Abstract
Skin cancer is a significant global health concern, and accurate and timely diagnosis is crucial for successful treatment. However, manual diagnosis can be challenging due to the subtle visual differences between benign and malignant lesions. This study introduces Skin-DeepNet, a novel deep learning-based framework designed for the automated early diagnosis and classification of skin cancer lesions from dermoscopy images. Skin-DeepNet incorporates a two-step pre-processing stage to enhance image contrast, followed by robust skin lesion segmentation using Mask R-CNN and GrabCut algorithm to achieve near-perfect segmentation accuracy (IOU up to 99.93%). Then a dual-feature extraction strategy is performed using a combination of a pre-trained high-resolution network (HRNet) model and attention block, which serve as feature descriptors. A deep belief networks (DBN) model is then trained on their outputs to capture high-level discriminative features. Finally, robust decision fusion strategies are employed to integrate the predictions of the proposed models using boosting and stacking to enhance overall Skin-DeepNet’s accuracy. The Skin-DeepNet’s performance has been validated on two challenging datasets: ISIC 2019 and HAM1000. The Skin-DeepNet system has outperformed the existing state-of-the-art systems by achieving an accuracy rate of 99.65% Precision of 99.51%, AUC of 99.94% on the ISIC 2019 dataset. Similarly, on the HAM1000 dataset, the Skin-DeepNet system demonstrated an accuracy rate, precision, and AUC of 100%, 99.92%, and 99.97%, respectively. These findings indicate that the developed Skin-DeepNet system can exhibit outstanding proficiency in accurately classifying skin lesions while aiding physicians in early diagnosis and treatment tasks in clinical settings.
Similar content being viewed by others
Introduction
One of the most common types of cancer is skin cancer, which begins in the epidermis, the outer layer of the skin. Skin cancer, one of the most common types of malignancy, is a global health challenge associated with uncontrolled skin cell proliferation caused by overexposure to sunlight or tanning beds1. More than 97,160 people in the United States were newly diagnosed in 2023, representing roughly 5.0% of new cancer cases2. Sadly, 7,990 individuals succumbed to skin cancer in the same year, highlighting its significant impact with 1.3% of all cancer-related deaths attributed to this condition2. Skin cancer is classified into four main types: melanoma, melanocytic nevi, basal cell carcinoma, and squamous cell carcinoma. Among these, melanoma is the most aggressive form due to its rapid metastasis to other organs. It is responsible for over 10,000 deaths annually in the United States. Early detection of melanoma is critical for effective treatment, as the five-year relative survival rate is approximately 92% when diagnosed in its early stages3. The primary method for diagnosing skin cancer remains visual examination by dermatologists, though its accuracy is approximately 60%. A significant challenge in detecting melanoma is the visual similarity between benign and malignant skin lesions, making diagnosis difficult even for highly trained specialists. Distinguishing lesions based on visual inspection alone is particularly challenging. Consequently, various imaging techniques, such as dermoscopy, have been introduced over the years. Dermoscopy is a non-invasive imaging method that enhances the visualization of the skin surface through magnification and the use of immersion fluids. This technique increases diagnostic accuracy for skin cancer to 89%. For example, dermoscopy has sensitivities of 82.6%, 98.6%, and 86.5% for melanocytic lesions, basal cell carcinoma, and squamous cell carcinoma, respectively2. Although dermoscopy greatly enhances the diagnosis of melanoma, accurately identifying certain lesions, especially early-stage melanomas with indistinct dermoscopic features, remains difficult. This calls for more developments aimed at further improving patient survival rates.
In order to assist dermatologists in overcoming the challenges of melanoma diagnosis and enhancing diagnostic accuracy, computer-aided diagnosis (CAD) systems have been developed. CAD systems are now a crucial component of everyday clinical practice for identifying abnormalities in medical imaging in different screening centers and healthcare facilities4. CAD systems normally consist of several components, such as image acquisition, image pre-processing, image segmentation, feature extraction, and disease classification. The first step in the diagnosis of pigmented cutaneous lesions is the taking of high-quality images with cutting-edge equipment like spectroscopes, dermatoscopes, or plain digital cameras. This is followed by the utilization of image pre-processing methods for the enhancement of image quality via artifact reduction and contrast improvement, which in turn enhances the robustness of the subsequent analytical steps. The segmentation phase delineates the borders of pigmented skin lesions within the image, usually using half-automatic algorithms requiring user interaction. The features extracted from the segmented areas are used as input to a classification system so that pigmented skin lesions can be differentiated into various diagnostic categories. Image segmentation is an important part in the diagnosis of skin cancer, as the features for classification are taken from the segmented area5. Skin cancer segmentation is very challenging because of the tremendous variations in color, texture, position, and size of the skin lesions as they appear in dermoscopic images. Moreover, low contrast between the images complicates the distinction of adjacent tissue types. Air bubbles, hair, dark borders, ruler marks, blood vessels, and color illumination variations are also among the other challenges for accurate lesion segmentation6.
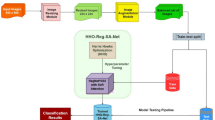
The development of deep learning methods within the area of machine learning has generated much interest in whether artificial intelligence can potentially be used, especially in the area of medicine7,8. It has been confirmed by recent research that deep features obtained using convolutional neural networks (CNNs) have better performance in detecting and classifying skin lesions than conventional feature extraction approaches9,10,11,12. Unlike traditional approaches that rely on separate extraction of shape, color, and texture features, deep features incorporate both local and global image information. Convolutional layers capture local details, while fully connected layers and global average pooling extract global context. This holistic representation has contributed to the improved accuracy and efficiency of deep learning models in medical image analysis. This study presents a fast and fully-automated system, named as Skin-DeepNet system for skin lesion classification that incorporates several innovative techniques. Firstly, an enhanced image pre-processing pipeline is developed to enhance contrast and a novel hair removal algorithm to improve image quality. Secondly, a robust segmentation algorithm is suggested using Mask R-CNN and GrabCut to accurately delineate lesion boundaries. Thirdly, the Skin-DeepNet system employs a dual-feature learning strategy, where pre-processed segmented images are processed through an HRNet backbone to extract hierarchical multi-scale features. These features are enhanced by an attention mechanism, then split into two pathways: one passes through a DRBM and Softmax layer to produce skin cancer class probabilities, while the other uses a DBN to refine and strengthen feature discriminability. Finally, robust decision fusion strategies are utilized to fuse the predictions obtained from the suggested HRNet and DBN models through boosting (e.g., XGBoost) and stacking (e.g., LR, RF, and ET) approaches to improve the overall accuracy of the Skin-DeepNet system. The key contributions of this study are as follows:
-
1.
In the study, Adaptive Gamma Correction with Weighting Distribution (AGCWD) approach is applied for contrast enhancement of images with preservation of important details and artifacts suppression. This is an important step for better visualization of small lesion features. An efficient algorithm is then proposed for hair removal from images. This is done through morphological operations in detecting the hair region boundary, followed by inpainting for filling the erased areas without leaving gaps, resulting in cleaner and more useful images for further analysis.
-
2.
An effective skin lesion segmentation algorithm is developed by merging the advantages of both the Mask R-CNN and the GrabCut algorithm. The two algorithms collaborate to improve the segmentation results through the removal of background noise and extraction of only the lesion area.
-
3.
We introduce an efficient dual feature extraction strategy to derive discriminative deep feature representations from dermoscopy images. Initially, the segmented images are processed through an HRNet backbone, which extracts features in a hierarchical, multi-scale manner. These features are refined by an attention mechanism, whose outputs are directed into two distinct pathways: one channel feeds into a nonlinear classifier composed of a DRBM and a Softmax layer to generate probability distributions across skin cancer categories, while the other channel leverages a DBN to further enhance and stabilize the discriminative quality of the feature representations.
-
4.
Robust decision fusion strategies are proposed to integrate the predictions of the proposed (HRNet and the attention block) and DBN models with boosting (e.g., XGBoost) and stacking (e.g., logistic regression (LR), random forest (RF), and extra trees (ET)). These ensemble learning techniques enhance predictive accuracy by aggregating the strengths of these two models. Boosting iteratively enhances predictions by giving heavy weight to misclassified instances, while stacking trains a meta-learner to optimally combine the multiple model outputs. This strategy drastically improves accuracy, stability, and generalizability.
-
5.
The developed Skin-DeepNet system performs better than state-of-the-art methods on the ISIC 2019 and HAM10000 datasets with higher accuracy, precision, recall, and F1-score. These outcomes demonstrate its potential for clinical application in real-world scenarios, helping dermatologists in the early and accurate diagnosis of skin lesions.
This paper is organized as follows: “Related works” reviews existing research on skin cancer classification. Section “Materials and methods” presents the skin cancer dataset used and provides a detailed description of the developed Skin-DeepNet system. Section “Experimental results” presents and discusses the experimental results, followed by concluding the paper and outlining the future research directions in “Conclusions and future work”.
Related works
Skin cancer classification using machine learning and deep learning has gained importance for its potential in early and accurate diagnosis, essential for effective treatment. This section examines recent advancements, particularly in CNNs, transfer learning, federated learning, and ensemble methods. For instance, Maiti et al.13 suggested a skin cancer classification system using quantized color features and generative adversarial networks (GANs) for improved diagnostic accuracy. The method adopts color quantization for discriminative feature extraction from dermoscopic images, and GANs are used for dataset augmentation and class imbalance correction to improve the generalization of the classification model. The technique was tested on benchmark datasets and showed better performance in the classification between benign and malignant lesions than state-of-the-art methods, indicating the promise of fusing color-based features and synthetic data generation for efficient skin cancer detection. Maiti and Chatterjee14 explored the influence of different feature selection techniques on melanoma classification performance using a dataset transformed by various feature selection methods, including filter, wrapper, and embedded approaches. The authors implemented the techniques in combination with machine learning classifiers to compare their performance in enhancing classification accuracy, minimizing dimensionality, and removing irrelevant or redundant features. The authors’ findings indicated that some feature selection methods, especially wrapper- and embedded-based approaches, greatly outperformed classification models without feature selection, underscoring the need for effective feature selection in melanoma detection and offering recommendations for optimizing diagnostic models in medical imaging studies. Tahir et al.1 introduced DSCC_Net, a deep learning-based skin cancer classification network utilizing a CNN and tested on ISIC 2020, HAM10000, and DermIS datasets. DSCC_Net achieved an Area Under the Curve (AUC) of 99.43%, accuracy of 94.17%, recall of 93.76%, precision of 94.28%, and an F1-score of 93.93% across four skin cancer types. Comparatively, ResNet-152, VGG-19, MobileNet, VGG-16, EfficientNet-B0, and Inception-V3 reported accuracies between 89.12% and 92.51%. DSCC_Net outperformed these models, providing valuable assistance for skin cancer diagnosis. Sharma et al.15 presented a cascaded model that combines the strengths of handcrafted feature extraction techniques with deep learning. By integrating deep convolutional networks (ConvNets) with handcrafted features, such as color moments and texture features, the model enhances skin disease image classification accuracy. Referred to as a cascaded ensembled deep learning model, the proposed architecture significantly outperforms the ConvNet al.one, improving classification accuracy from 85.3 to 98.3%, as demonstrated by simulation results. Naeem et al.16 introduced SCDNet, a model combining VGG16 with CNN for skin cancer classification. The model’s accuracy was compared with four state-of-the-art pre-trained classifiers in the medical field: ResNet50, Inception V3, AlexNet, and VGG19. Using the ISIC 2019 dataset, SCDNet achieved an accuracy of 96.91% for multiclass skin cancer classification, outperforming ResNet50 (95.21%), AlexNet (93.14%), VGG19 (94.25%), and Inception V3 (92.54%). The results demonstrate that SCDNet surpasses these existing classifiers.
Bechelli et al.17 evaluated ML and DL models for skin tumor classification. ML methods (logistic regression, k-nearest neighbors, etc.) underperformed with accuracies below 0.72, while DL models, using custom CNNs or transfer learning (VGG16, Xception, ResNet50), reached up to 0.88 accuracy. Ensemble learning improved ML accuracy to 0.75. On a larger, imbalanced dataset, fine-tuned pre-trained DL models, particularly VGG16, achieved strong results, with an F-score and accuracy of 0.88 on a smaller dataset, and 0.70 and 0.88 on the larger one. Imran et al.18 employed convolutional deep neural networks for skin cancer detection using the ISIC dataset. Recognizing the inherent limitations of individual machine learning models in the sensitive context of cancer detection, where misdiagnosis can have severe consequences, this study explores the benefits of ensemble learning. By combining the predictions of multiple diverse learners, ensemble methods can enhance accuracy and robustness. To this end, an ensemble of deep learners was developed, incorporating VGG, CapsNet, and ResNet models, for improved skin cancer detection. The ensemble approach exhibited superior performance compared to individual models across all evaluation metrics, including sensitivity, accuracy, specificity, F-score, and precision. The proposed method achieved an average accuracy of 93.5% with a training time of 106 s, suggesting its potential applicability for other disease detection tasks. Afza et al.19 introduced a new multi-class skin lesion classification method that integrates deep learning feature fusion with an extreme learning machine. Their approach comprises five stages: image acquisition and contrast enhancement, deep learning feature extraction using transfer learning, feature selection employing a hybrid whale optimization and entropy-mutual information (EMI) method, feature fusion through a modified canonical correlation approach, and final classification using an extreme learning machine. The feature selection enhances computational efficiency and accuracy. Tested on HAM10000 and ISIC 2018 datasets, the method achieved accuracies of 93.40% and 94.36%, outperforming state-of-the-art techniques while maintaining computational efficiency. Bassel et al.20 introduced a method for melanoma and benign skin cancer classification based on stacking classifiers in three folds. The system was trained on 1,000 skin images, with 70% used for training and 30% for testing. Feature extraction was conducted using ResNet50, Xception, and VGG16. Performance was evaluated using accuracy, F1 score, AUC, and sensitivity metrics. The Stacked CV method trained models using deep learning, support vector machine (SVM), RF, neural networks (NN), KNN, and logistic regression. Xception-based feature extraction achieved 90.9% accuracy, outperforming ResNet50 and VGG16. Further optimization with larger datasets could enhance the system’s reliability and robustness. Hamida et al.21 developed a hybrid system combining RF and deep neural networks (DNN) to improve skin disease classification. Using the HAM10000 dataset, the RF component diagnoses based on symptoms, while the DNN analyzes skin lesion images for more accurate results. The system achieves a 96.8% accuracy, performing well across six of seven skin disease classes. Though sensitivity variations and data quality issues are noted, the system shows promise for advancing skin disease diagnosis. Dahdouh et al.22 developed an advanced system integrating the internet of things (IoT) with a Raspberry Pi, camera, and a deep learning model based on a CNN for real-time melanoma detection and classification. The model’s process includes data pre-processing (cleaning, normalization, and augmentation) to minimize overfitting, feature extraction using the deep CNN algorithm, and classification with a Sigmoid activation function. Experimental results demonstrate the system’s effectiveness, achieving 92% accuracy, 91% precision, 91% sensitivity, and an AUC-ROC of 91.33% using a dataset of 3,297 images.
While the reviewed works show excellent advancement in skin cancer classification with machine learning and deep learning, there are several limitations in these studies. The majority of the developed models are trained on small or class-imbalanced datasets, which can restrict their generalizability to real-world, heterogeneous clinical settings and lead to overfitting despite high reported accuracy. For instance, though methods like those of Bechelli et al.17 and Dahdouh et al.22 report high performance numbers, their evaluations are often conducted on limited datasets, which questions their scalability and consistency in diverse populations. Besides, various approaches, including those using hand-engineered feature extraction (e.g., Sharma et al.15, can be biased and restrict scalability as they depend on manually chosen features that may fail to describe the complete intricacy of skin lesions. Moreover, computational efficiency and inference latency dimensions continue to be inadequately addressed, with limited research focusing on the actual deployment of such systems within clinical practice. Lastly, interpretability is an issue; most models are not decision-transparent, discouraging dermatologists from using them due to the need for explainable AI for usability and trust.
Materials and methods
The architecture of the Skin-DeepNet system, depicted in Fig. 1, starts with raw skin lesion images undergoing an image pre-processing stage, where AGCWD approach is applied to enhance image contrast without losing vital details and removing artifacts. Image segmentation is then carried out by the Mask R-CNN and GrabCut algorithms for delineating regions of interest. The pre-segmented images are then fed through a HRNet backbone, which is responsible for feature extraction in hierarchical fashion. These features are enhanced by an attention block that feeds its outputs into two parallel streams: one stream feeds information into a non-linear classifier formed by a DRBM coupled with the Softmax layer for outputting the probability distribution across various skin cancer classes, and the other stream passes through a DBN for further enhancing and consolidating the feature representations. The model integrates predictions from both pathways using advanced decision fusion techniques, including boosting (e.g., XGBoost) and stacking (e.g., LR, RF, and ET), which combine the complementary strengths of the HRNet-attention block with the DBN model to augment the overall diagnostic accuracy. The ensemble approach ensures robustness and precision in distinguishing between various skin cancer categories. This ensemble approach guarantees reliability along with accuracy in differentiating among the different classes of skin cancer.
Datasets description
The accurate identification of skin cancer relies heavily on robust and diverse datasets for training deep learning models capable of reliable prediction. However, acquiring datasets with a sufficient number of diverse and high-quality labeled images for effective deep model training remains a significant challenge. In this work, we leverage two established datasets, ISIC 201923 and HAM1000024, to address this critical need. The publicly accessible ISIC 2019 dataset comprises a diverse collection of 25,331 dermoscopic images, sourced from various institutions23. These images represent eight distinct categories of skin lesions, with varying sample sizes: melanoma (4,522), melanocytic nevi (12,875), basal cell carcinoma (3,323), squamous cell carcinoma (628), vascular lesions (253), dermato-fibromas (239), actinic keratosis (867), and benign keratosis (2,624). This dataset presents a challenging classification task due to its imbalanced class distribution and the visual similarities between certain lesion types. All images are stored in RGB format with varying resolutions. Figure 2 provides a few example images of ISIC 2019 dataset.
The HAM10000 dataset24, also known as the “Human Against Machine with 10,000 training images” dataset, is among the largest collections available for identifying pigmented skin lesions, with a total of 10,015 dermoscopic images accessible via the ISIC repository. The dataset is composed of seven distinct classes: melanocytic nevus (nv), dermatofibroma (df), actinic keratosis (akiec), vascular lesions (vacs), basal cell carcinoma (bcc), benign keratosis (bkl), and melanoma (mel). The dataset includes 6,705 nv images, 327 akiec images, 115 df images, 514 bcc images, 115 vacs images, 1,099 bkl images, and 1,113 mel images. The gender distribution within the dataset is 54% male and 45% female. Due to substantial intra-class similarities and minimal inter-class distinctions among some skin lesion types, this dataset presents significant challenges for accurate classification, with a high risk of misclassification. Figure 3 provides a few example images of HAM1000 dataset.
Image pre-processing stage
To enhance tumor boundary detection and segmentation accuracy, a two-step pre-processing technique is employed to mitigate the disruptive influence of skin artifacts (See Fig. 4-a). The initial step involves applying AGCWD approach to amplify the image’s contrast25. This approach, renowned for its effectiveness in various image processing applications (e.g., medical imaging, remote sensing, etc.), is chosen to accentuate subtle details, textures, and low-contrast areas within the input image. Through adaptively changing the gamma correction parameter, AGCWD approach can increase contrast in under- and overexposed regions, preserve fine details, and minimize the introduction of artifacts, such as noise or artificial color transitions (see Fig. 4b). The main steps to use AGCWD approach can be summarized as follows: (i) The input image histogram is examined to know the intensity level distribution, through which areas with low contrast or over-saturation can be identified, (ii) The weighting distribution function (WDF) is calculated from the histogram of the input image to assign weights to various intensity levels, promoting areas to be enhanced and suppressing areas that have sufficient contrast already, (iii) The cumulative distribution function (CDF) (e.g., the cumulative probability of pixel intensities at or below a particular level), is calculated for the input image, (iv) The gamma correction factor, γ, is adaptively computed for each pixel based on the intensity and the WDF such that the enhancement process can be accurately controlled, and (v) Adaptive gamma correction rescales original pixel intensities through the WDF. This has the effect of increasing low-contrast area intensity range and decreasing high-contrast area range, making the image overall clearer.
In the second step, the hair is effectively removed, allowing for enhanced clarity and accuracy in subsequent image analysis tasks, such as image segmentation or feature extraction. The input image is converted to grayscale. This transformation simplifies the data by reducing color complexity, which is beneficial for subsequent processing. Then, the grayscale image undergoes a morphological Blackhat operation using a structural element of size (11 × 11). This operation highlights dark regions (e.g., hair) against a lighter background, enhancing the visibility of hair structures for further processing. This is followed by applying a binary threshold to the result of the Blackhat operation to create a mask that isolates the hair regions. Pixels with intensity values above 10 are set to 255 (white), while those below are set to 0 (black), producing a binary image of hair locations (Fig. 4d). Finally, the original image is then inpainted using the thresholded binary mask. Inpainting reconstructs the areas identified as hair by filling them in using information from surrounding pixels. The inpainting algorithm used is the Telea method, which effectively removes hair artifacts while preserving image integrity. Figure 4c shows some hair removal results.
Image segmentation stage
In this stage, a fully automated and efficient image segmentation algorithm is introduced to detect skin lesions and improve the discriminative power of feature representations in the subsequent stage of the Skin-DeepNet system (See Fig. 5). Specifically, Mask R-CNN is utilized to automatically generate pixel-wise masks, effectively segmenting foreground objects from the complex background26,27. Mask R-CNN, an advancement of Fast R-CNN, is capable of both detecting objects within an image and generating pixel-wise segmentation masks for each detected instance. This model operates in two stages: initially, it generates proposals for potential objects in the image; then, it predicts each object’s class, refines the bounding box, and produces the corresponding binary mask27. In this study, the ResNet50 backbone network is used to extract feature maps, which are further processed by a feature pyramid network (FPN) to capture multi-scale features for precise object localization. The FPN leverages CNNs’ inherent multi-scale capabilities, enhancing skin lesion detection across different scales. A sliding window technique is applied on the feature maps to identify skin lesion regions in the form of bounding boxes. Since these bounding boxes vary in size, the ROIAlign method is utilized to standardize feature maps, ensuring uniformity. These standardized maps are then processed by a network head to classify objects and generate bounding boxes, while additional convolutional layers within the mask component create the segmentation mask for skin lesions. Mask R-CNN generates object class labels, bounding boxes, and binary masks as output. However, the generated masks can sometimes include background elements, leading to inaccuracies in the subsequent analysis. To mitigate this issue, the GrabCut algorithm is employed. Using the Mask R-CNN-generated mask as an initial seed, GrabCut effectively refines the segmentation by minimizing background noise interference and improving the accuracy of skin lesion detection within the developed Skin-DeepNet system.
The GrabCut algorithm is a robust segmentation technique designed to eliminate unwanted, complex background edges from skin lesion images, preserving only the lesion as the foreground28. GrabCut algorithm improves the initial lesion segmentation from Mask R-CNN through the application of graph cut operations. It begins by applying the process with the drawing of a bounding box around the lesion as indicated in the Mask R-CNN result. Then, it uses a gaussian mixture model (GMM) for the estimation of color distributions for the foreground (lesion) and background parts of the image. Using this GMM, the GrabCut algorithm assigns class labels (foreground or background) to each pixel based on its color characteristics. This process involves representing the image as a graph, where pixels are nodes and the connections between them are edges (as illustrated in Fig. 6). It iterates over each pixel, breaking weaker connections and assigning pixels to either the foreground or background accordingly. Integrating GrabCut with Mask R-CNN notably reduces background interference, improving segmentation accuracy and the precision of contour extraction in skin lesion analysis. To enhance the accuracy of the final segmentation outcomes, a post-processing procedure is conducted to eliminate irrelevant background elements, remove isolated regions, and merge the remaining areas. This procedure begins with the application of a morphological dilation operation using a (3 × 3) spherical kernel to smooth the mask boundaries. Lastly, a morphological reconstruction algorithm is employed to fill potential gaps within the mask, specifically addressing background regions that are inaccessible from the image edges. Figure 4 shows some examples of the image segmentation outcomes.
Feature extraction and classification stage
After identifying the ROI, the next step is the feature extraction, which focuses on isolating the most relevant information from the images while discarding details irrelevant to classification. In CAD systems for skin cancer in dermoscopic images, selecting appropriate features is essential, as it can enhance the accuracy of dermatologists’ diagnoses. Previous studies have highlighted that features can be extracted on a global or local scale; however, it is crucial to ensure that the selected features contain sufficient information to effectively differentiate between multiple classes1,2. Based on this fact, the proposed feature extraction algorithms are designed to capture the relevant features from the segmented skin lesions.
In this study, two deep learning-based algorithms are utilized to effectively capture robust feature representations from segmented skin lesions. The initial algorithm employs a transfer learning approach, utilizing different CNN architectures, to extract powerful feature representations from the input image. The comparative performance of these architectures is analyzed in Sect. 4. As illustrated in Fig. 7, the pre-trained HRNet model trained on the ImageNet dataset, has been chosen as the backbone CNN module because of its exceptional performance on the datasets utilized in this study. HRNet’s first sub-network begins at a high resolution, and additional sub-networks are introduced progressively, transitioning from high- to low-resolution branches. This parallel configuration of multi-resolution sub-networks allows for comprehensive data fusion across different scales, and enhancing the high-resolution feature representations. HRNet maintains high-resolution features across four stages, corresponding to four branches and four resolution levels. Following the initial step, two convolutional layers (3 × 3) with strides increase the channel width to 64 while reducing the resolution to one-quarter. The channel number, denoted as C, is set to 32 for HRNet-W32 (where W indicates width) across branches, with configurations of C, 2 C, 4 C, and 8 C for each successive branch. The resolution is gradually down-sampled in four stages, resulting in feature maps at different resolutions. These multi-scale feature maps are then combined to create a richer representation of the input image, which is subsequently processed by the attention block, as depicted in Fig. 8.
The attention block generates distinctive feature attention maps by utilizing features extracted from the HRNet network. Initially, the HRNet network generates feature maps, represented by \(\:{{F}}^{{l}}\). Then, these feature maps are subsequently transformed into a new representation, \(\:{{P}}^{{l}}\) using a (3 × 3) depthwise separable convolution (DSConv) followed by a (1 × 1) pointwise convolution (PConv). DSConv, which is computationally efficient and has a lesser number of parameters, increases the representational power compared to standard convolutions. The output of the PConv layer is then fed through Batch Normalization (BN) to improve training stability and enable faster convergence. Finally, a rectified linear unit (ReLU) is applied in order to introduce non-linearity. The attention block computes the final feature map \(\:{{P}}^{{l}}\) by fusing the feature maps of DSConv and PConv as follows:
Here, “Cat” represents the concatenation operation. This attention mechanism is employed to make the model pay more attention to useful features in the dermoscopy image. This mechanism selectively amplifies informative features and suppresses noisy or irrelevant areas, improving the accuracy and robustness of skin lesion classification. The process is implemented as follows:
-
1.
The convolutional layers of the attention block convert the input image to a series of feature maps representing information at different levels of complexity and detail (See Fig. 8).
-
2.
During the training process, the attention mechanism can learn attention maps that put greater emphasis on the most important spatial areas in the feature maps, thereby augmenting the model to concentrate on important information and boost classification outcomes.
Finally, to enable precise classification of dermoscopic images, the DRBM as a non-linear classifier is employed with two separate sets of visible units29. One set represents the input feature vectors, and the other set interacts with a Softmax layer to produce a probability distribution over various skin cancer classes30. This strategy effectively models the joint distribution between the input features as well as the corresponding target classes31. During the training process of DRBM classifier, the stochastic gradient descent algorithm will maximize the log-likelihood of the training set. Consequently, the following weight-update rules is be applied:
In this context, \(\varepsilon\) represents the learning rate, while \(\left\langle \cdot \right\rangle\) data and \(\left\langle \cdot \right\rangle\) model correspond to the positive and negative phases, respectively. The term \({b_i}\) refers to the bias for the visible units, and \({c_i}\) denotes the bias for the hidden units. As is well established, computing \(\:{\langle {v}_{i}{h}_{j}\rangle }_{model}\:\)in Eq. (2) is a challenging task. To address this, the contrastive divergence (CD) method32 is utilized to update the parameters of a specific RBM by performing k steps of Gibbs sampling from the probability distribution. The procedure for executing a single CD algorithm sample is as follows:
-
1.
The training data is used as input (\(\:{v}_{i}\)) to estimate the probability distribution of the hidden units (\(\:{h}_{j}\)). The estimated distribution is then used to draw a sample for the hidden activation vector (\(\:{h}_{j}\)).
-
2.
During the positive phase, the outer product of vectors (\(\:{v}_{i}\)) and (\(\:{h}_{j}\)) is calculated.
-
3.
The visible units (\(\:{v}_{i}\)’) are reconstructed by sampling from (\(\:{h}_{j}\)) using the conditional probability \(\:P({h}_{j}=1|v)\). Subsequently, the hidden unit activations (j′) are resampled from (\(\:{v}_{i}\)’) in a single Gibbs sampling step.
-
4.
In the negative phase, the outer product of (\(\:{v}_{i}\)) and (\(\:{h}_{j}\)) is recalculated.
-
5.
Finally, the weight matrix and biases are updated using Eqs. (2)–(4).
In many applications of the CD learning algorithm, the value of k is typically set to 1. In this study, weights were initialized with small random values drawn from a normal distribution with zero mean and a standard deviation of 0.02. During the positive phase of training, the probabilities of the hidden units were calculated based on the visible units and weights. This phase is characterized by a high probability of observing the training data. Subsequently, in the negative phase, the model generates samples, and the probability of generating these samples decreases. A complete cycle of positive and negative phases constitutes one training epoch. Weight updates in the training process are performed at the end of each epoch. This involves calculating the difference between the generated samples and the actual data vectors. The gradient of the visible unit probability with respect to the weights is then computed. This gradient represents the expected difference in the contributions of the positive and negative phases, which is subsequently used to adjust the model’s weights.
The second deep learning algorithm employs a DBN trained from scratch. We argue that applying DBNs to local feature representations, as opposed to raw image data, can guide the learning process, leading to more effective feature learning and faster training. Local features extracted from the attention block of the first algorithm are fed into a DBN model to learn additional and complementary feature representations, as illustrated in Fig. 1. The DBN architecture employed in this study consists of a stack of three restricted boltzmann machines (RBMs), resulting in three hidden layers (see Fig. 9). The first 2 RBMs are trained as generative models, while the final RBM, integrated with Softmax units, serves as a discriminative model for multi-class classification. The DBN is trained layer-wise in a greedy fashion, with each layer trained sequentially from bottom to top. The training of the DBN model is a 3 stage process: pre-training, supervised training, and fine-tuning stage.
-
1.
Pre-training stage: This stage involves training the first two RBMs sequentially in an unsupervised manner using a greedy layer-wise algorithm. This involves training each RBM individually, treating the outputs of one layer as the inputs to the next. This unsupervised pre-training phase leverages a massive amount of unlabeled data to improve the model’s generalization ability and prevent overfitting. Additionally, it simplifies the training process and accelerates convergence.
-
2.
Supervised Training phase: It involves training the ultimate RBM as a non-linear classifier on labeled data. Performance evaluation is conducted at the end of each epoch to monitor the model’s progress and assess its classification accuracy.
-
3.
Fine-tuning stage: In the final fine-tuning phase, a top-down backpropagation algorithm is employed to optimize the weights of the entire network for optimal classification performance.
Decision fusion stage
The final classification decision of the Skin-DeepNet system is done by fusing the outputs of the introduced algorithms explained in Sect. 3.4. When a dermoscopic image is fed into Skin-DeepNet system, multiple probability scores are calculated for each class. The most likely class is assigned to the input image. To enhance the classification performance even more, this study explores different fusion methods to combine the outputs of the introduced classification algorithms. Herein, ensemble learning strategy is employed as a machine learning paradigm in which a number of models (referred to as base learners or weak learners) are combined to produce a stronger, and more robust predictive model. The principle is that averaging predictions from multiple models can give enhanced performance compared to relying on one single model. In this stage, two well-known strategies based on boosting (e.g., XGBoost), and stacking (e.g., LR33, RF34, and ET35 are applied. While both boosting and stacking belong to the ensemble learning strategy, their operational methodologies are significantly different. Boosting follows a sequential model building strategy, successively enhancing predictions by rectifying previous errors. Stacking, on the contrary, employs a parallel process, aggregating predictions from independently trained models through a meta-learner to achieve greater predictive precision. These fusion techniques are applied in this study as below:
-
1.
Data Preparation: For every training instance, both the HRNet-attention block and DBN models predict probability scores (for all classes). These two sets of scores are merged together into one feature vector.
-
2.
Training Phase: The new dataset (combined features) is utilized to train either the XGBoost classifier or a meta-learner (e.g., LR, RF, and ET) that learns to combine the base model outputs optimally. The XGBoost classifier can enhance predictions sequentially by focusing on errors produced from the HRNet-attention block and DBN models. Rather than having to give weights manually to the HRNet-attention block and DBN models, XGBoost decide the optimal weighting. Alternatively, the meta-learner model identifies the optimal weights for a particular classifier based on actual-world classification performance.
-
3.
Inference Phase: The HRNet-attention block and DBN models produce their probability scores for a new sample. They are concatenated and fed into the XGBoost classifier or a meta-learner, which produces the final prediction. Table 1 lists the hyper-parameters’ values of the employed fusion techniques.
Experimental results
This section gives an extensive description of the experimental arrangement and methodology, along with performance measure metrics under which the developed Skin-DeepNet system has been tested. Comparative analysis is also drawn to compare the effectiveness and reliability of the developed Skin-DeepNet system with the current state-of-the-art skin cancer classification systems.
Implementation details
The Skin-DeepNet system is developed using the Python programming language and implemented within a Google Colab environment equipped with a 69 K GPU, 16GB RAM, and on a 64-bit Windows 10 operating system with an Intel Core i7 processor. TensorFlow is utilized as the deep learning framework for the development of models. To maintain uniformity and compatibility across diverse deep neural network architectures, all initial images underwent a pre-processing stage that involved resizing to a standardized resolution of (224 × 224) pixels, which is a prevalent input dimension endorsed by numerous pre-trained models. For the ISIC 2019 Challenge, the typical dataset split involves allocating 70% of the data for training (17,732 images), 15% for validation (3,799 images), and the remaining 15% for testing (3,799 images). The same split rates were applied to the HAM10000 dataset. This division strikes a balance between providing sufficient data for model training and maintaining representative subsets for validation and testing. Due to the substantial imbalance in the number of images across classes, the classification model may exhibit bias towards the majority class. To mitigate this issue, data augmentation techniques such as vertical and horizontal shifts, flips, and rotations were employed on the smaller classes. Herein, the proposed training methodology involved utilizing the Adam optimizer to train all the employed deep learning models. Training parameters were set as follows: an initial learning rate of 0.001, a batch size of 16, weight decay of 0.0002, a dropout rate of 0.5, and momentum of 0.95. To accelerate the convergence of the DRBM, its learning rate is increased to 0.01. To prevent overfitting, an early stopping procedure is implemented, terminating training when the validation error began to increase. While a maximum of 100 epochs is set, the actual number of training epochs varied based on the early stopping procedure. In this study, the DBN model employs CD learning algorithm, a structure comprising three RBMs, and a training process of 300 epochs for each RBM. The values of other hyper-parameters include a momentum of 0.9, a weight-decay of 0.0002, a dropout rate of 0.5, and a batch size of 64. The learning rate is set to 0.001, and the entire network is trained top-down using the back-propagation algorithm for 500 epochs. These settings collectively contribute to the model’s efficacy, balancing computational efficiency with learning capacity, and ensuring robust feature extraction and classification capabilities essential for accurate skin cancer segmentation and classification.
Evaluation metrics
The primary goal of the image segmentation experiment was to identify and localize skin lesions within dermoscopic images. This required an automated segmentation process to ensure accurate lesion localization, enabling accurate diagnostic classification. We employed several metrics to evaluate the performance of the proposed image segmentation procedure, including the Intersection over Union (IOU), Dice coefficient (Dic), Jaccard index (Jac), and Accuracy Rate (AR). The Dice coefficient and Jaccard index are commonly used metrics to measure the similarity between the segmented lesion and the ground truth annotation. While both metrics evaluate overlap, the Jaccard index is optimally used with non-regular segmentation borders whereas the IOU is optimally utilized for localized regions with rectangular borders. The following formulas are used to compute the evaluation metrics discussed above.
Then, the accuracy of the developed Skin-DeepNet system in classifying the dermoscopic images into various classes was evaluated. In this assessment process, five popular quantitative metrics were calculated: AR, Precision, F1-score, Recall, and AUC.
-
1.
Precision: Precision quantifies the accuracy of positive predictions, indicating the likelihood that a sample classified as positive is indeed positive. The Precision is calculated as follow:
-
2.
Recall: Recall measures the sensitivity of the model, assessing its ability to correctly identify positive samples among all actual positive cases. The Recall is calculated as follow:
-
3.
F1-score: The F1-score provides a balanced metric that considers both precision and recall, achieving higher values when both metrics are high. It is calculated as follows:
-
4.
AUC: The AUC evaluates the model’s overall classification accuracy, with higher values indicating better performance. It is particularly useful for assessing model stability. The AUC is calculated as follow:
Here, TP represents True Positives, TN represents True Negatives, FP represents False Positives, and FN represents False Negatives.
Skin lesion segmentation assessment
In this stage, Mask R-CNN was employed to accurately segment skin lesions within input images. While Mask R-CNN demonstrated promising results, it occasionally included background regions in the segmented images, which could negatively impact the system’s accuracy. To address this issue, Mask R-CNN’s output was used as an initial seed for the GrabCut algorithm, followed by applying some morphological reconstructions to eliminate background noise and fill potential gaps in the segmented mask. Herein, a set of experiments was carried out to evaluate the performance of different network backbones (e.g., ResNet34, ResNet50, DenseNet121, InceptionV3, and VGG19) with both the standalone Mask R-CNN and the combined Mask R-CNN and GrabCut approach. The results summarized in Tables 2 and 3, demonstrate the effectiveness of the proposed skin lesion segmentation algorithm. The tables present a comparative analysis of skin lesion segmentation results on the ISIC 2019 and HAM1000 datasets, evaluating the performance of various neural network backbones with and without the GrabCut algorithm. Across both datasets, the integration of GrabCut algorithm consistently enhances segmentation accuracy. For the ISIC 2019 dataset, ResNet34’s IOU improves from 83.12 to 96.11% with GrabCut algorithm, while ResNet50’s IOU increases from 88.23 to 99.94%, demonstrating near-perfect segmentation results. DenseNet121 and InceptionV3 also show considerable improvements, with IOU values rising from 78.34 to 86.56% and from 79.56 to 86.78%, respectively. VGG19, which was at an IOU of 68.92% in the first place, is improved to 79.65 using GrabCut. Similarly, on the HAM1000 dataset, ResNet34’s IOU rises from 87.72 to 97.34%, and ResNet50’s IOU from 89.93 to 99.91%. DenseNet121 and InceptionV3 improve, respectively, from 86.84 to 90.86% and from 87.78 to 91.34%, whereas VGG19’s IOU increases from 89.12 to 93.56%. Such findings are an indication of GrabCut’s robustness in enhancing segmentation results, as evidenced in the universal uplift in Dice and Jaccard coefficients, besides accuracy, in both datasets. This underscores the GrabCut algorithm’s effectiveness in significantly improving the accuracy of deep learning models (e.g., Mask R-CNN) particularly for skin lesion segmentation tasks, within the medical imaging domain.
Figure 10 illustrates some results of the proposed skin lesion segmentation algorithm on the ISIC 2019 and HAM1000 datasets, thereby validating its efficacy through comprehensive visual assessments. Each row contains a series of images that include: the original dermoscopic image, the reference mask, the resulting mask, and the final resulting segmented image. On the ISIC 2019 dataset, the masks generated exhibit high consistency with ground truth masks, leading to segmented images with high adherence to lesion boundaries. The accuracy is perceptible with regard to different lesion types and sizes, showing the efficacy of the proposed segmentation algorithm. Similarly, the HAM1000 dataset demonstrates that the resulting masks are very accurate in comparison to the ground truth, generating segmented images that are very representative of the lesion regions. The consistency of performance across the two datasets demonstrates the effectiveness and reliability of the proposed segmentation algorithm, thereby helping to highlight its potential for precise and efficient evaluation of skin lesions in clinical settings.
Table 4 compares the proposed skin lesion segmentation algorithm with other recently published state-of-the-art methods on the ISIC 2019 and HAM1000 datasets. The proposed skin lesion segmentation algorithm achieves higher results with an IOU of 99.84%, a Dic coefficient of 98.78%, a Jac index of 98.93%, and an accuracy of 99.94% on the ISIC 2019 dataset. The metrics here show better performance than other methods, such as those proposed by Banerjee et al.36 (IOU of 91%, Dic of 97.13%) and Singh et al.37 (IOU of 97.79%, Dic of 95.97%). Similarly, on the HAM1000 dataset, the novel algorithm has an IOU of 99.93%, a Dic of 99.88%, a Jac index of 99.97%, and an accuracy of 99.98%, improving over the work of Himel et al.38 (IOU of 96.07%, Dic of 98.14%) and Firdaus et al.39 (IOU of 90.37%, accuracy of 95.89%). The reported high performance in both datasets highlights the consistency and accuracy of the proposed algorithm for skin lesion segmentation, thus indicating its potential to improve diagnostic accuracy in dermatology.
Evaluating skin lesion classification models
To overcome the difficulties involved in multi-class classification of dermoscopic images, a transfer learning approach was used. It takes pre-trained CNN models as a starting point, and this is a less computationally intensive choice than training a CNN model from scratch. To optimize performance, we are precisely fine-tuned the hyper-parameters of these pre-trained CNN models. We evaluated the efficacy of numerous pre-trained CNN architectures, originally trained on the ImageNet dataset, as feature extractors for classifying dermoscopic images into multiple classes. Table 5 demonstrates the performance of the first classification algorithm, which integrates diverse pretrained CNN models, an attention block, and a DRBM classifier. The findings demonstrate that the HRNet model utilized in this study delivers the highest overall performance on both datasets. On the ISIC 2019 dataset, HRNet model attains an accuracy of 97.54%, Precision of 97.73%, Recall of 98.14%, F1-score of 97.93%, and AUC of 97.28%, outperforming other backbones like ResNet101 (accuracy 95.94%) and EfficientNetB0 (accuracy 96.74%). Similarly, on the HAM1000 dataset, HRNet model again outperforms with an accuracy of 98.34%, Precision of 98.56%, Recall of 98.51%, F1-score of 98.53%, and an AUC of 98.23%, demonstrating its robustness and superior classification ability. EfficientNetB0 model has also performed well by recording Precision of 98.13% and 98.89% on ISIC 2019 and HAM10000 datasets, respectively. In contrast, the worst performance on both datasets is recorded by ResNet34, with accuracy of 75.67% and AUC of 79.99% on ISIC 2019 dataset. These findings highlight HRNet’s superior performance in skin lesion classification tasks, indicating its potential for precise and trustworthy diagnosis in medical images. In contrast, the performance of the second proposed classification algorithm was assessed using four distinct DBN architectures, with results presented in Table 6. Training a DBN model from scratch is a complex process, requiring careful tuning of numerous hyper-parameters, such as the number of RBMs, the units per RBM, the training epochs, and the learning rate. These hyper-parameters are inter-dependent, and thus the fine-tuning is computationally expensive. For hyper-parameter optimization, a coarse search procedure was conducted using the training method outlined in Sect. 3.4. The DBN model was trained greedily layer wise, with each RBM being trained individually and its weights preserved. Then, the activations of the trained RBM were used as input to train the next RBM in the stack such that there is layer-by-layer learning process. Table 6 presents the comparative evaluation of skin lesion classification performance on various DBN architectures on ISIC 2019 and HAM1000 datasets. Among the architectures experimented, the DBN ([1024-2048-1024]) model performed better and competitive results across five key metrics were obtained. Specifically, this model obtained nearly perfect F1-scores (98.51% on ISIC 2019 and 99.15% on HAM1000) and good AUC values (98.88% and 99.57%, respectively), pointing to very good discriminative power and generalizability. Even though higher configurations (e.g., DBN [1024-2048-2048]) had slight improvements in some measures, the (DBN [1024-2048-1024]) model generally struck a balance between performance and complexity, avoiding overfitting while maintaining high diagnostic accuracy. This validation supported its designation as the ultimately selected model for subsequent classification tasks, thereby guaranteeing dependable and effective detection of skin cancer across various datasets.
Table 7 provides a comparative analysis of various fusion approaches that have been utilized for skin lesion classification on the ISIC 2019 and HAM1000 datasets. The results demonstrate the efficacy of fusing the decisions of various classification models for obtaining overall greater accuracy. Most notably, the XGBoost approach consistently provides the better performance in both datasets. On the ISIC 2019 dataset, XGBoost approach achieves an accuracy, precision, recall, F1-score, and AUC of 99.65%, 99.51%, 99.56%, 99.54%, and 99.94%, respectively. Similarly, on the HAM1000 dataset, XGBoost achieves a perfect accuracy rate of 100% and near-optimal values for all other metrics. It is interesting to note that RF also performs incredibly well, with a recall rate of 99.68% on ISIC 2019, which demonstrates its capability in detecting true positives. Conversely, less complex fusion approaches, such as LR and ET exhibit inferior performance, particularly for recall and F1 scores. These findings emphasize the effectiveness of ensemble-based fusion approaches, such as XGBoost and RF, for enhancing classification accuracy and stability in dermatological image analysis tasks. Overall, the results suggest that advanced fusion approaches are extremely suitable for boosting diagnostic accuracy in skin lesion classification, with XGBoost being the most promising approach in both datasets.
To give a clear view of class-wise performance and determine if there is any remaining bias, we assessed the proposed Skin-DeepNet system using per-class precision, recall (sensitivity), specificity, and F1-score, presented in Tables 8 and 9. Normalized confusion matrices for both the ISIC 2019 and HAM10000 datasets are also shown in Fig. 11. On the ISIC 2019 dataset, the proposed Skin-DeepNet system performed highly on most classes. Melanocytic nevi and benign keratosis, for instance, obtained recall values of 99.9% and 97.4%, respectively. This is consistent with their overwhelming representation in the dataset. However, performance was relatively low on under-represented classes. For instance, Squamous cell carcinoma (SCC) obtained a recall of 79.2%, and vascular lesions obtained 81.5%. These results are consistent with the inherent difficulty of recognizing rare classes with few samples and inter-class visual similarities. The confusion matrix indicates that SCC was sometimes confused with AKIEC or BKL, which are morphologically similar (See Fig. 11a). Vascular lesions were confused into DF, SCC, and BKL, indicating overlapping features in dermoscopic appearance. In spite of these difficulties, precision was still high for both SCC (96.5%) and vascular lesions (94.6%), suggesting that the model does not have a tendency for frequent false positives for minority classes. The results overall demonstrate that the use of class-balanced focal loss and ensemble fusion (through XGBoost) is effective in enhancing performance on all classes without sacrificing good generalization.
On the HAM10000 dataset, the proposed Skin-DeepNet system performed almost perfect classification. All classes had recall and precision over 98.9%, with melanoma, vascular lesions, and melanocytic nevi having 100% recall. The confusion matrix shows small off-diagonal values, indicating consistent classification accuracy for all types of lesions (See Fig. 11b). This was due to the more even class distribution in HAM10000 than in ISIC 2019. The class-wise in-depth analysis corroborates that although high overall accuracy is retained, biases at a class level remain, especially under highly imbalanced conditions. The results validate the incorporation of per-class analysis within performance measurement and direct future research towards enhancing the representation of rare classes through focused synthetic augmentation and data-aware sampling techniques.
Table 10 provides the comparative analysis of the developed Skin-DeepNet system with various state-of-the-art systems on the ISIC 2019 and HAM1000 datasets. On the ISIC 2019 dataset, the Skin-DeepNet system achieves outstanding results, with an accuracy of 99.65%, precision of 99.51%, recall of 99.56%, an F1 score of 99.54%, and an AUC of 99.94%. These metrics surpass those reported by most competing approaches, such as Singh et al.37 (accuracy of 98.04%, recall of 96.67%, and F1 score of 96.24%) and Radhika and Chandana43 (accuracy of 98.77%, recall of 98.42%, and F1 score of 98.76%). On the HAM1000 dataset, the Skin-DeepNet system demonstrates near-perfect performance, achieving perfect accuracy of 100%, precision of 99.92%, recall of 100%, an F1 score of 99.96%, and an AUC of 99.97%. This outperforms other methods like Monica et al.41 (accuracy of 99.98%, precision of 99.97%, and F1 score of 99.90%). The consistent and near-perfect metrics of the developed Skin-DeepNet system highlight its robustness and reliability, making it a highly advantageous system for real clinical practice. Its precision and accuracy are high, which can significantly boost diagnostic confidence, reduce the rates of misdiagnosis, and improve patient outcomes by providing clinicians with a very reliable tool for skin lesion classification. Finally, its improved performance can be attributed to its novel architectural design, which is assumed to enhance feature extraction and decision-making capability compared to existing state-of-the-art systems.
Conclusions and future work
This research proposes Skin-DeepNet, a new deep learning framework with notable efficacy in the automatic early detection and classification of skin cancer lesions from dermoscopy images. Through the incorporation of cutting-edge methodologies, including AGCWD approach for image enhancement, hybrid Mask R-CNN and GrabCut segmentation algorithm for an accurate skin lesion detection, and a dual-feature extraction approach combining HRNet-attention block and DBN models, the Skin-DeepNet system realizes state-of-the-art results on the ISIC 2019 and HAM1000 datasets. Novel decision fusion methodologies of the system, including XGBoost, offer excellent classification metrics with approximately perfect rates in accuracy, precision, recall, F1-score, and AUC on both datasets. These outstanding results demonstrate the potential of the developed Skin-DeepNet system to significantly enhance the diagnostic precision and consistency in a clinical setting and offer dermatologists a powerful tool for skin cancer detection and treatment planning. All future research must focus on the comparison of novel deep learning architectures, the development of attention mechanisms, the creation of user-friendly applications, and conducting large-scale clinical trials to establish real-world effectiveness, thereby leading to the evolution of more precise and available AI-supported healthcare interventions.
Data availability
In this study, the employed datasets from these links: (ISIC 2019 dataset) https://challenge.isic-archive.com/data/#2019 and (HAM10000 dataset) https://datasetninja.com/skin-cancer-ham10000 (accessed on 25 June 2024).
References
Tahir, M. et al. DSCC_Net: multi-classification deep learning models for diagnosing of skin cancer using dermoscopic images. Cancers (Basel). 15, 7. https://doi.org/10.3390/cancers15072179 (2023).
Naqvi, M., Gilani, S. Q., Syed, T., Marques, O. & Kim, H. Skin cancer detection using deep learning—a review. Diagnostics 13 (11), 1–26 (2023).
Ünver, H. M. & Ayan, E. Skin lesion segmentation in dermoscopic images with combination of Yolo and Grabcut algorithm. Diagnostics 9 (3). https://doi.org/10.3390/diagnostics9030072 (2019).
Hasan, K., Dahal, L., Samarakoon, P. N. & Islam, F. DSNet: Automatic dermoscopic skin lesion segmentation, Comput. Biol. Med. 120 (2020).
Melarkode, N., Srinivasan, K., Qaisar, S. M. & Plawiak, P. AI-Powered diagnosis of skin cancer: A contemporary review, open challenges and future research directions. Cancer 15 (4), 1–36 (2023).
Adjed, F. Skin cancer segmentation and detection using total variation and multiresolution analysis, Signal and Image Processing. Univeristé Paris-Saclay; Université d’Evry-Val-d’Essonne; UTP Petronas, (2017).
Idlahcen, F., Idri, A. & Goceri, E. Exploring data mining and machine learning in gynecologic oncology. No 2 Springer Neth. 57 https://doi.org/10.1007/s10462-023-10666-2 (2024).
Nakach, F. Z., Idri, A. & Goceri, E. A comprehensive investigation of multimodal deep learning fusion strategies for breast cancer classification. Artif. Intell. Rev. 57 (12). https://doi.org/10.1007/s10462-024-10984-z (2024).
Goceri, E. & Karakas, A. A. Comparative evaluations of CNN based networks for skin lesion classification. In 14th International Conference on Computer Graphics, Visualization, Computer Vision and Image Processing (CGVCVIP), Zagreb, Croatia, pp. 1–6. (2020).
Goceri, E. Impact of deep learning and smartphone technologies in dermatology: automated diagnosis. 2020 10th Int. Conf. Image Process. Theory Tools Appl. IPTA 2020. no. c https://doi.org/10.1109/IPTA50016.2020.9286706 (2020).
Rathod, J., Wazhmode, V., Sodha, A. & Bhavathankar, P. Diagnosis of skin diseases using Convolutional Neural Networks. In Proc. 2nd Int. Conf. Electron. Commun. Aerosp. Technol. ICECA, no. Iceca, pp. 1048–1051, 2018 (2018). https://doi.org/10.1109/ICECA.2018.8474593.
Alzamili, A. H. & Ruhaiyem, N. I. R. A comprehensive review of deep learning and machine learning techniques for early-stage skin cancer detection: challenges and research gaps. J. Intell. Syst. 34 (1). https://doi.org/10.1515/jisys-2024-0381 (2025).
Maiti, A., Chatterjee, B. & Santosh, K. C. Skin cancer classification through quantized color features and generative adversarial network. Int. J. Ambient Comput. Intell. 12 (3), 75–97. https://doi.org/10.4018/IJACI.2021070104 (2021).
Maiti, A. & Chatterjee, B. The Effect of Different Feature Selection Methods for Classification of Melanoma. Int. Symp. Signal Image Process. 123–133. https://doi.org/10.1007/978-981-33-6966-5 (2021).
Sharma, A. K., Member, S. & Tiwari, S. Dermatologist-Level Classification of Skin Cancer Using Cascaded Ensembling of Convolutional Neural Network and Handcrafted Features Based Deep Neural Network. IEEE Access 10, 17920–17932. https://doi.org/10.1109/ACCESS.2022.3149824 (2022).
Naeem, A., Anees, T., Fiza, M. & Naqvi, R. A. SCDNet: A deep Learning-Based framework for the multiclassification of skin cancer using dermoscopy images. Sensors 22 (15), 1–18 (2022).
Bechelli, S. & Delhommelle, J. Machine learning and deep learning algorithms for skin cancer classification from dermoscopic images. Bioengineering 9 (3), 1–18 (2022).
Imran, A. et al. Skin Cancer Detection Using Combined Decision of Deep Learners. IEEE Access 10, 118198–118212. https://doi.org/10.1109/ACCESS.2022.3220329 (2022).
Cha, J. Multiclass skin lesion classification using hybrid deep features selection and extreme learning machine. Sensors 22 (3), 1–22 (2022).
Bassel, A. et al. Automatic malignant and benign skin cancer classification using a hybrid deep learning approach. Diagnostics 12, 10 (2022).
Hamida, S., Lamrani, D., El Gannour, O., Saleh, S. & Cherradi, B. Toward enhanced skin disease classification using a hybrid RF-DNN system leveraging data balancing and augmentation techniques. Bull. Electr. Eng. Inf. 13 (1), 538–547. https://doi.org/10.11591/eei.v13i1.6313 (2024).
Dahdouh, Y., Anouar, A. B. & Ben Ahmed, M. Embedded artificial intelligence system using deep learning and raspberrypi for the detection and classification of melanoma. IAES Int. J. Artif. Intell. 13 (1), 1104–1111. https://doi.org/10.11591/ijai.v13.i1.pp1104-1111 (2024).
Kassem, M. A., Hosny, K. M. & Fouad, M. M. Skin lesions classification into eight classes for ISIC 2019 using deep convolutional neural network and transfer learning. IEEE Access. 8, 114822–114832. https://doi.org/10.1109/ACCESS.2020.3003890 (2020).
Tschandl, P., Rosendahl, C. & Kittler, H. Data descriptor: the HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data. 5, 1–9. https://doi.org/10.1038/sdata.2018.161 (2018).
Huang, S. C., Cheng, F. C. & Chiu, Y. S. Efficient contrast enhancement using adaptive gamma correction with weighting distribution. IEEE Trans. Image Process. 22 (3), 1032–1041. https://doi.org/10.1109/TIP.2012.2226047 (2013).
Bello, R. W., Mohamed, A. S. A. & Talib, A. Z. Contour extraction of individual cattle from an image using enhanced mask R-CNN instance segmentation method. IEEE Access. 9, 56984–57000. https://doi.org/10.1109/ACCESS.2021.3072636 (2021).
Mohammed, H. J. et al. ReID-DeePNet: A hybrid deep learning system for person Re-Identification. Mathematics 10 (19), 3530 (2022).
Abed, S. H., Al-Waisy, A. S., Mohammed, H. J. & Al-Fahdawi, S. A modern deep learning framework in robot vision for automated bean leaves diseases detection. Int. J. Intell. Robot Appl. 5 (2), 235–251. https://doi.org/10.1007/s41315-021-00174-3 (2021).
Al-Waisy, A. S., Qahwaji, R., Ipson, S. & Al-Fahdawi, S. A multimodal deep learning framework using local feature representations for face recognition. Mach. Vis. Appl. 29 (1), 35–54. https://doi.org/10.1007/s00138-017-0870-2 (2017).
Khodaverdian, Z., Sadr, H. & Edalatpanah, S. A. A shallow deep neural network for selection of migration candidate virtual machines to reduce energy consumption. In 2021 7th Int. Conf. Web Res. ICWR 2021, pp. 191–196, (2021). https://doi.org/10.1109/ICWR51868.2021.9443133.
Al-Waisy, A. S., Qahwaji, R., Ipson, S. & Al-fahdawi, S. A Multimodal Biometric System for Personal Identification Based on Deep Learning Approaches. In Seventh International Conference on Emerging Security Technologies (EST), pp. 163–168 (2017).
Hinton, G. E. Training products of experts by minimizing contrastive divergence. Neural Comput. 14 (8), 1771–1800. https://doi.org/10.1162/089976602760128018 (2002).
Patil, B. U., Ashoka, D. V. & Prakash, B. V. A. Data integration based human activity recognition using deep learning models. Karbala Int. J. Mod. Sci. 9 (1), 106–121. https://doi.org/10.33640/2405-609X.3286 (2023).
Weiss, G. M., Yoneda, K. & Hayajneh, T. Smartphone and Smartwatch-Based biometrics using activities of daily living. IEEE Access. 7, 133190–133202. https://doi.org/10.1109/ACCESS.2019.2940729 (2019).
Pagliaro, A. Forecasting significant stock market price changes using machine learning: extra trees classifier leads. Electron 12, 1–23. https://doi.org/10.3390/electronics12214551 (2023).
Banerjee, S. et al. Diagnosis of melanoma lesion using neutrosophic and deep learning. Trait Du Signal. 38 (5), 1327–1338. https://doi.org/10.18280/ts.380507 (2021).
Singh, S. K., Abolghasemi, V. & Anisi, M. H. Skin cancer diagnosis based on neutrosophic features with a deep neural network. Sensors 22 (16). https://doi.org/10.3390/s22166261 (2022).
Himel, G. M. S., Islam, M. M., Al-Aff, K. A., Karim, S. I. & Sikder, M. K. U. Skin cancer segmentation and classification using vision transformer for automatic analysis in Dermatoscopy-Based noninvasive digital system. Int. J. Biomed. Imaging. 1, 1. https://doi.org/10.1155/2024/3022192 (2024).
Firdaus, F. et al. Segmentation of skin lesions using convolutional neural networks. Comput. Eng. Appl. J. 12 (1), 58–67. https://doi.org/10.18495/comengapp.v12i1.466 (2023).
Banerjee, S., Singh, S. K., Chakraborty, A., Das, A. & Bag, R. Melanoma diagnosis using deep learning and fuzzy logic. Diagnostics 10 (8), 1–26 (2020).
Monica, K. M. et al. Melanoma skin cancer detection using mask-RCNN with modified GRU model. Front. Physiol. 14, 1–12. https://doi.org/10.3389/fphys.2023.1324042 (2023).
Krishnan, V. G. et al. A prediction of skin cancer using Mean-Shift algorithm with deep forest classifier. Iraqi J. Sci. 63 (7), 3200–3211. https://doi.org/10.24996/ijs.2022.63.7.39 (2022).
Radhika, V. & Sai Chandana, B. MSCDNet-based multi-class classification of skin cancer using dermoscopy images. PeerJ Comput. Sci. 9 (1). https://doi.org/10.7717/peerj-cs.1520 (2023).
Adegun, A. A. & Viriri, S. FCN-Based densenet framework for automated detection and classification of skin lesions in dermoscopy images. IEEE Access. 8, 150377–150396. https://doi.org/10.1109/ACCESS.2020.3016651 (2020).
Naeem, A. & Anees, T. DVFNet: A deep feature fusion-based model for the multiclassification of skin cancer utilizing dermoscopy images. PLoS One. 19 (3), 1–27. https://doi.org/10.1371/journal.pone.0297667 (2024).
Alwakid, G., Gouda, W., Humayun, M. & Sama, N. U. Melanoma detection using deep Learning-Based classifications. Healthc 10 (12). https://doi.org/10.3390/healthcare10122481 (2022).
Ali, K., Shaikh, Z. A., Khan, A. A. & Laghari, A. A. Multiclass skin cancer classification using EfficientNets – a first step towards preventing skin cancer. Neurosci. Inf. 2 (4), 100034. https://doi.org/10.1016/j.neuri.2021.100034 (2022).
Kousis, I., Perikos, I., Hatzilygeroudis, I. & Virvou, M. Deep learning methods for accurate skin cancer recognition and mobile application. Electron 11 (9), 1–19. https://doi.org/10.3390/electronics11091294 (2022).
Author information
Authors and Affiliations
Contributions
Alaa S. Al-Waisy: Conceptualization, Methodology, Software, Validation, Formal analysis, Writing – original draft. Shumoos Al-Fahdawi: Conceptualiza-tion, Methodology, Software, Validation, Writing – original draft, Supervision, Project administration. Mohammed I. Khalaf: Formal analysis, Resources, Visualization, Investigation, Writing – review & editing. Mazin Abed Mohammed: Formal analysis, Re-sources, Visualization, Investigation, Writing – review & editing. Bourair AL-Attar and Mohammed Nasser Al-Andoli: Visualization, Investigation, Writing – review & editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Al-Waisy, A.S., Al-Fahdawi, S., Khalaf, M.I. et al. A deep learning framework for automated early diagnosis and classification of skin cancer lesions in dermoscopy images. Sci Rep 15, 31234 (2025). https://doi.org/10.1038/s41598-025-15655-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-15655-9
Keywords
This article is cited by
-
Advancing skin cancer diagnosis with deep learning and attention mechanisms
Scientific Reports (2025)