Abstract
Dengue and leptospirosis are prevalent diseases in tropical and subtropical regions, posing significant public health challenges. These illnesses exhibit overlapping symptoms, including fever, muscle pain, and headaches, which complicates diagnosis and delays appropriate treatment. This study explores the use of attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR) combined with multivariate analysis to distinguish between the two infections by analyzing blood plasma in both liquid and dry states. A total of 114 patient samples at varying infection stages (n = 43 for leptospirosis and n = 71 for dengue) were examined using linear discriminant analysis (LDA), quadratic discriminant analysis (QDA), and support vector machine (SVM) in conjunction with genetic algorithms (GA), successive projection algorithms (SPA), and principal component analysis (PCA) for feature selection and extraction. The SPA-QDA model applied to dried plasma delivered exceptional results, achieving 100% sensitivity, specificity, and accuracy in distinguishing the two diseases using only 30 spectral variables. ANOVA calculations, conducted with a 95% confidence level, identified four key wavenumbers (1601 cm−1, 1735 cm−1, 1747 cm−1, and 1870 cm−1) as critical for class differentiation.
Similar content being viewed by others
Introduction
Dengue and leptospirosis are prevalent in tropical and subtropical regions, where they pose significant public health challenges. Both diseases exhibit similar symptoms, including fever, muscle pain, and headaches, making diagnosis difficult and delaying appropriate treatment1. Dengue is a systemic and dynamic infection caused by the dengue virus (DENV), which consists of five serotypes (DENV-1 to DENV-5), with the most recent, DENV-5, identified in recent years. The illness can present in varying degrees of severity, ranging from an asymptomatic infection to a febrile condition (39–40 °C) without complications or, in severe cases, increased vascular permeability, which may lead to hemorrhagic manifestations and potentially fatal outcomes. Its clinical presentation closely resembles other illnesses associated with febrile syndrome2.
Leptospirosis is a zoonotic disease caused by the bacterium Leptospira. In its early acute phase, it presents with nonspecific symptoms such as fever, myalgia, and headache, lacking distinctive features for immediate diagnosis. Due to its diverse clinical manifestations, leptospirosis can resemble various infectious diseases, including influenza, hepatitis, dengue, hantavirus infections, yellow fever, malaria, other viral hemorrhagic fevers and brucellosis3. Currently, laboratory-based diagnostic techniques are the primary methods for detecting dengue and leptospirosis, as clinical evaluation alone is prone to misinterpretation4. Conventional approaches include pathogen isolation, genome detection, and serological or antigen-antibody immunoassays; however, these methods may suffer from reduced specificity, particularly in distinguishing between related viruses such as dengue and Zika5,6.
For leptospirosis, diagnostic tools are significantly impacted by sensitivity limitations, with rates dropping as low as 40%7,8. This variability is influenced by the type of sample used (typically blood or urine) and the stage of the disease at the time of collection. Currently, no single gold-standard test exists for definitively diagnosing either disease, as detection relies on a combination of laboratory tests and clinical or epidemiological evaluation. Therefore, the development of alternative diagnostic methods is crucial, particularly in endemic regions such as the Caribbean and Latin America, where outbreaks and high mortality rates remain a persistent challenge.
Biospectroscopy refers to the application of spectroscopic techniques for analyzing biological samples. This approach has garnered significant attention in the medical field as a promising tool for disease detection and diagnosis9,10,11,12. In this context, attenuated total reflection-Fourier transform infrared spectroscopy (ATR-FTIR) provides a simple and rapid method for analyzing various biological samples, including biofluids, cells, and tissues, without requiring sample pretreatment. The infrared spectra arise from molecular vibrations within the sample, where specific components interact with the infrared beam and absorb portions of it. The 1800–900 cm− 1 range, known as the biofingerprint region, offers valuable insights into sample composition, capturing key vibrational bands associated with lipids (~ 1750 cm− 1), protein amide groups (amide I at ~ 1650 cm− 1, amide II at ~ 1550 cm− 1, and amide III at ~ 1260 cm− 1), carbohydrates (~ 1150 cm− 1), and nucleic acids (RNA/DNA at ~ 1080 cm− 1 and ~ 1225 cm− 1)13.
Although highly informative, infrared spectra of biological samples can be complex to interpret due to the multitude of molecular contributions to the obtained signal, posing a challenge for real-world applications. To address this issue, multivariate analysis is often integrated with ATR-FTIR spectroscopy, utilizing various chemometric methods for data processing. In the context of disease detection and diagnosis, multivariate classification techniques such as linear discriminant analysis (LDA), quadratic discriminant analysis (QDA), and support vector machines (SVM) have proven effective in enhancing the diagnostic potential of spectroscopic and chemometric approaches across various applications14,15,16,17.
Santos et al.9. conducted a pioneering study to differentiate healthy individuals from those with dengue, Zika or chikungunya infections by analyzing liquid phase blood samples placed directly on the ATR-FTIR glass with a piece of aluminum paper acting as substrate at room temperature (22 °C) and using air measurements as background. By using chemometric algorithms, they were able to classify samples with a remarkable sensitivity and specificity of 100% for healthy, dengue and chikungunya samples, and values close to 90% for Zika samples9.
Naseer et al.18 introduced an innovative method for dengue diagnosis, using freeze-dried blood serum samples to remove water while preserving the structural integrity of biomolecules. ATR-FTIR spectroscopy was employed to identify biochemical markers of dengue infection and differentiate between healthy and infected samples. Using principal component analysis combined with linear discriminant analysis (PCA-LDA), the study achieved classification sensitivity and specificity of 89% and 95%, respectively18.
Expanding on their methodology, Ali et al.11 explored the use of ATR-FTIR spectroscopy to differentiate between dengue and hepatitis C infections in human blood by analyzing lyophilized serum samples. Through multivariate analysis, they achieved an impressive classification accuracy of 99.2%, demonstrating the potential of this approach as a reliable alternative for the differential diagnosis of viral diseases11.
These recent studies highlight the effectiveness of ATR-FTIR spectroscopy combined with multivariate analysis in analyzing biological samples, particularly blood plasma and serum. This approach not only facilitates disease detection but also supports the development of alternative methods for differential diagnosis. The primary aim of this pilot study was to establish a simple, rapid, and cost-effective methodology based on ATR-FTIR spectroscopy19,20 and multivariate classification to accurately distinguish between leptospirosis and dengue using liquid and dried blood plasma samples. Given that both diseases exhibit nonspecific acute symptoms, which can lead to misdiagnosis21 there is a critical need for improved diagnostic methods. Enhancing early-stage detection will aid in accurate medical management, ultimately reducing mortality rates and the risk of outbreaks, particularly in endemic regions.
Methods
Collection and sample preparation
This study included patients from the Department of Atlántico, Colombia, who were admitted to various hospitals for medical evaluation and treatment of infection-related symptoms. The Public Health Laboratory of the Department of Atlántico processed, diagnosed, and collected 95.53% of the total samples, while the remaining 4.47% were handled by the Tropical Diseases Research Group at Universidad del Norte (Colombia). The study was approved by the medical ethics committee of Universidad del Atlántico (Colombia), with official approval granted in August 2023 (acta nº 007). Informed consent was obtained from all participants, and all procedures adhered to relevant ethical guidelines and regulations, including the Declaration of Helsinki. Liquid blood plasma samples were obtained from 114 patients at different stages of infection (n = 43 for leptospirosis, and n = 71 for dengue), comprising both male and female participants aged five to sixty years. Blood was collected via venipuncture, transferred to EDTA-containing tubes as an anticoagulant, and processed within two hours. Plasma separation was performed by density gradient centrifugation using a TDL-4 model centrifuge (Zenith Lab Inc., China) at 3500 rpm for 10 min at room temperature, after which aliquots were stored in cryogenic tubes at −80 °C for subsequent analysis in both liquid and dried phases. Leptospirosis was diagnosed using the microscopic agglutination test (MAT) as the reference serological method, along with IgM ELISA testing. Dengue was diagnosed using the ELISA tests for IgM and IgG antibodies as well as for the NS1 antigen.
ATR-FTIR spectroscopy
ATR-FTIR spectral measurements were performed using a JASCO 4700 FTIR spectrometer (JASCO Corporation, Tokyo, Japan) with an ATR attachment containing a diamond crystal internal reflective element at a 45° incidence angle from the IR beam. The instrument was set up to perform a total of 16 scans with 4 cm−1 spectral resolution on both the background and sample. Although the resolution was set to 4 cm⁻¹, the final exported spectra contained 1557 data points, corresponding to a wavenumber interval of approximately 0.964 cm⁻¹. This denser point spacing results from the instrument’s internal interpolation and zero-filling processes. Pure isopropanol (98%) was used for cleaning the ATR diamond crystal. Each sample was placed directly on the ATR diamond crystal using a micropipette with a 20 µL aliquot, then they were allowed to dry for exactly 15 min using a portable USB mini fan placed vertically over the sample under temperature conditions (between 20 and 25 °C) and average humidity of 50%. Additionally, an aluminum foil sheet was placed on the sample, acting as a substrate before spectral measurements. Each sample was analyzed in triplicate, resulting in a total of 342 spectra for both liquid and dried blood plasma samples (n = 129 for leptospirosis, and n = 213 for dengue). Furthermore, randomization was applied when performing the measurements of dengue and leptospirosis samples to minimize any systematic bias.
Data analysis and chemometric methods
All computational analyses, including data import, preprocessing, and multivariate classification models development, were conducted in the MATLAB® 2022 environment (MathWorks Inc., Natick, MA, USA) using the PLS Toolbox version 9.2 (Eigenvector Research Inc., USA) along with custom laboratory-made routines. Each plasma sample was represented by its mean spectrum, calculated from the three spectral replicates per sample, resulting in 43 mean spectra for leptospirosis and 71 for dengue. The ATR-FTIR mean spectra were preprocessed by selecting the fingerprint region between 1900 and 1000 cm− 1, yielding a 114 × 935 data matrix. Baseline correction was performed using automatic weighted least squares, and normalization was applied to the amide I peak (~ 1650 cm− 1). For model development, samples were divided into training (~ 70%, n = 77), validation (~ 15%, n = 18), and prediction (~ 15%, n = 18) sets using the Kennard-Stone sample selection algorithm on the ATR-FTIR mean spectra22. Before construction of discrimination models, one leptospirosis sample from the liquid plasma dataset and one dengue sample from the dried plasma dataset were excluded due to an outlier spectral behavior. The training set was used for model construction, the validation set for model optimization by assessing the validation performance, and the prediction set for evaluating the optimized models in real-case scenarios involving unknown samples.
The algorithms used for model construction included Principal Component Analysis (PCA), which reduces data dimensionality by projecting the original variables onto a new orthogonal feature space23. Additionally, the Successive Projections Algorithm (SPA) and Genetic Algorithm (GA) were applied to select spectral variables that maximize class differentiation24,25. These dimensionality reduction techniques were combined with linear discriminant analysis (LDA) and quadratic discriminant analysis (QDA) to enhance class separation by analyzing variance relationships both between and within classes15. This resulted in the following model combinations: PCA-LDA, SPA-LDA, GA-LDA, PCA-QDA, SPA-QDA, and GA-QDA. Furthermore, a nonlinear classifier, Support Vector Machines (SVM)26 was also tested in combination with PCA, SPA, and GA for data discrimination, leading to additional models: PCA-SVM, SPA-SVM, and GA-SVM17.
A detailed description of these algorithms has been previously provided by our research group and can be found elsewhere12,27. For GA-LDA/QDA calculations, 40 generations were used, each containing 80 chromosomes. The one-point crossover and mutation probabilities were set at 60% and 10%, respectively. Additionally, the algorithm was run three times, each starting from a different random population. The best solution, determined by the highest fitness value among the three GA iterations, was selected for further analysis.
The calculations of figures of merit, including sensitivity (probability that a test result will be positive when the class is actually positive), specificity (probability that a test result will be negative when the class is actually negative), precision (the proportion of positive samples correctly classified), accuracy (number of samples correctly classified considering true and false negatives) and both the area under the ROC curve (AUC) and F-Score, were calculated to evaluate the model performance, since these are important quality metrics to assess the models discriminant ability when they are applied to unknown/test samples28. These parameters have a maximum value of 1 (100%) and a minimum of 0 (0%), and can be obtained by using the following equation29:
where FN is defined as the number of false negatives, FP as the number of false positives, TP as the number of true positives, and TN as the number of true negatives. Herein, for all calculations, leptospirosis samples were considered the positive class, and dengue samples were considered the negative class.
Results
In this pilot study, a total of 114 liquid and dry phase blood plasma samples from patients suffering from acute symptoms of dengue (n = 71) or leptospirosis (n = 43) were collected. The blood plasma samples were analyzed by infrared spectroscopy using the ATR mode. The raw spectra of all liquid and dry phase samples in the biofingerprint region between 1900 and 1000 cm−1 are shown in Fig. 1A,B, where major biomolecules show significant absorption bands.
Biofingerprint spectra for dengue and leptospirosis samples in liquid (A) and dry (B) blood plasma samples; pre-processed spectra (baseline correction and amide I normalization) for dengue and leptospirosis in liquid (C) and dry (D) blood plasma samples; averaged pre-processed spectrum for dengue and leptospirosis in liquid (E) and dry (F) blood plasma samples.
For construction of multivariate classification models, the spectra were preprocessed with baseline correction and normalization to the amide I peak to eliminate physical interferences and highlight chemical signals between classes, as shown in Fig. 1C,D. The mean preprocessed spectra of both classes, dengue and leptospirosis, are presented in Fig. 1E,F, showing similarities in their absorption bands. However, the difference in the wavenumber at 1550 cm− 1 between the spectra in liquid and dry phase is evident. This band is minimal in height in liquid plasma, but as the drying process is carried out until completing the continuous time of 15 min, the band reaches its maximum absorbance value.
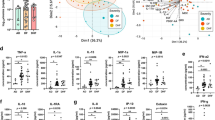
Figure 2A,B show the PCA scores for liquid and dry blood plasma samples, respectively. Based on the PCA scores for the liquid samples (Fig. 2A), the score plot contains 95% of the explained variance within the first two principal components. In general, the PCA analysis of liquid samples did not reveal a clear separation between the samples belonging to the dengue and leptospirosis classes. However, there is an important observation in this plot related to the presence of an atypical leptospirosis sample that is far from the confidence level limits. This sample was considered an outlier and removed from the data before the construction of multivariate classification models. In Fig. 2B, the PCA scores of dry plasma samples contain 91.6% of the total variance within the dataset using the first two principal components. Similarly, there is no clear separation between the samples from both classes. In addition, an atypical value is evident, but for dengue. This sample was also eliminated from the data before performing multivariate classification.
Classification models based on LDA, QDA and SVM discrimination algorithms (PCA-LDA, PCA-QDA, SPA-LDA, SPA-QDA, GA-LDA GA-QDA, PCA-SVM, SPA-SVM and GA-SVM) were built for the pre-processed data. Table 1 shows the confusion matrix for each model, considering the prediction set samples for both liquid and dry phase blood samples.
In the LDA-based models, GA-LDA and SPA-LDA showed poor classification performance for leptospirosis, whereas PCA-LDA, using the first three principal component scores, misclassified only one sample in this class. For distinguishing dengue patients, SPA-LDA and GA-LDA outperformed PCA-LDA. QDA did not improve the classification accuracy based on the PCA-QDA results obtained for either disease; however, it yielded better results with SPA-QDA for leptospirosis samples. Notably, GA-QDA demonstrated a substantial enhancement in the classification performance for both diseases, achieving 100% accuracy for all prediction samples while requiring only 31 spectral variables for the samples in the liquid phase.
Using the same models applied to dry phase samples, all LDA models substantially improved the classification results when compared to the liquid samples. The results of the SPA-QDA model for both classes are particularly notable, with the model achieving 100% correct classification for all prediction samples using 30 spectral variables.
The SVM-based models (PCA-SVM, SPA-SVM and GA-SVM) did not provide better classification in general. The results show a better classification for the negative class (dengue samples), and the classification of the positive class (leptospirosis) improved when changing the phase to dry, but with a non-optimal percentage. To provide a better evaluation of the models performance, some figures of merit, including sensitivity, specificity, accuracy, F-Score and AUC calculations, are depicted in Table 2.
Considering leptospirosis as the positive class, the sensitivity values for the prediction set ranged from 28.5 to 100%, while the specificity varied from 36.3 to 100%, and the accuracy ranged from 38.8 to 100%. The best models for both liquid and dry phases were the GA-QDA and SPA-QDA models, both showing 100% accuracy and an F-score and AUC of 1.
The discriminant functions showing the separation between leptospirosis and dengue samples in liquid and dry phases is shown in Fig. 3A,B for the best performing GA-QDA and SPA-QDA models, respectively. The spectra with the selected subset of 31 (GA-QDA) and 30 (SPA-QDA) variables out of the 935 wavenumbers for both models is shown in Fig. 3C, highlighting the main spectral regions used for correct discrimination. The ROC curve is shown in Fig. 3D for the GA-QDA and SPA-QDA models.
Discrimination function (DF) plot for GA-QDA in liquid phase (A) and SPA-QDA in dried phase (B) models (the blue circles indicate dengue, and the red circles indicate leptospirosis); (C) selected variables for both models (GA-QDA in liquid phase – blue; SPA-QDA in dried phase – red); (D) ROC curves for both GA-QDA in liquid phase and SPA-QDA in dried phase models.
Analysis of variance (ANOVA) was applied to the selected variables by GA-QDA and SPA-QDA to verify significant differences between the absorption signals of dengue and leptospirosis samples. At a 95% confidence level, a set of 10 out of 31 variables selected by the GA-QDA model, and 4 out of 30 variables by the SPA-QDA model, presented significant differences (P < 0.05), as shown in Fig. 4A,B.
According to the box plots of plasma samples in liquid phase (Fig. 4A), the mean absorbance values of dengue samples were greater than those of leptospirosis for all the selected wavenumbers, with the exception of the wavenumber at 1612 cm− 1. Additionally, more extreme values were identified for dengue samples, while for leptospirosis, a more evenly distribution was observed, with the exception of the wavenumber at 1296 cm− 1. The box plots for the same plasma sample in dry phase are shown in Fig. 4B. Overall, the mean absorbance values of leptospirosis are greater than dengue.
Discussion
Leptospirosis and dengue are infectious diseases that present very similar initial acute symptoms, where high fever, headache, muscle pain, nausea and chills are common symptoms in most patients21; but, some patients are asymptomatic. In the study developed by Rodriguez-Villamarin et al.30 they indicate that the symptomatic similarity between leptospirosis and dengue diseases in their initial phase has caused difficulty for a timely diagnosis, for the adequate clinical care of the patients, as well as for their notification and classification. In this sense, many patients suspected of dengue that test negative for this infection may be positive for Leptospira30. Therefore, the similarity in the acute phase may result in additional risks for patients, since the differential diagnosis of both diseases can be challenging and medical care must be specific for each disease when dealing with viral infections (dengue) and bacterial infections (leptospirosis). Currently, there is no clinical test that can establish with a single analysis which of the two diseases the patient has.
The use of spectroscopic techniques to analyze biological samples, known as biospectroscopy, has gained great attention in the last decades as it allows the investigation of diseases based on biochemical changes that occur as a consequence of pathological events17. Vibrational spectroscopy techniques have been successfully applied in many studies in medicinal chemistry31.They provide alternative methods for the detection, identification and diagnosis of diseases through simple, cost-effective and accurate methodologies. For example, ATR-FTIR combined with multivariate analysis techniques has been used to detect AIDS virus, where blood plasma spectra from 80 healthy individuals and 40 HIV-positive patients were analyzed. The GA-LDA model achieved 83% sensitivity and 92% specificity in the discrimination of the two classes32.
Herein, we show the results of using multivariate analysis techniques to analyze ATR-FTIR spectral data for differential diagnosis of leptospirosis and dengue in patients from Colombia, an endemic country for both diseases. Firstly, some outlier samples were identified after exploratory analysis by PCA. For plasma samples in liquid phase, the unique outlier refers to a non-pathogenic strain of Leptospira known as saprophytic, as confirmed by the reference MAT test. On the other hand, for dried blood plasma samples, one dengue outlier was found corresponding to a male patient, age 67 years and without alarming symptoms, confirmed being infected with dengue by the ELISA test for IgM. These findings demonstrate the utility to identify patterns and improve the understanding of the dataset. Furthermore, classification models based on LDA, QDA and SVM were built to allow the differential diagnosis of leptospirosis and dengue.
The GA-QDA model in liquid plasma samples achieved outstanding results, showing maximum sensitivity, specificity and accuracy for classifying both diseases (100% classification). The model was built using only 31 spectral variables indicated below: 1077, 1106, 1143, 1175, 1246, 1260, 1296, 1315, 1345, 1386, 1425, 1430, 1434, 1436, 1461, 1494, 1495, 1538, 1612, 1673, 1677, 1678, 1689, 1694, 1711, 1713, 1719, 1745, 1753, 1765 and 1793 cm−1. The ANOVA test, at a 95% confidence level, highlighted a set of 10 variables selected by the GA-QDA model that were statistically significant to distinguish the two classes (1296, 1612, 1673, 1677, 1678, 1689, 1694, 1711, 1713 and 1719 cm−1). The tentative assignment for these 10 variables were: 1290–1334 cm−1 as the amine III band; 1620–1690 cm−1 as the amide I band; and, 1700–1720 cm−1 as the C=Ostretching vibrations of ketones and aldehydes33,34. However, in the region between 1500 and 1700 cm−1, where amide I and amide II are present, it is clear that the contribution of water is very high. It is possible that the water spectrum masks the biological information of the sample due to the strong similarity in the absorption bands obtained in both classes (leptospirosis and dengue), reason that led us to carry out the drying process of these same samples following the drying time reported in the literature for plasma and serum samples35. The best model for dried plasma was the SPA-QDA model, which achieved outstanding results in terms of maximum sensitivity, specificity and accuracy (100%) for both classes by using only 30 spectral variables indicated as follows: 999, 1016, 1034, 1064, 1100, 1121, 1152, 1222, 1291, 1353, 1392, 1410, 1456, 1491, 1507, 1510, 1530, 1541, 1550, 1575, 1601, 1624, 1633, 1637, 1650, 1672, 1714, 1735, 1747, 1870. The ANOVA test, with a 95% confidence level, highlighted a set of 4 variables selected by the SPA-QDA model that are statistically significant (1601, 1735, 1747 and 1870 cm−1). The region where the wavenumber at 1601 cm−1 is located is characteristic of amide I bands in proteins (C=O stretching vibration weakly coupled to the CN stretching and NH bending vibrations). The wavenumber at 1735 cm−1 is assigned to the C= O stretching in lipids, 1747 cm−1 to vibrations of ester groups (C= O) in triglycerides, lipids and organic acids36; and, 1870 cm−1 is assigned to the C=O bonds in carbonyl compounds37. The results were overall better in dry samples since the analytes are pre-concentrated on the crystal surface and the measurement sensitivity is improved. Unfortunately, the drying step implies in giving up the short initial time for direct measurement and cleaning of the ATR accessory38, thus increasing the analysis time and reducing the analytical frequency.
When observing the results for the unbalanced blood samples in liquid and dry phases (since the leptospirosis and dengue classes do not have the same number of samples), the quadratic discrimination is the most appropriate tool. The differential diagnosis between leptospirosis and dengue in liquid and dry phase using infrared spectroscopy and multivariate analysis techniques is based on the biochemical variations that occur as consequences of the pathological process. The spectral regions responsible for sample discrimination cannot be directly associated with specific molecules, such as biomarkers; however, they represent important information about the classes of biomolecules mostly present in these diseases. Therefore, other techniques, such as mass spectrometry, should be more suitable for the investigation of potential biomarkers, and a larger number of samples should be included in subsequent studies. We recognize that this study has some limitations, such as the absence of paired samples and the unbalanced class sizes. Additionally, it lacks of detailed clinical information regarding patient comorbidities, medication use, or other physiological conditions that may influence biospectroscopic profiles. Due to ethical constraints and data protection protocols, only anonymized demographic data such as age range and sex were accessible. This restricts the capacity to evaluate potential confounding factors that may affect spectral variation beyond the presence of dengue or leptospirosis. Future studies should incorporate a more comprehensive clinical profile of each patient to assess the impact of comorbidities and to enhance the diagnostic specificity and the generalizability of the proposed model. Nevertheless, this analytical approach, particularly when applied to dried blood plasma, offers a valuable alternative to address the challenges associated with the diagnosis of leptospirosis and dengue. It represents a simple, cost-effective, and accurate methodology for differential diagnosis in resource-limited or endemic settings.
Conclusion
In this study, we demonstrate the use of ATR-FTIR spectroscopy combined with multivariate analysis for the differential diagnosis of two infectious diseases, leptospirosis and dengue, using liquid and dry phase blood plasma samples. By using quadratic discriminant analysis, GA-QDA and SPA-QDA models achieved maximum correct classification rates, with sensitivity, specificity and accuracy values of 100% when analyzing the test (unknown) samples of the current datasets. Results were overall better in dry samples since the analytes are pre-concentrated on the crystal surface, improving the measurement sensitivity. In liquid phase samples, it is possible that the water spectrum masks the biological information, since a strong similarity in the absorption bands is observed in both classes (leptospirosis and dengue). The outstanding results obtained in this pilot study highlight the feasibility of this methodology to be applied in clinical practice to serve as a rapid and accurate test to discriminate between the two diseases, applied especially to blood plasma samples through a drying process. Nevertheless, future studies are needed to incorporate a broader and more diverse cohort, including healthy individuals and patients with other febrile conditions, to support the development of a multiclass diagnostic model with enhanced and immediate clinical utility.
Data availability
The datasets used and or analysed during the current study available from the corresponding author on reasonable request .
References
Veena, R. K. et al. Epidemiological analysis of leptospirosis, dengue, and Co-infection rates among febrile illness cases in Dakshina Kannada, Karnataka. Indian J. Med. Microbiol. 51, 100698 (2024).
Rodríguez-Salazar, C. A. et al. Manifestaciones clínicas y hallazgos de laboratorio de una serie de casos febriles agudos con diagnóstico presuntivo de infección por el virus dengue. Quindío (Colombia). Infection 20, 1–9 (2015).
Hartskeerl, R. A., Collares-Pereira, M. & Ellis, W. A. Emergence, control and re-emerging leptospirosis: Dynamics of infection in the changing world. Clin. Microbiol. Infect. 17, 494–501. https://doi.org/10.1111/j.1469-0691.2011.03474.x (2011).
Clemen G, Angel J, Montes C, Jr, T. & Osorio L. Contribución de la prueba rápida NS1 e IgM al diagnóstico de dengue en Colombia en el periodo pre-zika. (2019).
Peeling, R. W. et al. Evaluation of diagnostic tests: Dengue. Nat. Rev. Microbiol. 8, S30–S38 (2010).
Gárcia, J. D., Agûero, J., Parra, J. A. & Santos, M. F. Enfermedades infecciosas Concepto. Clasificación Aspectos generales y específicos de las infecciones. Criterios de sospecha de enfermedad infecciosa. Pruebas diagnósticas complementarias. Criterios de indicación. Medicina - Programa de formaciòn Medica Continuada acreditado 10, 3251–3264 (2010).
Pinto, G. V. et al. Current methods for the diagnosis of leptospirosis: Issues and challenges. J. Microbiol. Methods https://doi.org/10.1016/j.mimet.2022.106438 (2022).
Suwannin, P. et al. Enhancing leptospirosis control with nanosensing technology: A critical analysis. Compar. Immunol. Microbiol. Infect. Dis. https://doi.org/10.1016/j.cimid.2023.102092 (2024).
Santos, M. C. D. et al. ATR-FTIR spectroscopy with chemometric algorithms of multivariate classification in the discrimination between healthy: Vs. dengue vs. chikungunya vs. zika clinical samples. Anal. Methods 10, 1280–1285 (2018).
Naseer, K., Ali, S. & Qazi, J. ATR-FTIR spectroscopy based differentiation of typhoid and dengue fever in infected human sera. Infrared Phys. Technol. 114, 103664 (2021).
Ali, S., Naseer, K., Hussain, I. & Qazi, J. ATR-FTIR spectroscopy-based differentiation of hepatitis C and dengue infection in human freeze-dried sera. Infrared Phys. Technol. 118, 1350–4495 (2021).
Morais, C. L. M., Lima, K. M. G., Singh, M. & Martin, F. L. Tutorial: multivariate classification for vibrational spectroscopy in biological samples. Nat. Protoc. 15, 2143–2162. https://doi.org/10.1038/s41596-020-0322-8 (2020).
Santos, M. C. D., Morais, C. L. M., Nascimento, Y. M., Araujo, J. M. G. & Lima, K. M. G. Spectroscopy with computational analysis in virological studies: A decade (2006–2016). TrAC Trends Anal. Chem. 97, 244–256. https://doi.org/10.1016/j.trac.2017.09.015 (2017).
Kawakami, R. et al. Aspects of the successive projections algorithm for variable selection in multivariate calibration applied to plasma emission spectrometry. Anal. Chim. Acta 443, 107–115 (2001).
Dixon, S. J. & Brereton, R. G. Comparison of performance of five common classifiers represented as boundary methods: Euclidean distance to centroids, linear discriminant analysis, quadratic discriminant analysis, learning vector quantization and support vector machines, as dependent on data structure. Chemom. Intell. Lab. Syst. 95, 1–17 (2009).
Morais, C. L. M. & Lima, K. M. G. Principal component analysis with linear and quadratic discriminant analysis for identification of cancer samples based on mass spectrometry. J. Braz. Chem. Soc. 29, 472–481 (2018).
da Silva, T. G. et al. Spectrochemical analysis of blood combined with chemometric techniques for detecting osteosarcopenia. Sci. Rep. 13, 9686 (2023).
Naseer, K., Ali, S., Mubarik, S., Hussain, I. & Mubarik, S. FTIR spectroscopy of freeze-dried human sera as a novel approach for dengue diagnosis. Infrared Phys. Technol. 102, 1–5 (2019).
Fadlelmoula, A. Review Fourier transform infrared (FTIR) spectroscopy to analyse human blood over the last 20 years: A review towards lab-on-a-chip devices.
Araújo, R., Ramalhete, L., Ribeiro, E. & Calado, C. Plasma versus serum analysis by FTIR spectroscopy to capture the human physiological state. BioTech 11, 56 (2022).
Bezerra, L. F. M. et al. Serological evidence of leptospirosis in patients with a clinical suspicion of dengue in the State of Ceará Brazil. Biomedica 35, 557–562 (2015).
Kennard, R. W. & Stone, L. A. Computer aided design of experiments. 11, 137–148 (1969).
Bro, R. & Smilde, A. K. Principal component analysis. Anal. Methods 6, 2812–2831. https://doi.org/10.1039/c3ay41907j (2014).
Soares, S. F. C., Gomes, A. A., Araujo, M. C. U., Filho, A. R. G. & Galvão, R. K. H. The successive projections algorithm. TrAC Trends Anal. Chem. 42, 84–98. https://doi.org/10.1016/j.trac.2012.09.006 (2013).
McCall, J. Genetic algorithms for modelling and optimisation. J. Comput. Appl. Math. 184, 205–222 (2005).
Cortes, C., Vapnik, V. & Saitta, L. Support-vector networks editor. Mach. Leaming 20, (1995).
da Silva, T. G. et al. Spectrochemical analysis of blood combined with chemometric techniques for detecting osteosarcopenia. Sci. Rep. 13, 9686 (2023).
Neves, A. C. O. et al. ATR-FTIR and multivariate analysis as a screening tool for cervical cancer in women from northeast Brazil: A biospectroscopic approach. RSC Adv. 6, 99648–99655 (2016).
Morais, C. L. M. & Lima, K. M. G. Comparing unfolded and two-dimensional discriminant analysis and support vector machines for classification of EEM data. Chemom. Intell. Lab. Syst. 170, 1–12. https://doi.org/10.1016/j.chemolab.2017.09.001 (2017).
López, V., Fernández, A., García, S., Palade, V. & Herrera, F. An insight into classification with imbalanced data: Empirical results and current trends on using data intrinsic characteristics. Inf. Sci. (N Y) 250, 113–141 (2013).
Rodríguez-Villamarín, F. R., Prieto-Suárez, E., Escandón, P. L. & Restrepo, F. H. Proporción de leptospirosis y factores relacionados en pacientes con diagnóstico presuntivo de dengue, 2010–2012. Revista de Salud Publica 16, 597–609 (2014).
Alkhuder, K. Fourier-transform infrared spectroscopy: A universal optical sensing technique with auspicious application prospects in the diagnosis and management of autoimmune diseases. Photodiagn. Photodyn. Ther. https://doi.org/10.1016/j.pdpdt.2023.103606 (2023).
Silva, L. G. et al. ATR-FTIR spectroscopy in blood plasma combined with multivariate analysis to detect HIV infection in pregnant women. Sci. Rep. 10, 20156 (2020).
Walsh, M. J. et al. IR microspectroscopy: Potential applications in cervical cancer screening. Cancer Lett. 246, 1–11. https://doi.org/10.1016/j.canlet.2006.03.019 (2007).
Purandare, N. C. et al. Infrared spectroscopy with multivariate analysis segregates low-grade cervical cytology based on likelihood to regress, remain static or progress. Anal. Methods 6, 4576–4584 (2014).
Hughes, C. et al. Assessing the challenges of Fourier transform infrared spectroscopic analysis of blood serum. J Biophoton. 7, 180–188 (2014).
Movasaghi, Z., Rehman, S. & Rehman, I. U. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 43, 134–179. https://doi.org/10.1080/05704920701829043 (2008).
Nandiyanto, A. B. D., Oktiani, R. & Ragadhita, R. How to read and interpret ftir spectroscope of organic material. Indonesian J. Sci. Technol. 4, 97–118 (2019).
Perez-Guaita, D., Garrigues, S. & de La, M. Infrared-based quantification of clinical parameters. TrAC Trends Anal. Chem. 62, 93–105. https://doi.org/10.1016/j.trac.2014.06.012 (2014).
Acknowledgements
The authors would like to thank the Chemistry Postgraduation Program of the Federal University of Rio Grande do Norte (PPGQ-UFRN). Kássio M. G. Lima thanks the National Council for Scientific and Technological Development (CNPq) grant (Ref. 305562/2020-7). Likewise, the authors also thank the Doctoral Scholarship agreement No. 004-2020 granted within the framework of the “Convocatoria del Fondo de Ciencia, Tecnología e Innovación del Sistema General de Regalías”, Colombia.
Funding
The APC is funded by the Ministry of Science, Technology and Innovation(MINCIENCIAS) – Colombia, grant number 111691892296 (agreement 799–2023), and the Universidad del Atlántico.
Author information
Authors and Affiliations
Contributions
Alejandra Zambrano: carried out the research, data curation, review and was in charge of writing the original manuscript draft.Jorge Trilleras: director of research, conceptualization, data curation and review.Victoria A. Arana: co-director of research, methodology, conceptualization, data curation and review.Kássio M. G. Lima: formal analysis, review and editing.Ana C. O. Neves: formal analysis, data curation and treatment.Camilo L. M. Morais: data treatment and manuscript revisions.Claudia Romero: worked on the supply, treatment and reference analyses of blood samples. Andrew K. I. Falconar: worked on the supply, treatment and reference analyses of blood samples. Boanergue Salas: conceptualizationRoberto García: worked on the supply, treatment and reference analyses of blood samples. Carlos Carmona: worked on the supply, treatment and reference analyses of blood samples.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zambrano, A., Trilleras, J., Arana, V.A. et al. ATR-FTIR and multivariate analysis for differential diagnosis of dengue and leptospirosis: a feasibility study. Sci Rep 15, 34092 (2025). https://doi.org/10.1038/s41598-025-14594-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-14594-9