Abstract
Pain is a multifaceted phenomenon that significantly affects a large portion of the global population. Objective pain assessment is essential for developing effective management strategies, which in turn contribute to more efficient and responsive healthcare systems. However, accurately evaluating pain remains a complex challenge due to subtle physiological and behavioural indicators, individual-specific pain responses, and the need for continuous patient monitoring. Automatic pain assessment systems offer promising, technology-driven solutions to support and enhance various aspects of the pain evaluation process. Physiological indicators offer valuable insights into pain-related states and are generally less influenced by individual variability compared to behavioural modalities, such as facial expressions. Skin conductance, regulated by sweat gland activity, and the heart’s electrical signals are both influenced by changes in the sympathetic nervous system. Biosignals, such as electrodermal activity (EDA) and electrocardiogram (ECG), can, therefore, objectively capture the body’s physiological responses to painful stimuli. This paper proposes a novel multi-modal ensemble deep learning framework that combines electrodermal activity and electrocardiogram signals for automatic pain recognition. The proposed framework includes a uni-modal approach (FCN-ALSTM-Transformer) comprising a Fully Convolutional Network, Attention-based LSTM, and a Transformer block to integrate features extracted by these models. Additionally, a multi-modal approach (CrossMod-Transformer) is introduced, featuring a dedicated Transformer architecture that fuses electrodermal activity and electrocardiogram signals. Experimental evaluations were primarily conducted on the BioVid dataset, with further cross-dataset validation using the AI4PAIN 2025 dataset to assess the generalisability of the proposed method. Notably, the CrossMod-Transformer achieved an accuracy of 87.52% on Biovid and 75.83% on AI4PAIN, demonstrating strong performance across independent datasets and outperforming several state-of-the-art uni-modal and multi-modal methods. These results highlight the potential of the proposed framework to improve the reliability of automatic multi-modal pain recognition and support the development of more objective and inclusive clinical assessment tools.
Similar content being viewed by others
Introduction
Pain is a complex and common sensory experience that indicates actual or potential tissue damage or resembles the sensation of such damage, requiring attention and treatment1. From a duration perspective, it is typically classified as either acute or chronic2. Acute pain serves as a vital warning mechanism that helps avoid harm, while chronic pain can persist beyond the initial injury. If left untreated, acute pain can transition into chronic pain, which leads to significant problems that affect quality of life and increases social costs. Chronic pain incurs higher costs than cancer, heart disease, and Human Immunodeficiency Virus (HIV) combined3. Therefore, effective pain management is not only essential for immediate relief but also for mitigating the risk of chronic pain and its associated long-term health consequences.
Accurate pain recognition is essential for effective management. Traditionally, pain assessment has relied on self-report methods (e.g., Visual Analogue Scale (VAS), Numerical Rating Scale (NRS), McGill Pain Questionnaire (MPQ)4. However, these methods are subjective and unsuitable for non-verbal patients, such as those on mechanical ventilation, infants, or individuals with cognitive impairments5. For these patients, pain assessment depends on clinicians observing and reporting behavioural signs (e.g., facial expressions, body movements) and physiological indicators, including Electrodermal Activity (EDA) and Electrocardiogram (ECG). Yet, relying on human observers presents several challenges, including limited sensitivity, difficulty in continuous monitoring, and potential biases6,7. Furthermore, real-time and continuous pain monitoring is crucial for timely intervention, as untreated pain can escalate quickly and negatively impact patient outcomes.
Numerous promising approaches have been developed under the scope of automatic pain assessment, ranging from traditional handcrafted feature engineering8 to more recent deep learning-based methods9. Among these, video-based approaches–particularly those analysing facial expressions10–have been extensively explored. Despite their engineering innovation, their clinical applicability is often limited by practical challenges such as patient movement, privacy concerns, and cross-cultural variability in expressive behaviour11,12,13,14,15. These limitations underscore the value of physiological signals, such as EDA and ECG, which offer non-behavioural, sensor-based alternatives that are less intrusive and better suited for continuous monitoring. In this context, recent studies have focused on enhancing domain generalisation in physiological signal analysis using various strategies. For instance, Tao et al.16 proposed a dual-scale convolutional model with self-attention to enhance cross-session electroencephalography decoding. Zhong et al.17 introduced a framework that aligns both marginal and conditional distributions to learn domain-invariant representations without requiring access to the target domain, while the authors in18 developed an adversarial learning method guided by spatiotemporal pattern differences in SPD-matrix features to address inter-subject variability. Nevertheless, most physiologically based methods remain uni-modal, which constrains their ability to model the complex and multidimensional nature of pain19,20. This limitation highlights the need for multi-modal systems that can integrate complementary physiological cues to enhance robustness in pain assessment applications.
A growing body of work highlights the advantages of multi-modal approaches for pain recognition. These methods emphasise the integration of complementary signals to enable more comprehensive and accurate assessment. To enhance performance and robustness, studies have integrated physiological signals with other data sources, aiming to address the limitations of individual modalities. Such multi-modal systems show promise in improving generalisability and reliability, particularly in real-world clinical contexts21. For example, Werner et al.22 presented a system to combine video information (facial landmarks and 3D distances) with physiological signals (EDA, ECG, and EMG) for automatic pain detection. This multi-modal approach achieved significant improvement in performance compared to previous video-based methods alone. Lopez-Martinez et al.23 proposed a novel approach that leverages multi-task learning with neural networks to account for individual differences in pain response. The authors utilised multi-modal physiological data (EDA and ECG) for personalised pain recognition. In a different study, Wang et al.24 utilised EDA, ECG and EMG, combining deep learning (biLSTM) for automatic feature extraction with traditional handcrafted features derived from expert knowledge. Collectively, these examples demonstrate the value of multi-modal fusion in advancing pain recognition systems by capturing complementary information from diverse sensing modalities.
While previous research has explored multi-modal approaches and physiological signals for pain recognition, many methods still face challenges in effectively integrating physiological modalities. Key limitations include difficulties in synchronising temporal patterns across modalities and capturing both short- and long-term dependencies within and between signal streams. Traditional deep learning models such as convolutional neural networks (CNNs) and long short-term memory (LSTM) networks provide strong inductive biases for local spatial features and short-range temporal patterns; however, their fixed receptive fields and sequential processing often limit their ability to model long-range and cross-modal dependencies, particularly in complex physiological data not explicitly designed for such architectures25,26. Hybrid CNN-LSTM models attempt to combine spatial and temporal learning but still rely on fixed-length sequential processing and may fail to capture global temporal dependencies effectively. In contrast, Transformer-based models offer a more powerful and flexible framework for sequence modelling by leveraging self-attention mechanisms that can simultaneously attend to relationships across all time steps. This enables the modelling of global dependencies without relying on recurrence, making Transformers particularly well-suited for capturing nuanced temporal and cross-modal patterns in physiological data27. These models have shown promise in broader affective computing tasks, such as multimodal depression recognition28, and are increasingly applied to physiological data for pain assessment. However, many current Transformer-based approaches focus on either intra-modal or inter-modal relationships in isolation19,29, which limits their capacity to represent the multidimensional and dynamic nature of pain. Combining traditional models with Transformers enables the extraction of local feature extraction while allowing the Transformer to capture global, cross-modal dependencies, thereby addressing the limitations of each approach. Consequently, there is a need for advanced architectures capable of jointly modelling intra- and inter-modal relationships, improve generalisability, and enable more accurate and clinically viable pain recognition systems.
To address the aforementioned challenges, this study proposes a novel multi-modal framework that integrates advanced DL techniques with transformer architectures. Drawing inspiration from Waligora et al.29, who utilised a Joint Multi-modal Transformer to fuse EDA and video data from the BioVid dataset, a similar strategy of using Transformer architectures to enhance multi-source integration was adopted. However, unlike their approach, which applies a single transformer to jointly process video and physiological features, our framework introduces a two-stage transformer architecture specifically designed for physiological signals. The first stage captures temporal patterns within each signal by combining fully convolutional networks (FCN) and attention-based LSTM (ALSTM) outputs, while the second stage fuses EDA and ECG representations across modalities. This decoupled design helps preserve modality-specific temporal dynamics and mitigates early-stage modality competition, which is particularly relevant for low-dimensional, noise-prone physiological inputs. This architecture extends the original concept by focusing exclusively on physiological signals, particularly EDA and ECG, to leverage their unique properties in the pain recognition task. By integrating these physiological signals, the proposed framework exploits their complementary characteristics to effectively model both intra- and inter-modality relationships, thereby capturing the complex, multi-dimensional nature of pain. The main contributions of this paper are summarised as follows: 1) A hybrid deep learning framework is introduced for physiological pain recognition, which first performs intra-modality fusion via FCN–ALSTM–Transformer blocks and then applies inter-modality fusion via a CrossMod-Transformer. This architecture enables modality-specific temporal encoding while mitigating modality interference during fusion; 2) The proposed multi-modal framework demonstrates superior performance over uni-modal baselines on Part A of the BioVid Heat Pain Database, validating the effectiveness of combining EDA and ECG signals for pain recognition; 3) To assess generalisability, cross-dataset validation was conducted using the AI4Pain 2025 dataset, which includes EDA and blood volume pulse (BVP) signals. Without architectural changes, the model maintained strong performance, highlighting its robustness and adaptability to different physiological signal combinations and experimental settings; and 4) attention maps are provided to enhance the interpretability of the results, offering insights into how the model prioritises information from EDA and ECG signals.
Methods
This section outlines the methods and processes employed in the study, beginning with a review of the BioVid Heat Pain Database and AI4PAIN, which were used for evaluation. It is important to note that the Biovid Heat Pain Database served as the primary dataset for designing and developing the proposed framework, while the AI4PAIN dataset was employed exclusively for cross-dataset evaluation to assess the generalisability of the framework. The section then details the data pre-processing techniques applied to the physiological signals, followed by a detailed description of the proposed deep learning architecture, which incorporates both uni-modal and multi-modal feature integration strategies. Lastly, the experimental setup and evaluation metrics are presented to provide a comprehensive assessment of the approach and its effectiveness.
BioVid heat pain database
In this research, we utilised two modalities from the publicly accessible BioVid Heat Pain Database (Part A)30, comprising electrodermal activity (EDA) and electrocardiography (ECG) data. The dataset includes recordings from 87 healthy participants (44 males and 43 females) across three age groups: 20–35 years, 36–50 years, and 51–65 years. These modalities were employed to evaluate the proposed framework. Other modalities (e.g., frontal head videos and trapezius Electromyography (EMG) are excluded. As previously mentioned in the Introduction, video-based methods have limitations, such as privacy concerns, difficulties maintaining consistency during data capture, and the potential for patients to control and alter their expressions. Although EMG has been used in previous studies for pain recognition22, its practical application is limited and often noisy, making EDA and ECG signals more suitable for real-world applications and wearable sensor-based pain detection23.
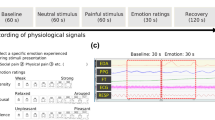
In BioVid, heat pain was induced on the right arm of the subjects using a thermode under controlled conditions. There are 4 different subject-specific pain intensities, and the procedure was repeated 20 times for each subject and level of pain. Each stimulation lasted 4 seconds, and there was a resting state lasting randomly between 8 and 12 seconds after each stimulus (see Fig. 1). Twenty baseline (BLN) measurements (\(32^\circ\)C) are also recorded, resulting in 100 samples per subject captured within a 5.5-second window with a 3-second delay after the pain stimulus began. The sampling frequency of all physiological signal recordings is equal to 512 Hz. Therefore, the data for each signal forms a channel with dimensions of 2,816 \(\times\) 20 \(\times\) 5 \(\times\) 87 for a time window of 5.5 seconds. As the goal of this research is to detect pain, we only consider BLN (No Pain) and PA4 (Very Severe Pain).
(a) Representation of pain stimulation, illustrating the heat levels ranging from \(T_0\) to \(T_4\) and the intervals between each stimulus. The recordings are taken from the 5.5-second segment highlighted in grey (adapted from30). (b) Example of the raw physiological data corresponding to temperature level \(T_4\) (Severe Pain).
Despite its widespread use, the BioVid has some limitations that complicate the comparability of different research approaches. The first limitation is the inconsistency in the subject subsets that researchers use for their experiments. According to the results reported by the database’s creators in31, some subjects do not visibly respond to the stimulus. As a result, some studies, such as32, exclude 20 participants who showed minimal reactions to pain, while other researchers have used the entire dataset. Additionally, facial expressions responding to the highest pain stimuli begin after 2 seconds31, leading to different time window lengths when analysing the data. For instance, Thiam et al. in33 and34 employed a different segmentation approach, truncating the original time frame by 1 second and using only 4.5 seconds of the samples, with a shift from the elicitation’s onset of 4 instead of 3 seconds. Similarly, Waligora et al.29 cropped the original 5.5-second time window by 2 seconds and have utilised 3.5 seconds of the samples. Despite these limitations, we decided to preserve the integrity of the original data, first to enable comparison with most studies that have used all the available data, and second to conduct a more comprehensive analysis of the pain signal characteristics. Therefore, we compare our results only with studies that have used all the available data and exclude those that have truncated the data or the 20 noisy subjects.
AI4PAIN dataset
The AI4PAIN 2025 Grand Challenge dataset35 represents a recent and valuable resource specifically developed to support advanced research in automatic pain recognition. It includes physiological recordings from four biosignal modalities: electrodermal activity (EDA), blood volume pulse (BVP), respiration (RESP), and peripheral oxygen saturation (SpO2), all acquired using Biosignal Plux sensors at a sampling rate of 100 Hz. In addition, the dataset also includes modalities such as functional near-infrared spectroscopy (fNIRS) and facial video recordings36, however these were not employed in this study. The dataset comprises recordings from 65 participants, including 23 females, with ages ranging from 17 to 52 years. Pain stimuli were delivered using a transcutaneous electrical nerve stimulation (TENS) device, with electrodes placed on the inner forearm and the back of the hand. To minimise habituation and control for order effects, the stimulation site and intensity were counterbalanced across sessions. Each recording session began with a 60-second baseline period, used to represent the No Pain condition. This was followed by 12 pain-inducing stimuli–six repetitions each for the low-pain and high-pain conditions–administered at randomised anatomical locations (arm and hand). Each stimulus lasted approximately 10 seconds, followed by a 40-second rest interval to allow physiological signals to return toward baseline before the subsequent trial. The data acquisition protocol closely adheres to the procedures established in prior work37. In this study, we employed the EDA and BVP signals to evaluate the performance of the proposed method. The BVP signal was used in place of ECG to provide cardiac-related information, enabling a modality substitution while maintaining physiological relevance.
Data pre-processing
The BioVid database includes both raw and pre-processed signals, and for all our experiments, we utilised the pre-processed data. Specifically, the EDA signals were filtered using a Butterworth low-pass filter, while the ECG signals were processed with a Butterworth band-pass filter covering the range of 0.1 to 250 Hz. These filters are part of the original BioVid dataset preprocessing. Our pipeline begins with this preprocessed data and performs no additional filtering. Additionally, prior to classification, all physiological signals were downsampled to a frequency of 25 Hz. This was achieved by linearly interpolating each preprocessed signal to a fixed length of 138 points, ensuring uniform temporal resolution for model input. This step ensures consistency in input dimensions across trials and reduces computational complexity while retaining the critical temporal dynamics necessary for downstream modelling21. Therefore, in total, we have utilised 480,240 (138 \(\times\) 20 \(\times\) 2 \(\times\) 87) samples, with 240,120 samples for the No Pain (BLN) class and 240,120 samples for the Very Severe Pain (PA4) class. This binary setup is fully balanced, with equal numbers of samples from both classes. To normalise the data, a Robust Scaler38 was used per subject, which is less sensitive to outliers compared to standard scaling methods. This scaler centres the data using the median and scales according to the inter-quartile range (IQR).
Regarding the processing of signals from the AI4PAIN dataset, we utilised the raw biosignals as provided by the dataset creators. The primary motivation for this decision was to assess the performance of the proposed method on unfiltered data, in contrast to the BioVid dataset, which contains filtered signals. This allowed us to explore the model’s ability to handle raw physiological inputs and evaluate the potential of such data for pain recognition.
Proposed framework for pain detection
The proposed Cross-Mod Transformer is deep learning framework designed for pain recognition using physiological data. Figure 2 presents the proposed model architecture. It applies a hierarchical fusion mechanism that combines advanced DL models, including a hybrid architecture combining fully convolutional networks (FCN), attention-based long short-term memory (ALSTM) networks, and Transformers, enhanced with attention mechanisms. This design enables the model to learn complex patterns in time-series data, making them ideal for capturing detailed features without manual feature extraction.
The architecture operates in single-modality (uni-modal) and multi-modality streams. The Uni-modal Stream is responsible for feature extraction through the FCN and ALSTM blocks and feature fusion via a Transformer block. While the Multi-modal Stream performs multi-modal fusion. In the Uni-modal Stream, each modality (EDA and ECG) is processed independently, with the Transformer block capturing intra-modal dependencies. In the Multi-modal Stream, a separate Transformer block is applied to integrate features across EDA and ECG signals, effectively capturing inter-modal dependencies. The following paragraphs provide detailed descriptions of each architecture in these two streams.
Uni-modal stream
In the first stage, each physiological signal modality (EDA and ECG) is processed independently to capture its unique temporal patterns through a dedicated feature extraction stream. This stage utilises two parallel networks, an FCN and an ALSTM, for each modality. The rationale for using each component is described in the following paragraphs.
The FCN is used to capture detailed, short-term temporal patterns within the EDA and ECG signals. Originally introduced by Long et al. 39 for image analysis and later adapted by Wang et al. 25 for time-series classification (TSC), FCNs are effective at capturing short-term patterns across time steps. Unlike traditional CNNs, which downsample data with pooling layers, an FCN maintains the length of the time series throughout the network, preserving detailed temporal information. Each convolutional layer is followed by batch normalisation and a ReLU activation, allowing the network to learn robust feature maps. A key feature of FCN is the use of a Global Average Pooling (GAP) layer in place of fully connected layers; this drastically reduces the number of parameters, mitigates overfitting, and reveals which time steps contribute most to the classification decision. Consequently, the FCN retains temporal resolution while requiring fewer parameters than a CNN, making it well-suited to TSC tasks where sequential detail is critical. The architecture implemented in this study is illustrated in Fig. 2A and summarised in Table 1. Specifically, the FCN processes an input sequence x through the following steps:
where \(x\!\in \!\mathbb {R}^{C_{\textrm{in}}\times S}\) is the input sequence, k the kernel size, and p the zero-padding. The resulting feature maps are batch-normalised,
then passed through a ReLU non-linearity,
and–optionally–dropout,
After the final convolutional layer L, GAP collapses the temporal dimension,
producing the fixed-length descriptor \(g\!\in \!\mathbb {R}^{C_{\textrm{out}}}\) that is forwarded to subsequent modules.
In parallel to the FCN, the attention-based LSTM (ALSTM) extracts long-range dependencies in the EDA and ECG signals. Recurrent neural networks (RNNs)40 are well suited to sequential data, and the LSTM unit41 mitigates vanishing and exploding gradients, thus capturing long-term temporal patterns. At each time step t, the LSTM computes
where \(\textbf{x}_t\) is the input vector, \(\textbf{h}_t\) the hidden state, and \(\textbf{c}_t\) the cell state; \(\sigma (\cdot )\) is the logistic sigmoid and \(\odot\) denotes element-wise multiplication. To focus on the most relevant time steps in the sequence, a Bahdanau-style attention mechanism42 is applied over the hidden states \(\{\textbf{h}_t\}_{t=1}^{S}\). First, a relevance score is computed,
where \(\operatorname {ELiSH}(x)=x\) for \(x\!\ge 0\) and \((e^{x}-1)\bigl /\,\bigl (1+e^{-x}\bigr )\) otherwise. Scores are normalised to attention weights,
which yield the context vector
Finally, the context vector is concatenated with the last hidden state to form the ALSTM feature representation,
which is forwarded to the subsequent Transformer block. The architecture is depicted in Fig. 2B and outlined in Table 2.
Once the FCN and ALSTM networks extract their respective features, these features are integrated using a Transformer block. Transformers, popularised by Vaswani et al.27, are highly effective for capturing complex interactions between features, making them ideal for tasks that involve integrating different sources of information. In the proposed approach, a Transformer block (refer to Fig. 2C) is applied to each modality, enabling the model to capture intra-modal dependencies. Specifically, the Transformer focuses on the relationships between different time steps and features within the same modality (EDA or ECG). Each Transformer encoder operates on a sequence of input embeddings \(X\in \mathbb {R}^{S\times d}\), where S is the sequence length and d the feature dimension. The self-attention mechanism begins by computing queries, keys, and values through learned projections:
The attention weights are then computed using scaled dot-product attention:
where \(d_k\) is the dimension of the key vectors. In the multi-head version, this is performed in parallel H times and concatenated:
where each \(h_i\) is a head-specific self-attention output. The result passes through a residual connection, layer normalisation, and a feed-forward network:
where \(\sigma (\cdot )\) denotes a ReLU activation.
Building on the architecture in43, which used concatenation to combine features from a parallel FCN-ALSTM model, the Transformer block offers a more refined fusion process. Unlike simple concatenation, the Transformer captures both long- and short-term dependencies through self-attention and cross-attention mechanisms, allowing the architecture to prioritise the most relevant aspects of each feature set and thereby enhancing performance29.
For a more detailed description of the Transformer block, Fig. 2C provides an illustration, which includes two Transformer encoders and two cross-attention blocks. In the uni-modal stream, Transformer encoders are applied to the output of each feature extractor–one to the FCN-derived features and another to the ALSTM-derived features. These encoders enhance the representations by focusing on the most relevant aspects of each feature set, effectively capturing complementary information from both short-term (FCN) and long-term (ALSTM) perspectives. After the Transformer encoders, the integration of features is further refined through a cross-attention mechanism. This allows the representation from one stream to be used as a query, and the other as key/value. For example, if \(\tilde{\mathscr {F}}\) and \(\tilde{\mathscr {L}}\) denote the encoded FCN and ALSTM features, respectively, then the cross-attention outputs are:
This process enables each stream to attend to the most informative parts of the other. The resulting outputs are then concatenated:
and summarised via mean pooling over the sequence dimension:
The extracted feature vectors from the cross-attention blocks are concatenated and passed through a series of fully connected (FC) layers to form the final uni-modal representation. From this sequence, 128-dimensional features are obtained from the third FC layer of each physiological signal, representing high-level embeddings. These features are subsequently forwarded to the multi-modal integration stage. The complete architecture of the Transformer block is detailed in Table 3.
Multi-modal stream
In the final stage of the proposed framework (Fig. 2D), a separate Transformer block is used to fuse the uni-modal feature representations extracted from the EDA and ECG streams. Although this Transformer block is structurally similar to the one used in the uni-modal stream, its functionality here is to focus on learning inter-modal dependencies. The Transformer block is designed to capture how these signals interact and complement each other during pain-related episodes by processing the EDA and ECG features together. For instance, EDA and ECG responses might exhibit patterns that are strongly indicative of pain when considered in combination, but could appear ambiguous or less significant if analysed individually. The multi-modal Transformer performs joint attention across the two modalities, effectively integrating the distinct temporal patterns captured from EDA and ECG data. Let \(\textbf{m}_{\text {EDA}}, \textbf{m}_{\text {ECG}} \in \mathbb {R}^{d}\) denote the uni-modal feature vectors from each stream. These are fused via cross-attention and projected through FC layers to produce the final output:
The integration process within this Transformer block follows the same mechanism as in the uni-modal Stream, consisting of Transformer encoders and cross-attention blocks. The 128-dimensional feature vectors extracted by the uni-modal architecture serve as input, which is then refined through the cross-attention mechanism to emphasise the most relevant features across both EDA and ECG modalities. After the cross-attention blocks, the resulting features from both EDA and ECG are concatenated and passed through four FC layers. These layers process the fused features and produce the final classification output for the multi-modal Stream, concluding the proposed framework’s architecture. In summary, the proposed multi-modal framework leverages both intra-modal and inter-modal dependencies to improve pain recognition accuracy. By combining features from EDA and ECG signals, the model can provide a more comprehensive representation of physiological responses to pain, highlighting the value of a multi-modal approach over uni-modal analysis.
Overview of the proposed model architecture. The input signal (EDA or ECG) is processed by an ALSTM block (green) and an FCN block (purple). The 128-dimensional feature vectors extracted by these two blocks are cross-attended using a Transformer block. The resulting uni-modal features (extracted from the FC3 layer) are then cross-attended using a separate Transformer block.
Experimental setup
All experiments have been conducted using a system equipped with an NVIDIA GeForce RTX 4080 GPU (Laptop variant) featuring 7424 CUDA Cores, a boost clock speed ranging from 1350 to 2280 MHz, and 12 GB of GDDR6 memory. The CPU utilised was a 13th Gen Intel(R) Core(TM) i7-13650HX with a base clock speed of 2.60 GHz, supported by 16 GB of RAM. The framework for the experiments was implemented using Python version 3.8.8 and PyTorch version 2.1.2.
We adopted a multi-step training procedure where each sub-network (i.e., FCN, ALSTM, Transformer) is trained separately. This strategy involves training each network block individually, with final FC layers temporarily added for training and optimisation, using varying hyperparameters suited to their specific architecture. For clarity, these final FC layers are not shown in Fig. 2 and are discarded after training, while the extracted features are saved and then passed to the subsequent sub-network. While we also experimented with end-to-end training, it produced inferior results compared to the multi-step approach. Since these features were originally trained separately to address the same classification problem, their integration simplifies the training process. Moreover, separate training enables each modality to learn robust features without interference. This decision was motivated by the phenomenon of modality competition, where dominant modalities can hinder learning from weaker ones during joint training44. At no point, the validation or test labels were used during the training of any sub-network, thereby ensuring a strict separation of training and evaluation data and avoiding data leakage.
For hyperparameter tuning, some parameters, such as the learning rate and dropout rate, were optimised using Optuna (Version 3.6.1)45, a Bayesian optimisation Python package, while others were set empirically. The optimisation process was performed independently for each sub-network and modality (EDA and ECG), utilising a 10-fold CV. The optimal hyperparameters derived from this process were then fixed and used for both evaluation schemes. The hyperparameters used for each sub-network are summarised in Table 4. Moreover, the average training time per epoch across all sub-networks and modalities was approximately 10.38 seconds.
The evaluation protocols were designed to align with previous studies and facilitate fair comparisons. For the BioVid dataset, we employed both 10-fold cross-validation (CV) and leave-one-subject-out (LOSO) cross-validation. In the 10-fold CV setup, the dataset was partitioned into 10 non-overlapping folds based on subject IDs (i.e., each fold was stratified by subject). In each iteration, nine folds were used for training, and one fold was reserved for testing. The process was repeated 10 times so that each fold served as the test set once. LOSO was conducted across all 87 subjects, using one subject as the test set in each iteration and the remaining 86 for training. The final performance was computed by averaging the classification results across all folds or test subjects. For the AI4PAIN dataset, we followed the original dataset split and adopted the standard hold-out validation scheme, separating the data into training and validation sets.
In terms of evaluation metrics, we used accuracy, F1 score, sensitivity, and specificity for the BioVid dataset. Accuracy reflects the overall proportion of correct predictions, while the F1 score–defined as the harmonic mean of precision and recall–provides a balanced measure of performance. Precision quantifies the proportion of true positive predictions among all predicted positives, and recall (or sensitivity) measures the proportion of actual positives that were correctly identified. Specificity captures the model’s ability to correctly identify negative (no-pain) instances, thus reflecting its effectiveness in reducing false positives. For the AI4PAIN dataset, we report accuracy, precision, and F1 score to assess model performance.
In addition to these quantitative metrics, we further explored the model’s interpretability by visualising attention maps. These maps offer valuable insights into how the model processes and prioritises information from both EDA and ECG signals during classification. Specifically, this interpretability analysis was conducted using the BioVid dataset only, as it served as the primary dataset for model development and detailed evaluation. By examining the self-attention, cross-attention, and embedding attention maps, we gained a deeper understanding of the model’s decision-making process and its reliance on different temporal regions of the signals. The attention maps provide a qualitative layer to our evaluation, enhancing the transparency and trustworthiness of the model.
Results
This section presents the experimental results and compares the performance of the proposed models with that of existing methods using the BioVid (used for model development and evaluation) and AI4PAIN (used exclusively for cross-dataset validation to assess generalisability) databases. Results are reported for both uni-modal and multi-modal approaches under 10-fold cross-validation, leave-one-subject-out (LOSO) cross-validation, and hold-out validation schemes.
For the uni-modal setting, three separate experiments are conducted for each sensing modality: one using attention-based long short-term memory (ALSTM) network, another with a fully convolutional (FCN) network, and a third combining these two networks using a Transformer block. For the multi-modal approach (EDA & ECG), the multi-modal architecture (CrossMod-Transformer) was employed. These results are presented in Table 5 for the 10-fold CV and in Table 6 for the LOSO evaluation. In both tables, the best-performing model in each evaluation scheme is highlighted. As can be seen, the CrossMod-Transformer model, which integrates both EDA and ECG signals, achieved the highest accuracy in both 10-fold CV (85.92%) and LOSO CV (87.52%). Notably, the ECG modality performed relatively poorly when used as a single source of information. The FCN model on this modality showed the weakest performance, with an average accuracy of 61.82% in 10-fold cross-validation and 62.27% in LOSO. However, when combined with EDA, the ECG provided complementary information, leading to improved performance compared to EDA alone. Additionally, the CrossMod-Transformer model demonstrated relatively high specificity, around 90% in both evaluation schemes, minimising the risk of unnecessary interventions for non-painful conditions.
Moreover, Fig. 3 presents a direct comparison of the average accuracy obtained with different modalities in 10-fold cross-validation and LOSO cross-validation. Overall, the LOSO CV demonstrated higher accuracy across all models and modalities, likely due to its use of a larger portion of the dataset for training in each fold compared to 10-fold CV. Additionally, it provides a more robust evaluation of the model’s generalisation capabilities. Regarding individual sensing modalities, the results indicate that the EDA modality performs better than the ECG modality in all architectures. The effectiveness of the hybrid FCN-ALSTM-Transformer architecture (i.e., FAT in Fig. 3) is also evident. For instance, in the EDA modality, the best-performing model is the hybrid architecture, with an accuracy of approximately 85% in 10-fold CV and 86% in LOSO CV, improving the FCN results by about 5% and ALSTM by 3%.
To evaluate the impact of input sequence length, we also trained models using the full-length physiological signals (2816 points at 512 Hz). This resulted in only marginal accuracy improvements (1.22% for ECG, 0.44% for EDA, and 0.76% for the Crossmod-Transformer), while substantially increasing computational cost. These findings support our use of the 138-point downsampled sequences as a more efficient and practical alternative with minimal compromise in classification performance.
The Wilcoxon signed-rank test was additionally used to analyse the statistical significance of performance improvements, and the results are summarised in the footnotes of Table 5 and Table 6. These comparisons assess the intra-modality improvements, which is the effect of increased model complexity within a single modality, and also inter-modality improvements, which measures the benefit of fusing EDA and ECG signals. For both ECG and EDA, the proposed hybrid FCN-ALSTM-Transformer significantly outperformed the FCN and ALSTM baselines (Wilcoxon \(p<0.05\) or \(p<0.01\), depending on the modality and evaluation schemes). In the multi-modal setting, although the Crossmod-Transformer did not demonstrate a statistically significant improvement over the strongest EDA baseline (Wilcoxon \(p>0.05\)) in the 10-fold evaluation scheme, it significantly outperformed all uni-modal baselines in the LOSO setting (Wilcoxon \(p \ll 0.01\)), highlighting its robustness and superior generalisation across subjects. To further examine the performance distributions across validation folds and subjects, the F1-score distributions are visualised using violin plots (Fig. 4). These plots provide insight into the variance of each model’s performance and help contextualise the reported confidence intervals and standard deviations.
Average accuracy obtained with different modalities evaluated in 10-fold cross-validation and leave-one-subject-out cross-validation classifying No Pain versus Severe Pain. This comparison highlights intra-modality improvements (due to increased model complexity) and inter-modality gains (from fusing EDA and ECG).
Violin plots of F1-score distributions for each model across validation folds (left: 10-fold CV) and subjects (right: LOSO CV). The plots highlight model stability and variance across cross-validation strategies. Notably, EDA-based models and the Crossmod-Transformer exhibit both high performance and lower variability compared to ECG-based counterparts.
In addition, experiments conducted using the AI4PAIN database evaluated the modalities both individually and in combination, following a similar protocol. The evaluation included uni-modal setups using individual models, as well as a multi-modal configuration that integrated the two modalities and model architectures. Table 7 presents the corresponding results. The findings follow similar trends observed in Tables 5 and 6, where EDA outperforms the cardiac-based modality. In this case, we use the BVP signal, a different form of cardiac information. Specifically, the FCN model applied to BVP achieved an accuracy of 68.51%, ALSTM reached 70.52%, and the combined FCN-ALSTM-Transformer achieved the highest accuracy for BVP at 71.27%. A similar performance trend is observed for the EDA signal, where the FCN-ALSTM-Transformer achieved the best result with an accuracy of 73.17%. Finally, the Crossmod-Transformer, which integrates both modalities, delivered the highest overall performance with an accuracy of 75.83%, a precision of 77.09%, and an F1 score of 74.63%.
Attention maps and interpretation
Improving the interpretability of DL models is critical for understanding their decision-making process and enhancing their reliability in research and clinical applications. In this study, interpretability analysis was conducted using the BioVid Heat Pain Database, which served as the primary dataset for model development. The final transformer block is utilised to generate three types of attention maps: 1) self-attention maps, 2) cross-attention maps, and 3) embedding attention maps, each offering distinct insights into how the model processes and prioritises information. Examples of these visualisations are presented in Fig. 5, 6, 7, highlighting the alignment of model attention with signal features from EDA and ECG. The attention maps use a colour gradient to visualise attention intensities, where red indicates high attention, blue represents low attention, and yellow and green signify medium levels of attention.
The overlayed signal-attention maps (Fig. 5) are derived from the diagonal elements of the self-attention weights. These weights, which represent the model’s focus on each sequence step relative to itself, are overlayed on the original signals to visualise the correlation. For EDA (Fig. 5a), the model demonstrates an evenly distributed attention pattern across the signal, with an increase at key inflection points. The smooth nature of the attention distribution correlates with the gradual changes inherent in EDA signals. The higher attention weights near peak and inflection reflect the physiological importance of these transitions. For ECG (Fig. 5b), attention weights peak at prominent features such as R-peaks, especially at the end of the session. This suggests that the model assigns higher importance to sharp transitions and high-amplitude events, which are indicative of cardiac activity. Attention diminishes in relatively stable regions, reflecting a focus on dynamic patterns.
Cross-attention maps (Fig. 6), which capture the interaction between ECG and EDA, highlighting the dependencies and demonstrating how information from one signal (e.g., ECG) influences the processing of the other (e.g., EDA). These maps are generated using the weights from the cross-attention layers, with ECG serving as the query for EDA in one direction (Fig. 6a), and EDA querying ECG in the other direction (Fig. 6b). The ECG to EDA cross-attention map, shown in Fig. 6a, highlights that a specific region of the EDA signal (key) is consistently attended to in response to the ECG signal (query). This implies that certain features or time steps in the EDA signal are crucial for interpreting or representing the ECG. The map similarity to the EDA self-attention map reinforces this interpretation, suggesting that the model consistently assigns significant importance to the same region of the EDA signal across both self-attention and cross-attention scenarios. The EDA to ECG cross-Attention Map (Fig. 6b) exhibits multiple vertical lines, indicating that the model consistently attends to distinct regions of the ECG signal (key) in response to various parts of the EDA signal (query). This pattern highlights the reliance of EDA on a broader temporal context from ECG, where specific features or time steps in the ECG contribute meaningfully to interpreting the slower and smoother dynamics of the EDA signal. The model assigns significant importance to similar ECG regions, indicating that these segments are crucial not only for ECG self-representation but also for supporting the interpretation of EDA signals. This alignment reinforces the model’s ability to leverage complementary information across modalities.
The embedding attention maps (Fig. 7) highlight the regions of the input signals prioritised by the model during its final decision-making process. These maps were generated from the contributions of the weights in the last FC layer of the model, with embeddings for EDA, ECG, and their fused representations processed and interpolated to enhance visual clarity. The EDA embedding map (Fig. 7a) is dominated by horizontal lines, suggesting that certain embedding dimensions consistently focus on similar time steps throughout the sequence. This reinforces the global, contextual role of EDA in the model. The presence of occasional high-intensity horizontal lines suggests that specific features or time steps in the EDA signal are particularly influential. The ECG embedding map (Fig. 7b) exhibits a more grid-like pattern, highlighting localised areas of high activation. These activations suggest that the ECG signal contributes to the final prediction through distinct temporal segments, potentially capturing transient but critical physiological changes. The localised nature of the activations reflects the model’s selective focus on specific ECG features. The fused embedding attention map (Fig. 7c) demonstrates an integration of the EDA and ECG attention patterns. It combines the horizontal dominance of the EDA with the localised contributions of the ECG, resulting in a complex but balanced representation. This map illustrates how the fusion mechanism leverages complementary strengths from both modalities.
The attention maps presented in this study provide a comprehensive understanding of how the model processes and prioritises information from EDA and ECG signals. By visualising self-attention, cross-attention, and embedding attention maps, we gain insights into the distinct and complementary roles of these physiological signals in the decision-making process of the proposed Crossmod-Transformer. The self-attention maps highlight the intrinsic dynamics within each signal, with EDA capturing gradual variations and ECG emphasising sharp transitions. The cross-attention maps reveal the interaction between the two signals, showing how each modality relies on distinct temporal features from the other. Finally, the embedding attention maps illustrate how the model integrates information from both EDA and ECG, leveraging their unique strengths to form a cohesive and insightful representation. These visualisations not only enhance the interpretability of the model but also highlight the importance of integrating complementary modalities for robust and clinically relevant predictions.
Cross-attention maps illustrating interdependence between ECG and EDA. (a) ECG-to-EDA cross-attention map, revealing consistent focus on specific EDA regions (keys) in response to ECG queries. (b) EDA-to-ECG cross-attention map, showing distinct ECG regions (keys) attended to by EDA queries, indicating reliance on complementary temporal features.
Embedding attention maps derived from the contributions of the final fully connected layer. (a) EDA embedding attention map, dominated by horizontal lines, reflecting consistent attention across embedding dimensions. (b) ECG embedding attention map, exhibiting a grid-like pattern with localised high activations, highlighting selective focus on specific temporal features. (c) Fused embedding attention map, integrating EDA and ECG attention patterns into a balanced representation, combining global and local features.
Comparative analysis with existing methods
The performance of the proposed approach is compared with state-of-the-art methods that have also utilised all the available data from 87 subjects of the BioVid Heat Pain Database (Part A). We categorise these studies into ECG-based studies, EDA-based studies, and multi-modal approaches, regardless of the number of modalities or their type that are utilised. Table 8 summarises the corresponding results. In comparing ECG-based methods, our model performed competitively. However, the method proposed by Gkikas et al.46 performed better than ours, achieving an accuracy of 67.04% in LOSO compared to our 65.17%. The superior performance of Gkikas et al.’s method can be attributed to their advanced ECG preprocessing technique. The authors applied the Pan-Tompkins Algorithm, which is one of the most well-known methods for detecting the QRS complex in ECG signals. Including these additional preprocessing steps, Gkikas et al. could probably extract more relevant features from the ECG signals, which likely contributed to the performance of their method for the ECG-based pain recognition task. In contrast, for the EDA modality, our proposed model (FCN-ALSTM-Transformer) achieved the highest accuracy among all comparative studies. For example, our approach obtained 85.21% and 86.12% in 10-fold and LOSO CV, respectively. These results highlight the robust capability of our model to independently analyse data for pain recognition, establishing it as a leading approach in EDA-specific contexts. Finally, in the multi-modal setting, our Crossmod-Transformer model also outperformed all other published methods. achieving an accuracy of 87.52% in LOSO and 85.92% in 10-fold CV. This result validates the effectiveness of our hybrid architecture in leveraging complementary information from both physiological signals for enhanced pain recognition.
Regarding the results obtained on the AI4PAIN dataset, Table 9 presents a comparison across relevant studies. We observe that several studies utilise behavioural data from facial videos, brain activity signals from fNIRS or a combination of both. In contrast, our approach utilises physiological modalities–specifically BVP and EDA–which demonstrate promising performance using the Crossmod-Transformer and can serve as a viable alternative or complementary source of information. Among the referenced works, the best test set result was reported by Bargshady et al.9 reported the highest accuracy of 66.96% using a Transformer-based approach applied to facial video data, while Gkikas et al.47 achieved 55.69% using a combination of video and fNIRS. Among the physiological approaches, the best result before our work was 56.25% accuracy obtained using BVP alone with a Gaussian SVM classifier 35. Our proposed Crossmod-Transformer achieved a significantly higher accuracy of 75.83% on the validation set using only physiological signals (EDA and BVP). This demonstrates not only the effectiveness of the proposed framework but also the strong potential of autonomic signals to serve as robust and scalable inputs for pain recognition.
Computational efficiency
To assess the practical feasibility of the proposed framework for real-world deployment, we evaluated its computational efficiency in terms of both theoretical and empirical measures. This evaluation was conducted using the BioVid Heat Pain Database, as it served as the primary dataset for model development. Figure 8 illustrates the floating-point operations (FLOPS) and inference time per sample for each sub-network and modality. Lightweight uni-modal models such as FCN and ALSTM exhibit low computational demands (73.28 MFLOPS and 55.82 MFLOPS, respectively), while the noticeable increase for the multi-modal Crossmod-Transformer (442.76 MFLOPS) reflects the added complexity of feature fusion across modalities.
In total, the combined FLOPS of all sub-networks (including the uni-modal models and the Crossmod-Transformer) is approximately 1.03 GigaFLOPS (GFLOPS), with an average inference time of 134 milliseconds per sample on our hardware (Intel i7-13650HX CPU). These results indicate that the proposed framework is computationally efficient and potentially suitable for deployment in resource-constrained environments, such as wearable devices.
Discussion
The results obtained in this work demonstrate the effectiveness of the proposed CrossMod-Transformer multi-modal deep learning model for pain recognition using both ECG and EDA signals. In addition, the fusion of ECG and EDA modalities within the model outperformed other published methods across both cross-validation schemes. These findings highlight the value of integrating these two modalities in pain recognition, as their combination provides richer and more informative features compared to single modalities. These results align with previous research in pain assessment, which has highlighted the importance of multi-modal approaches (e.g., combining ECG and EDA signals)12,23.
Preprocessing techniques played a critical role in ECG-based pain recognition. While the CrossMod-Transformer model demonstrated superior performance in combining EDA and ECG signals, the ECG-specific accuracy in this study was slightly lower than that reported by Gkikas et al.46, who achieved 67.04% in LOSO. A key factor contributing to their superior performance appears to be the use of advanced preprocessing methods for ECG signals. Specifically, we believe that the application of the Pan-Tompkins Algorithm enhanced the detection of QRS complexes, enabling the extraction of more precise pain-relevant features. This highlights the importance of incorporating robust preprocessing techniques in refining ECG feature extraction and improving model accuracy.
The attention maps generated in this study provide a detailed understanding of how the model processes and prioritises information from EDA and ECG signals. The self-attention maps revealed the model’s ability to focus on key temporal features within each modality, such as inflection points in EDA and R-peaks in ECG, reflecting their physiological significance. The cross-attention maps demonstrated the complementary interaction between the two signals, with ECG focusing on distinct EDA regions and EDA relying on broader ECG contexts, highlighting their synergistic roles. The embedding attention maps further illustrated how the model integrates these signals in its final decision-making, with EDA contributing global context and ECG offering dynamic, localised insights. These visualisations emphasise the interpretability of the model and its ability to integrate multi-modal information for reliable predictions, which is especially valuable in clinical applications.
The selection of EDA and ECG signals for this study was motivated by their compatibility with wearable technologies. These devices offer a cost-effective and non-intrusive method for pain recognition in practical applications21. Unlike video-based approaches, which often require controlled environments and encounter challenges such as occlusion and privacy concerns11, wearable devices like smartwatches or sensor patches can collect data continuously in real-world settings. In addition, these devices support decentralised healthcare by reducing reliance on specialised equipment, thereby broadening access to pain monitoring systems. Studies have demonstrated that patients with chronic pain are willing to use wearable health technologies over extended periods, indicating their feasibility for long-term pain management applications57.
Using the second dataset, AI4PAIN, we evaluated the generalisation capabilities of the proposed method. While the primary dataset comprised EDA and ECG signals, the AI4PAIN dataset offers a different yet physiologically relevant combination (EDA and BVP). Notably, although CrossMod-Transformer was initially developed for EDA–ECG fusion, it was applied to the AI4PAIN dataset without any architectural modifications or with only minimal adaptation. In this evaluation, BVP was used as the cardiac input in place of ECG. The model demonstrated strong performance in both uni-modal and multi-modal settings. Consistent with the BioVid results, EDA outperformed the cardiac modality. The combination of the two modalities further improved performance, achieving 75.83% accuracy, compared to 71.27% and 73.17% for BVP and EDA, respectively. This cross-dataset validation demonstrates the versatility of the CrossMod-Transformer in integrating autonomic signals beyond its original EDA and ECG combination, confirming its applicability to pain detection across independent experimental settings.
While the CrossMod-Transformer model has demonstrated strong performance in integrating ECG, EDA, and BVP signals for pain recognition, several limitations remain. One key limitation is the reliance on a limited set of three physiological modalities, which may not fully capture the complex and multifaceted nature of pain responses. The inclusion of additional biosignals, such as photoplethysmography (PPG), respiration, or functional near-infrared spectroscopy (fNIRS), could further enrich the feature space and enhance the model’s discriminative power37,58. These modalities offer complementary insights into cerebral and peripheral physiological processes associated with pain perception. Integrating such signals would allow the model to learn from a broader and more diverse range of autonomic and cortical responses, thereby increasing its sensitivity to pain-specific patterns and potentially improving both accuracy and robustness in real-world settings.
In conclusion, this paper introduces a hybrid deep-learning model for pain recognition. Experimental results demonstrate the effectiveness of the proposed approach using physiological signals from the BioVid Heat Pain Database and AI4PAIN. The model achieved high accuracy, particularly when combining ECG and EDA with FCN, ALSTM, and Transformer components. Similarly, the use of BVP and EDA in a multi-modal configuration further improved performance. These findings underscore the importance of utilising multiple modalities to capture the multidimensional nature of pain. Additionally, the proposed multi-step training strategy reduced computational demands while improving accuracy, enhancing the model’s flexibility and scalability for future integration of new deep learning components or modalities without requiring full retraining. Future work should investigate the inclusion of additional physiological signals, systematically assess their individual and combined performance, and analyse their respective advantages and limitations in multi-modal setups. Additionally, the available classes in the pain-related datasets provide opportunities to explore and evaluate various pain assessment scenarios under both binary and multiclass classification settings. Future work needs to investigate these scenarios more systematically. Furthermore, we believe that model interpretability is a critical factor for clinical adoption. Deeper analysis and clearer explanations of the model’s decision-making processes are crucial for building trust and facilitating integration into healthcare systems for patient monitoring.
Data availability
The Biovid dataset used in the manuscript is publicly available at https://www.nit.ovgu.de/BioVid.html and the AI4PAIN dataset is publicly available at https://sites.google.com/view/ai4pain2025/home.
Code availability
The implementation of our framework will be made publicly available at: https://github.com/JalehFar/CrossMod-Transformer.
References
Raja, S. N. et al. The revised international association for the study of pain definition of pain: concepts, challenges, and compromises. Pain 161, 1976–1982 (2020).
Turk, D. C. & Melzack, R. Handbook of pain assessment (Guilford Press, 2011).
Werner, P. et al. Automatic recognition methods supporting pain assessment: A survey. IEEE Trans. Affect. Comput. 13, 530–552 (2019).
Hawker, G. A., Mian, S., Kendzerska, T. & French, M. Measures of adult pain: Visual analog scale for pain (vas pain), numeric rating scale for pain (nrs pain), mcgill pain questionnaire (mpq), short-form mcgill pain questionnaire (sf-mpq), chronic pain grade scale (cpgs), short form-36 bodily pain scale (sf-36 bps), and measure of intermittent and constant osteoarthritis pain (icoap). Arthritis Care Res. 63, S240–S252 (2011).
Franck, L. S., Greenberg, C. S. & Stevens, B. Pain assessment in infants and children. Pediatr. Clin. N. Am. 47, 487–512 (2000).
asking the right questions. Kappesser, J. & Williams, A. C. d. C. Pain estimation. Pain 148, 184–187 (2010).
Mende-Siedlecki, P. et al. Seeing no pain: Assessing the generalizability of racial bias in pain perception. Emotion 21, 932 (2021).
Gkikas, S., Chatzaki, C. & Tsiknakis, M. Multi-task neural networks for pain intensity estimation using electrocardiogram and demographic factors. In Information and Communication Technologies for Ageing Well and e-Health, 324–337, https://doi.org/10.1007/978-3-031-37496-8_17 (Springer Nature Switzerland, 2023).
Bargshady, G., Joseph, C., Hirachan, N., Goecke, R. & Rojas, R. F. Acute pain recognition from facial expression videos using vision transformers. In 2024 46th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 1–4, https://doi.org/10.1109/EMBC53108.2024.10781616 (2024).
Gkikas, S. & Tsiknakis, M. A full transformer-based framework for automatic pain estimation using videos. In 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 1–6 (IEEE, 2023).
Pouromran, F., Radhakrishnan, S. & Kamarthi, S. Exploration of physiological sensors, features, and machine learning models for pain intensity estimation. Plos one 16, e0254108 (2021).
Phan, K. N., Iyortsuun, N. K., Pant, S., Yang, H.-J. & Kim, S.-H. Pain recognition with physiological signals using multi-level context information. IEEE Access 11, 20114–20127 (2023).
Gkikas, S. et al. Automatic pain intensity estimation based on electrocardiogram and demographic factors. In ICT4AWE, 155–162 (2022).
Kächele, M., Thiam, P., Amirian, M., Schwenker, F. & Palm, G. Methods for person-centered continuous pain intensity assessment from bio-physiological channels. IEEE J. Sel. Top. Signal Process. 10, 854–864 (2016).
Gkikas, S. & Tsiknakis, M. Synthetic thermal and rgb videos for automatic pain assessment utilizing a vision-mlp architecture. In 2024 12th International Conference on Affective Computing and Intelligent Interaction Workshops and Demos (ACIIW), 4–12, https://doi.org/10.1109/ACIIW63320.2024.00006 (2024).
Tao, W. et al. Adfcnn: Attention-based dual-scale fusion convolutional neural network for motor imagery brain-computer interface. IIEEE Trans. Neural Syst. Rehabil. Eng. 32, 154–165. https://doi.org/10.1109/TNSRE.2023.3342331 (2024).
Zhong, X.-C. et al. Eeg-dg: A multi-source domain generalization framework for motor imagery eeg classification. IEEE J. Biomed. Health Inform. 29, 2484–2495 (2025).
Wei, F., Xu, X., Li, X. & Wu, X. Bdan-spd: A brain decoding adversarial network guided by spatiotemporal pattern differences for cross-subject mi-bci. IEEE Trans. Ind. Inform. 20, 14321–14329. https://doi.org/10.1109/TII.2024.3450010 (2024).
Lu, Z., Ozek, B. & Kamarthi, S. Transformer encoder with multiscale deep learning for pain classification using physiological signals. Front. Physiol. 14, 1294577 (2023).
Gkikas, S. & Tsiknakis, M. Automatic assessment of pain based on deep learning methods: A systematic review. Comput. Methods Programs Biomed. 231, 107365 (2023).
Fernandez Rojas, R., Brown, N., Waddington, G. & Goecke, R. A systematic review of neurophysiological sensing for the assessment of acute pain. NPJ Digit. Med. 6, 76 (2023).
Werner, P. et al. Automatic pain recognition from video and biomedical signals. In 2014 22nd international conference on pattern recognition, 4582–4587 (IEEE, 2014).
Lopez-Martinez, D. & Picard, R. Multi-task neural networks for personalized pain recognition from physiological signals. In 2017 Seventh International Conference on Affective Computing and Intelligent Interaction Workshops and Demos (ACIIW), 181–184 (IEEE, 2017).
Wang, R., Xu, K., Feng, H. & Chen, W. Hybrid rnn-ann based deep physiological network for pain recognition. In 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 5584–5587 (IEEE, 2020).
Wang, Z., Yan, W. & Oates, T. Time series classification from scratch with deep neural networks: A strong baseline. In 2017 International joint conference on neural networks (IJCNN), 1578–1585 (IEEE, 2017).
Rojas, R. F., Romero, J., Lopez-Aparicio, J. & Ou, K.-L. Pain assessment based on fnirs using bi-lstm rnns. In 2021 10th International IEEE/EMBS Conference on Neural Engineering (NER), 399–402 (IEEE, 2021).
Vaswani, A. Attention is all you need. Advances in Neural Information Processing Systems (2017).
Tao, Y. et al. Depressive semantic awareness from vlog facial and vocal streams via spatio-temporal transformer. Digit. Commun. Netw. 10, 577–585 (2024).
Waligora, P. et al. Joint multimodal transformer for emotion recognition in the wild. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 4625–4635 (2024).
Walter, S. et al. The biovid heat pain database data for the advancement and systematic validation of an automated pain recognition system. In 2013 IEEE international conference on cybernetics (CYBCO), 128–131 (IEEE, 2013).
Werner, P., Al-Hamadi, A. & Walter, S. Analysis of facial expressiveness during experimentally induced heat pain. In 2017 Seventh international conference on affective computing and intelligent interaction workshops and demos (ACIIW), 176–180 (IEEE, 2017).
Subramaniam, S. D. & Dass, B. Automated nociceptive pain assessment using physiological signals and a hybrid deep learning network. IEEE Sens. J. 21, 3335–3343 (2020).
Thiam, P., Bellmann, P., Kestler, H. A. & Schwenker, F. Exploring deep physiological models for nociceptive pain recognition. Sensors 19, 4503 (2019).
Thiam, P., Hihn, H., Braun, D. A., Kestler, H. A. & Schwenker, F. Multi-modal pain intensity assessment based on physiological signals: A deep learning perspective. Front. Physiol. 12, 720464 (2021).
Fernandez Rojas, R., Hirachan, N., Joseph, C., Seymour, B. & Goecke, R. The ai4pain grand challenge 2025: Advancing pain assessment with multimodal physiological signals. In Proceedings of the 27th ACM International Conference on Multimodal Interaction (ICMI 2025) (ACM, Canberra, Australia, 2025).
Fernandez-Rojas, R., Joseph, C., Hirachan, N., Seymour, B. & Goecke, R. The ai4pain grand challenge 2024: Advancing pain assessment with multimodal fnirs and facial video analysis. In Proceedings of the 2024 12th International Conference on Affective Computing and Intelligent Interaction Workshops and Demos (ACIIW) (IEEE, 2024).
Fernandez Rojas, R. et al. Multimodal physiological sensing for the assessment of acute pain. Front. Pain Res. 4, 1150264 (2023).
Ahsan, M. M., Mahmud, M. P., Saha, P. K., Gupta, K. D. & Siddique, Z. Effect of data scaling methods on machine learning algorithms and model performance. Technologies 9, 52 (2021).
Long, J., Shelhamer, E. & Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition, 3431–3440 (2015).
Hopfield, J. J. & Tank, D. W. “neural’’ computation of decisions in optimization problems. Biol. Cybern. 52, 141–152 (1985).
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. 9, 1735–1780 (1997).
Bahdanau, D. Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv:1409.0473 (2014).
Karim, F., Majumdar, S., Darabi, H. & Chen, S. Lstm fully convolutional networks for time series classification. IEEE access 6, 1662–1669 (2017).
Huang, Y., Lin, J., Zhou, C., Yang, H. & Huang, L. Modality competition: What makes joint training of multi-modal network fail in deep learning?(provably). In International conference on machine learning, 9226–9259 (PMLR, 2022).
Akiba, T., Sano, S., Yanase, T., Ohta, T. & Koyama, M. Optuna: A next-generation hyperparameter optimization framework. In The 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, 2623–2631 (2019).
Gkikas, S. et al. Multimodal automatic assessment of acute pain through facial videos and heart rate signals utilizing transformer-based architectures. Front. Pain Res. 5, 1372814 (2024).
Gkikas, S., Rojas, R. F. & Tsiknakis, M. Painformer: a vision foundation model for automatic pain assessment (2025). arxiv:2505.01571.
Farmani, J., Giuseppi, A., Bargshady, G. & Fernandez Rojas, R. Multimodal automatic acute pain recognition using facial expressions and physiological signals. In International Conference on Neural Information Processing, 49–62 (Springer, 2025).
Kächele, M. et al. Bio-visual fusion for person-independent recognition of pain intensity. In Multiple Classifier Systems: 12th International Workshop, MCS 2015, Günzburg, Germany, June 29-July 1, 2015, Proceedings 12, 220–230 (Springer, 2015).
Gouverneur, P. et al. Comparison of feature extraction methods for physiological signals for heat-based pain recognition. Sensors 21, 4838 (2021).
Kächele, M. et al. Multimodal data fusion for person-independent, continuous estimation of pain intensity. In Engineering Applications of Neural Networks: 16th International Conference, EANN 2015, Rhodes, Greece, September 25-28 2015. Proceedings, 275–285 (Springer, 2015).
Gkikas, S. & Tsiknakis, M. Twins-painvit: Towards a modality-agnostic vision transformer framework for multimodal automatic pain assessment using facial videos and fnirs. In 2024 12th International Conference on Affective Computing and Intelligent Interaction Workshops and Demos (ACIIW), 13–21, https://doi.org/10.1109/ACIIW63320.2024.00007 (2024).
Prajod, P., Schiller, D., Don, D. W. & André, E. Faces of experimental pain: Transferability of deep-learned heat pain features to electrical pain*. In 2024 12th International Conference on Affective Computing and Intelligent Interaction Workshops and Demos (ACIIW), 31–38, https://doi.org/10.1109/ACIIW63320.2024.00009 (2024).
Khan, M. U. et al. Empirically transformed energy patterns: A novel approach for capturing fnirs signal dynamics in pain assessment. Comput. Biol. Med. 192, 110300. https://doi.org/10.1016/j.compbiomed.2025.110300 (2025).
Nguyen, M.-D., Yang, H.-J., Kim, S.-H., Shin, J.-E. & Kim, S.-W. Transformer with leveraged masked autoencoder for video-based pain assessment (2024). arxiv:2409.05088.
Vianto, J. et al. Multimodal model for automated pain assessment: Leveraging video and fnirs. Appl. Sci. 15, https://doi.org/10.3390/app15095151 (2025).
Han, J. J., Graham, J. H., Snyder, D. I. & Alfieri, T. Long-term use of wearable health technology by chronic pain patients. Clin. J. Pain. 38, 701–710 (2022).
Fernandez Rojas, R., Joseph, C., Bargshady, G. & Ou, K.-L. Empirical comparison of deep learning models for fnirs pain decoding. Front. Neuroinformatics 18, 1320189 (2024).
Author information
Authors and Affiliations
Contributions
Conceptualisation: G.B., R.F.R.; Methodology: J.F., G.B., R.F.R.; Data analysis: J.F., G.B., R.F.R., S,G.; Investigation: G.B, S.G., M.T., R.F.R.; Analysis of Results: J.F., G.B, S.G., M.T., R.F.R.; Writing – Original Draft: J.F.; Writing – Review & Editing: G.B, S.G., M.T., R.F.R.; Visualisation: J.F., S.G.; Supervision: G.B, M.T., R.F.R. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Farmani, J., Bargshady, G., Gkikas, S. et al. A CrossMod-Transformer deep learning framework for multi-modal pain detection through EDA and ECG fusion. Sci Rep 15, 29467 (2025). https://doi.org/10.1038/s41598-025-14238-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-14238-y
Keywords
This article is cited by
-
Machine learning methods on BioVid heat pain database for pain intensity estimation
Scientific Reports (2025)