Morszeck Foundation. FB was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—project number 538209205.
Author information
Authors and Affiliations
Contributions
FB: conceptualization, methodology, investigation, data curation, formal analysis, writing—original draft, visualization, project administration. MH: conceptualization, data curation, writing—review and editing. NW: conceptualization, data curation, writing—review and editing. MG: conceptualization, data curation, writing—review and editing. JK: conceptualization, data curation, methodology, writing—review and editing. EM: conceptualization, data curation, writing—review and editing. MSR: conceptualization, resources, data curation, writing—review and editing. SS: conceptualization, data curation, writing—review and editing. AJ: conceptualization, data curation, writing—review and editing. TFW: conceptualization, data curation, writing—review and editing. MD: conceptualization, data curation, writing—review and editing. BB: conceptualization, data curation, writing—review and editing. MH: conceptualization, data curation, writing—review and editing. SA: conceptualization, data curation, writing—review and editing. MH: conceptualization, data curation, writing—review and editing. JH: conceptualization, data curation, writing—review and editing. DK: conceptualization, data curation, writing—review and editing. UG: conceptualization, data curation, writing—review and editing. AR: conceptualization, data curation, writing—review and editing. JD: conceptualization, data curation, writing—review and editing. LU: conceptualization, data curation, writing—review and editing. HPS: conceptualization, resources, data curation, writing—review and editing. HG: conceptualization, resources, data curation, writing—review and editing. KMH: conceptualization, resources, data curation, writing—review and editing. SD: supervision, conceptualization, resources, data curation, writing—review and editing. EKM: conceptualization, resources, data curation, writing—review and editing. MW: supervision, conceptualization, methodology, data curation, writing—review and editing, visualization, project administration. PN: supervision, methodology, data curation, formal analysis, writing—review and editing, project administration.
Corresponding author
Ethics declarations
Competing interests
All authors have no conflict of interest regarding the submitted work. Outside of the submitted work, the following relationships are reported: MH: honoraria: Sanofi.MSR: consulting or advisory role: BMS, Amgen, GSK, Janssen, Sanofi, Pfizer, AbbVie, Novartis, and Roche; research funding: Sanofi; travel accommodation and expenses: BMS, AbbVie, Janssen, Sanofi, and GSK; Honoraria: BMS, Janssen, GSK, AbbVie, and Sanofi; receipt of equipment, materials, drugs, medical writing, gifts or other services: Novartis and Sanofi.SS: travel grants or honoraria: Celgene, BMS, Janssen, Takeda, and Amgen. BB: honoraria: Janssen, GSK, AMGEN, Pfizer, Oncopeptides, and Sanofi.UG: stock and other ownership interests: Biontech; honoraria: Boehringer Ingelheim, Amgen, AstraZeneca, Bristol-Myers Squibb, MSD Oncology, Sanofi Aventis GmbH, Fujifilm, Cellrion, Ipsen, and Novartis; consulting or advisory role: Amgen and MSD Oncology; research funding: Ipsen and Macrogenics; travel, accomodations or expenses: Boehringer Ingelheim and GSK.JD: advisory board and travel support: Amgen, BMS, Janssen, Sanofi, and Takeda; honoraria: Amgen, BMS, Janssen, and Sanofi.LU: advisory board: EasyRadiology; speaker: Siemens Healthineers and Bayer Healthcare. HPS: consulting fee or honoraria: Siemens, Curagita, Profound, and Bayer; travel support: Siemens, Curagita, Profound, and Bayer; board member: Curagita; consultancy: Curagita and Bayer; payment for lectures: Siemens, Curagita, Profound, and Bayer.HG: grants and/or provision of investigational medicinal products: Amgen, Array Biopharma/Pfizer, BMS, Celgene, Chugai, Dietmar-Hopp-Foundation, Janssen, Johns Hopkins University, Mundipharma GmbH, and Sanofi; research support: Amgen, BMS, Celgene, GlycoMimetics Inc., GSK, Heidelberg Pharma, Hoffmann-La Roche, Karyopharm, Janssen, Incyte, Millenium Pharmaceuticals Inc., Molecular Partners, Merck Sharp and Dohme, MorphoSys AG, Pfizer, Sanofi, Takeda, and Novartis; advisory board: Adaptive Biotechnology, Amgen, BMS, Janssen, and Sanofi; honoraria: Amgen, BMS, Chugai, GlaxoSmithKline, Janssen, Novartis, Sanofi, and Pfizer. EKM: consulting or advisory role: Amgen, Bristol Myers Squibb/Celgene, GlaxoSmithKline, Janssen-Cilag, Pfizer, Sanofi, Stemline, and Takeda; honoraria: Amgen, Bristol Myers Squibb/Celgene, GlaxoSmithKline, Janssen-Cilag, Pfizer, Sanofi, Stemline, and Takeda; research funding: Bristol Myers Squibb/Celgene, GlaxoSmithKline, Janssen-Cilag, Sanofi, and Takeda; travel accommodations and expenses: Bristol Myers Squibb/Celgene, GlaxoSmithKline, Janssen-Cilag, Sanofi, Stemline, and Takeda.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bauer, F., Hajiyianni, M., Weinhold, N. et al. Automated radiomics model for prediction of therapy response and minimal residual disease from baseline MRI in multiple myeloma. Sci Rep 15, 35400 (2025). https://doi.org/10.1038/s41598-025-13165-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-13165-2