Abstract
Light intensity and spectral quality are two important factors that affect the cryopreservation success of plant germplasm. In a previous study, we found that light emitting diodes (LEDs) with 90% red and 10% blue (RB) light have a positive effect on potato (Solanum tuberosum L.) shoot tip recovery during post-cryopreservation regeneration. In the present study, transcriptome profiles of non-cryopreserved (−) and cryopreserved (+) potato shoot tips exposed to RB, cool white fluorescent tubes (CW) and full darkness (D) were compared. Results indicated that light spectrum significantly affects transcript numbers in post-cryopreserved samples resulting in 3,284 differentially expressed genes of recovering shoot tips exposed to CW, RB and D. Gene ontology (GO) enrichment analysis revealed that several stress-related terms were more abundant in CW+, whereas morphogenesis-related processes were activated under RB+. Overall, cryopreservation had an extensive effect on gene expression, as 12,017 genes were differentially expressed in non-cryopreserved and cryopreserved shoot tips. Among all cryopreserved samples, enriched GO terms included various defense and stress responses to biotic stimuli and oxidative stress, whereas suppressed responses were mainly related to organ morphogenesis. The present study emphasizes that the optimization of light spectral properties to decrease light-related stress may greatly benefit the cryopreservation efficiency of plant germplasm.
Similar content being viewed by others
Introduction
Potato is the fourth most important crop globally, encompassing numerous cultivars and landraces that are typically propagated by vegetative means to maintain their distinct traits. While field and in vitro collections are suitable for short- or medium-term conservation of these resources, both methods require regular multiplication and maintenance, making them unsuitable for long-term preservation1,2. Cryopreservation is a powerful tool for safeguarding plant genetic resources, thereby ensuring the availability of the essential raw materials for food and agriculture. In cryopreservation, plant material (germplasm) is cooled to an ultra-low temperature (below − 150 °C), while maintaining viability and regeneration capability upon thawing. At an extremely low temperature, all metabolic processes are arrested, and thus the material can, in theory, be stored infinitely. Consequently, cryopreservation is the best and most economical method to ensure the long-term preservation and backup of clonally propagated species3,4. The major challenges of potato cryopreservation arise from several factors that can compromise recovery, including genotype-specific responses5,6,7, stress induced by cryo-treatments8, and physiological damage during the regeneration phase9. Moreover, most protocols are developed using a limited number of accessions, making them less broadly applicable across genetically diverse cultivars. As a result, further optimization is often required to accommodate cultivar-specific variation10. Among key factors affecting recovery outcomes, the environmental conditions during the regeneration phase play a critical role11,12.
Light, its intensity, duration, and spectral quality, are among the most important factors that affect plant growth and morphogenesis13. Light intensity, both before and after cryopreservation, significantly affects the outcome of cryopreservation14. Generally, initial post-cryopreservation recovery is carried out either in low light conditions or in darkness for some days to several weeks before transferring the plant materials to standard conditions under fluorescent tubes15,16,17. This is to avoid light-caused photo-oxidation18 and damage to the photosynthesis apparatus by excess light19.
Besides light intensity, we have previously17 found that light spectral quality significantly affects cryopreservation success of potato shoot tips in vitro and demonstrated that light emitting diodes (LEDs) consisting of 90% red and 10% of blue (RB) light promote shoot formation. Furthermore, regeneration percentages of five potato cultivars were doubled when recovered under RB LEDs instead of cool white fluorescent tubes (CW)17.
In the present study, we examined the biological processes behind the positive effect of RB LEDs on potato shoot tip recovery. We conducted a comprehensive transcriptome analysis of recovering shoot tips under the exact same RB LEDs and CW as previously17 in comparison to shoot tips recovering in darkness. Moreover, we analyzed the overall cryopreservation effect at the transcriptional level by comparing the cryopreserved potato shoot tips to non-cryopreserved controls.
Although transcriptome-level studies related to plant cryopreservation have greatly advanced our understanding of the molecular effects of cryoprotectants and ultra-low temperatures, they have primarily focused on model species Arabidopsis thaliana seed-derived explants20,21,22, grapevine zygotic embryos23, but also vegetatively propagated non-model species banana24. However, none of these studies have examined the influence of light conditions during the post-cryopreservation recovery phase. The present study addresses this important gap by providing the first comprehensive transcriptomic analysis of cryopreserved, clonally propagated potato shoot tips, with a specific focus on light spectral quality during recovery.
Materials and methods
Plant material
Disease-free in vitro plants of potato (Solanum tuberosum L. cv. Agrie Dzeltenie) were kindly provided by the potato genebank of Estonian Crop Research Institute (ECRI). Cultivar Agrie Dzeltenie was selected for this study because it had the highest survival and regeneration percentages among five tested cultivars under CW and RB light treatments17. Notably, RB light treatment doubled the regeneration percentage of ‘Agrie Dzeltenie’ from 25 to 53%. In addition, this cultivar consistently demonstrated stable regeneration rates across independent experiments17,25. For propagation, the plants were multiplied by nodal cuttings and placed into 250 ml jars (Combiness) containing Murashige and Skoog (MS) medium26 supplemented with 2% sucrose and 0.64% (w/v) agar (pH 5.7). Each jar contained 30 plants. The jars were closed with lids including 0.2 mm filters to enable gas exchange and placed in a growth chamber illuminated by cool white fluorescent tubes (Philips TL-D 840) with a 16/8-h light/dark photoperiod (40 µmol m-2s-1) at 22 ± 1 °C.
Cryopreservation
For cryopreservation, shoot tips (2–3 mm) of 3.5-week-old in vitro plants were aseptically dissected under a stereomicroscope. The non-cryopreserved control shoot tips were trimmed to 1 mm, which is approximately the size of a cryopreserved shoot tip two days post thawing. The explants were then placed onto 4.5-cm sterile filter papers moistened with 2 ml of liquid MS medium (with 3% sucrose and without growth hormones) and incubated overnight. Cryopreservation was carried out using the DMSO-droplet method as described by Kaczmarczyk et al.9. This method was chosen for the present study because it was also employed in previous experiments that revealed the influence of light quality on cryopreservation outcomes17,25, ensuring consistency and comparability with earlier findings. Briefly, before cryopreservation, shoot tips were treated with cryoprotectant solution (10% DMSO (v/v)) in MS medium containing 30% sucrose for 2 h at room temperature (RT) before being transferred into droplets of 2.5 µl cryoprotectant solution on aluminium foil strips (0.5 × 1.5 cm). After that, the shoot tips were flash-frozen by immersing the aluminium foil strips directly into cryotubes filled with liquid nitrogen (LN). The shoot tips were kept in LN for 1 h. After that, the shoot tips were rewarmed quickly by transferring the aluminium foil strips into liquid MS medium (50 ml) for 1.5 h at RT. After thawing, the cryopreserved samples (+) were placed on regeneration medium (MS with 0.5 mg/l zeatin riboside, 0.2 mg/l gibberellic acid (GA3), 0.5 mg/l indole acetic acid (IAA), and 30 g/l sucrose) on small Petri dishes (5.5 cm in diameter) and were allowed to regenerate for 48 h in total darkness. For non-cryopreserved (−) control samples, the procedure was the same as for the cryopreserved ones, except cooling in LN.
Light treatments
After the 48-h dark-incubation, the cryopreserved (+) samples were exposed to three different light conditions: total darkness (D), cool white fluorescent tubes (CW) (Philips TL-D 840) and a combination of blue and red LEDs (90% red, 10% blue) (RB) (Hangzhou Welthink Electronic Co.). The non-cryopreserved control samples (−) were only exposed to CW light. The light treatments were performed in growth chambers at 22 ± 1 °C. The spectral characteristics of CW and RB are provided in Supplementary Table S1. The period of light treatments for both (+) and (−) samples was 16 h, after which the samples (ten shoot tips per each treatment with three biological replicates) were collected into RNALater and stored at -80 °C until RNA extraction. The intensity of both RB and CW was 40 µmol m− 2s− 1. Dark-treated samples were harvested in infrared light (provided by Sony DCR-TRV320).
Experimental design
First, the overall effect of cryopreservation on the transcriptomic profile was analyzed by comparing cryopreserved shoot tips (CW+, RB + and D+) to the non-cryopreserved control shoot tips (CW− (Fig. 1). Then, to study the effect of light versus dark during post-cryopreservation recovery, gene expression under CW + and RB + was compared with D+. Finally, the effect of light quality was examined by comparing RB + to CW+ (Fig. 1).
Experimental design. Cryopreservation effect (grey arrows): the cryopreserved shoot tips (RB+, CW+ and D+) were compared to the non-cryopreserved control (CW−). Light effect (white arrows, grey outline): the cryopreserved shoot tips (CW+ and RB+) were compared to D+. Light quality effect (white arrows, violet outline): RB+ was compared to CW+. CW cool white fluorescent tubes, RB combination red and blue LEDs, D total darkness. Minus (−) denotes non-cryopreserved controls, and plus (+) cryopreserved shoot tips.
RNA extraction and sequencing
Three biological replicates (one biological replicate is 10 potato in vitro shoot tips grown in the same culture vessel) from each of the four different treatments were collected for RNA extraction. Tissue samples were homogenized using TissueLyser® (Qiagen) and total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Subsequently, genomic DNA was removed using RNase-free DNase Set (Qiagen) according to the manufacturer’s instructions. The quality and quantity of total RNA were analyzed using an Agilent Bioanalyzer 2100 (Agilent Technologies) in combination with the Agilent RNA 6000 Nano Kit. Approximately 1 µg total RNA (RIN > 7.4) was used for library preparation using Illumina’s TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. Quality of the libraries was assessed using an Agilent Bioanalyzer 2100 and concentrations with Qubit® Fluorometric Quantitation (Life Technologies). The obtained 12 libraries were pooled and run in one lane using an Illumina HiSeq2500 in high-output, paired-ended, 100 cycle mode, followed by read extraction (FastQ format) using bcl2fastq v1.8.4 (supported by Illumina).
Bioinformatic analyses
Quality control and mapping the reads
The quality of reads was assessed by FastQC v. 1.11.227 with default settings to ensure the per base pair quality score was above 20, and that there were no overrepresented sequences. TopHat (v2.0.8b) and bowtie1 (v 1.1.1) were used to map the reads to the S. tuberosum reference genome taking the annotation into account (genome and annotation: PGSC_DM_v4.03)28,29. Reads per gene were counted based on the TopHat output using featureCounts v1.5.030 and the annotation as mentioned above.
Differential expression analyses
The DESeq2 (v1.14.1)31 in Bioconductor/R package (https://www.bioconductor.org/help/workflows/rnaseqGene/) was used to identify differentially expressed genes (DEGs) between different treatments (control vs. treatments) and by using a negative binomial test32. Computing was done in the Linux shell using the taito.csc.fi platform (https://research.csc.fi). The parameters used in DESeq2 analysis were: mode=“Union”, singleEnd = FALSE, ignore.strand = FALSE, fragments = TRUE. False discovery rate-adjusted p-value (FDR) was estimated based on Benjamini Hochberg correction33, and significance levels were corrected accordingly during DESeq2 analysis. Changes in gene expression with an FDR (padj value) of < 0.05 were considered significant. Principal component analysis (PCA) and heatmap were applied to visualize the similarities and distances between samples31,34. To evaluate sample clustering, sample distances, and PCA in DESeq2, variance-stabilized transformation of the count data across the mean was performed, and regularized-logarithm transformation count data (rlog) were used. However, for DEG analysis, the raw and non-transformed counts were used31.
Gene ontology enrichment analysis
Gene Ontology (GO) enrichment analysis of differentially expressed genes was performed using the PlantRegMap35 GO enrichment tool, and was visualized by R package pheatmap v1.0.836 and BINGO v3.03 plug-in for Cytoscape 3.7.037. GO terms with corrected p-values (q-values) < 0.05 were considered significantly enriched among differentially expressed genes. For both tools, PRM GO term association file, downloaded from PlantRegMap, was used. To discover the overall processes affected by cryopreservation and light, enrichment of up- and down-regulated genes in each treatment comparison was performed, and the result of each list was visualized in a heatmap. The analysis was applied for DEGs with a 2-fold change (lfc > 1 and < -1) except for the detailed difference between RB + and CW + where enrichment analysis was performed for all DEGs.
Quantitative real-time PCR (qPCR) analysis
To verify the RNA sequencing (RNA-Seq) results, quantitative real-time PCR (qPCR) was carried out. Approximately 400 ng of total RNA from each sample (with the exception of 200 ng from one RB + sample) was reverse transcribed using SuperScript® III Reverse Transcriptase (Thermo Fisher Scientific) according to the manufacturer’s instructions. Based on significant expression differences in the RNA-Seq analysis, eight genes (Supplementary Table S2) were selected for qPCR. The qPCR reactions were performed according to the MIQE guidelines38 with newly designed primers (Supplementary Table S2). The amplifications were carried out with a LightCycler® 480 instrument (Roche Diagnostics, Espoo, Finland) using LightCycler® 480 SYBR Green I Master mix (Roche), 0.5 µM gene-specific primers, and 5 µl cDNA (1/20 dilution) in a total volume of 20 µl. Amplification was initiated by pre-incubation at 95 °C for 10 min, followed by 45 cycles of 10 s at 95 °C, 10 s at 58 °C, and 20 s at 72 °C. The amplification was followed by melting curve analysis to ensure that each primer pair amplifies a single product. For data analysis, the arithmetic mean of two technical replicates was used. Gene expression data was normalized by the combined average expression of two reference genes encoding adenosine kinase (ADK) and adenine phosphoribosyltransferase (APRT), and the relative expression of the target genes was calculated with the LightCycler® 480 software version 1.5.1.62 using the efficiency-adjusted advanced relative quantification method.
Results
Read alignment and DESeq2 analysis
The RNA-Seq analysis revealed that 27,438 genes in total were expressed in both control (−) and cryopreserved (+) samples. In general, the number of expressed genes was similar in control and cryopreserved samples exposed to CW, RB or D (Supplementary Table S3). The PCA analysis illustrated a clear and distinguished effect of cryopreservation and light on gene expression, as the samples were clustered by treatment (Fig. 2). Cryopreservation explained 73%, and light conditions (exposure to CW, RB or D) 16% of the variance between the two principal components.
Cryopreservation effect: CW+, RB + and D + compared to CW−
Number of DEGs
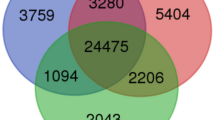
Cryopreservation and light had an extensive effect on gene expression as 11,977 genes were differentially expressed between cryopreserved and non-cryopreserved potato shoot tips exposed to different light conditions. In CW+, 8,639 genes were differentially expressed with 4,464 up- and 4,175 down-regulated DEGs compared to CW−. In RB + there were 7,947 DEGs compared to CW− with 4,103 up- and 3,844 down-regulated genes. The highest number of DEGs (9,353) was in D + compared to CW− with 4,708 up- and 4,645 down-regulated DEGs (Fig. 3A, Supplementary Table S4).
Approximately half of the DEGs were common to all cryopreserved samples. Specifically, among up-regulated genes, 3,047 DEGs were common to CW+, RB + and D+, while 586 DEGs were specific to CW+, 305 to RB+, and 859 to D+. Among down-regulated genes, 2,720 genes were common to CW+, RB+ and D + shoot tips, 680 were specifically down-regulated in CW+, 272 in RB+, and 1080 in D+ (Fig. 3A).
Gene ontology (GO) analysis
Cryopreservation effect
GO analysis of up- and down-regulated DEGs common to all cryopreserved shoot tips (identified by Venn diagram) revealed the biological processes of recovering shoot tips independent of light conditions. The majority of up-regulated processes were related to stress and defense responses to biotic stimulus (Fig. 4A, B, Supplementary Table S5). The top enriched GO terms were ‘protein phosphorylation’, ‘response to biotic stimulus’, ‘defense response’, ‘response to external biotic stimulus’, and ‘response to other organism’ (Fig. 4A).
A network of the top activated biological processes (BP) common to all cryopreserved samples independent of light conditions as revealed by GO enrichment analysis. (A) GO terms related to ‘response to stimulus’ are shown in blue, ‘secondary metabolite biosynthetic process’ in violet, and ‘protein phosphorylation’ in green. (B) A detailed network of GO terms related to ‘response to stimulus’. Only significantly enriched terms (padj < 0.05) are highlighted. The size of the node is relative to the number of genes associated with the term.
Most of the down-regulated processes common to all cryopreserved samples were related to morphogenesis. The top enriched GO terms were ‘shoot system development’, ‘reproductive shoot system development’, ‘anatomical structure morphogenesis, ‘post-embryogenic plant morphogenesis, and ‘regulation of organ morphogenesis’ (Supplementary Table S6).
Cryopreservation: light-specific differences
To reveal the general biological processes in each post-cryopreservation light treatment in comparison to the non-cryopreserved control, we performed the GO analysis for each comparison separately (CW + vs CW−, RB + vs CW− and D + vs CW−) and the results were visualised by a heatmap (Fig. 5). This approach confirmed the most common up- and down-regulated processes shared by all cryopreserved samples, independent of post-cryopreservation light conditions, but also demonstrated the effect of the different light qualities on potato gene expression, in general (Figs. 4, 5). In CW+, the number of GO terms related to stress and defense responses was higher than in RB + and D+. GO terms ‘response to chitin’, ‘cinnamic acid biosynthetic process’, and ‘response to wounding’ were enriched only in CW + and D + but not in RB+.
Heatmap comparison of top enriched GO terms related to biological processes (BP) based on up-regulated DEGs in cryopreserved vs. non-cryopreserved shoot tips exposed to different light conditions (CW + vs. CW−, RB + vs. CW− and D + vs. CW−). The stronger the red colour, the higher the enrichment score (-log10 q-value). Grey – not enriched.
Light effect: CW + and RB + in comparison with D+
Number of DEGs
The 16-h light-exposure had a significant effect on transcript accumulation during post-cryopreservation recovery as 3,259 genes were differentially expressed in CW + and RB + in comparison to D+. In CW+, the number of DEGs was double compared to DEGs between RB + and D+. Specifically, the number of DEGs in CW + was 2,882 with 1,915 up- and 967 down-regulated genes, while in RB+, the number of DEGs was 1,473 with 1,020 up- and 453 down-regulated genes (Fig. 3B). The majority of genes were specifically up-regulated in CW+ (1,051) while only 156 transcripts were specific to RB + compared to D+. Even the number of commonly up-regulated genes (864) was lower than the number of genes induced specifically by CW+. Similarly, among the down-regulated genes, the highest number of DEGs was specific to CW + with 735 DEGs, while both light conditions shared 232 DEGs, and 221 were specifically down-regulated in RB+ (Fig. 3B).
GO analysis
Among up-regulated DEGs in both CW + and RB+, numerous GO terms related to biological processes (BP) were activated. Concurrently, no significant GO enrichment was observed among the down-regulated DEGs. The majority of the activated GO terms common to both CW + and RB + were related to photosynthesis and abiotic stimuli (Fig. 6A, B). The most highly enriched photosynthesis-related GO terms were ‘photosynthesis’, ‘photosynthesis, light reaction’, ‘generation of precursor metabolites and energy’, ‘regulation of photosynthesis’ and ‘photosynthesis, light harvesting’. The terms related to light included ‘response to light stimulus’, ‘response to radiation’, ‘response to light intensity’, ‘response to far-red light’, ‘response to blue light’ and ‘response to red light’ (Fig. 6B). In addition, ‘response to high light intensity’, ‘photorespiration’, ‘photoinhibition’ and ‘photoprotection’ were activated in both CW + and RB+ (Fig. 6C). Several GO terms related to photosystem II assembly and repair were also upregulated in both light types, but in CW+, more terms with higher enrichment scores were identified (Fig. 6D). Activation of ‘flavonoid metabolic process’ (Fig. 6E) was identified in both light types compared to darkness. Several stress-related terms were more abundant in CW + than in RB+ (Fig. 6F). In general, the processes shared by both light conditions had higher enrichment scores in CW + than in RB+.
Up-regulated pathways in CW + and RB + in comparison to D+. The higher the number (-log10 q-value), the stronger the GO enrichment. The terms are grouped related to their function. (A) Photosynthesis. (B) Response to light stimulus. (C) Response to high light intensity. (D) Assembly and repair. (E) Secondary metabolite-/ pigment-related. (F) Defense response.
In addition, several stress and defense-related GO terms, such as ‘response to stress’ and ‘defense response to other organism’, were enriched only in CW + and not in RB + in comparison to D+. (Fig. 6F). There were no GO terms specifically up-regulated by RB + only.
Light quality effect: RB + compared to CW+
Number of DEGs
The recovering shoot tips in RB + had 499 DEGs compared to CW + with 155 up- and 344 down-regulated genes. However, the expression differences for most genes remained below 2-fold, as only 18 genes were up- and 114 were down-regulated more than 2-fold (lfc > + 1 or < -1), whereas 73 genes were up- and 280 down-regulated at the 1.5-fold level (lfc 0.6) (Supplementary Table S4).
GO analysis
GO analysis revealed that several processes related to photosynthesis and responses to stimuli were enriched among the down-regulated genes in RB + in comparison to CW+. The most enriched photosynthesis-related terms among DEGs with 1.5-fold down-regulation were ‘response to abiotic stimulus’, ‘response to stimulus’, ‘response to cytokinin’, ‘photosynthesis’ and ‘response to chemical’ (Fig. 7A). In addition, several responses to both abiotic and biotic stimuli were down-regulated in RB + compared to CW+. These were ‘response to abiotic stimulus’, ‘response to temperature stimulus’, ‘response to stress’, and ‘response to chemical’ including ‘response to cold’, ‘response to heat’, ‘defense response to other organism’, and ‘defense response to bacterium’ (Fig. 7B).
GO enrichment analysis of down-regulated DEGs in RB + in comparison to CW+. (A) The top 20 enriched GO terms related to biological processes (BP). Terms related to ‘photosynthesis’ are shown in green and ‘response to stimulus’ in red. (B) A detailed network of GO terms related to ‘response to stimulus’. Only significantly enriched GO terms (padj < 0.05) are highlighted. The size of the node is relative to the number of genes related to the GO term.
GO analysis of RB+-induced DEGs were enriched in terms related to developmental processes and hormones. Fourteen of the 34 enriched terms were related to development and ten to hormones. The top enriched terms were ‘regionalization’, ‘regulation of hormone levels’, ‘cytokinin metabolic process’, ‘pattern specification process’ and ‘xylem and phloem pattern formation’ (Supplementary Table S7).
Verification of the RNA-Seq results
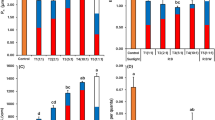
The RNA-Seq results were verified by a qPCR analysis of eight genes with significant expression differences between the treatments in the transcriptome analysis. The studied genes encoded e.g. stress and cell wall-related proteins and transcription factors specific to RB + treatment. The expression profiles resulting from RNA-Seq and qPCR were highly similar (Supplementary Fig. S1) indicating the reliability of the results in general.
Discussion
Light effect
Potato exhibits significant genetic diversity3, largely due to the high number of varieties. However, due to its highly heterozygous nature, its sexually produced seeds are not true-to-type and as a result, these varieties can only be preseved by vegetative propagation. Cryopreservation offers an ideal solution, allowing the stable, long-term storage of disease-free material without the need for continuous subculturing39.
The present study emphasizes the importance of light conditions during post-cryopreservation recovery of potato shoot tips in vitro. Both light conditions had an extensive effect on transcript accumulation during potato shoot tip recovery, as 3,259 genes were differentially expressed between RB+, CW + and D+. Interestingly, more genes (1,051) were induced by CW + alone than shared by CW + and RB+ (864), and only 156 genes were specifically up-regulated in RB + shoot tips of potato.
In general, during initial post-cryopreservation recovery, thawed shoot tips are placed into diminished light or darkness from a few days to weeks to reduce stress and the generation of reactive oxygen species (ROS) caused by light15,16,40,41. The present study suggests that some of the light-associated stress could depend on the light spectrum. Among the top down-regulated genes in RB + compared to CW + are several associated with stress response. For example, a chloroplast small heat shock protein (PGSC0003DMG400011628, log₂FC = -1.95) and heat shock protein 83 (PGSC0003DMG400005573, log₂FC = -1.73) are chaperones involved in protecting proteins during extreme temperature and oxidative stress42,43. Snakin-2 (PGSC0003DMG40001598, log₂FC = -2.52), an antimicrobial peptide from potato is induced by wounding and pathogen infection44 and plays a role in early defense signaling and wounding responses45. The downregulation of 24 K germin (PGSC0003DMG400031010, log₂FC = -3.18 ) which is involved in various abiotic and biotic stress responses as well as development and defense46 further reflects reduced activation of protective mechanisms. In addition, carbonic anhydrase (PGSC0003DMG40000493, log₂FC = -2.83) which functions in CO₂ conversion and pH homeostasis but is also known to respond to high illumination, osmotic and biotic stress47,48,49 was also repressed under RB + conditions. Together, these findings suggest that red-blue light may provide a less stressful environment for shoot tip recovery resulting in increased survival and regrowth following cryopreservation.
Light intensity in both light treatments was 40 µmol m− 2s− 1, and the only difference was the proportion of different wavelengths. CW had twice as much blue light and half as much red light as RB. In addition, in CW, approximately half of the given light contained green and yellow wavelengths. Takahashi et al.41. demonstrated that yellow wavelengths cause photodamage to photosystem II, which may explain the enrichment of photosystem II repair-related GO terms in CW+. One of the advantages of using LED-based illumination is their much lower heat-emission compared to cool white fluorescent tubes. Despite the fact that both light treatments were carried out in a climate-controlled growth chamber, it is possible that some of the elevated photoinhibition and photosystem repair processes were due to heat emission by cool white fluorescent tubes.
There was a relatively low number of specifically induced genes in RB+, and they were related to photomorphogenetic processes affecting organ and tissue development. The positive effect of RB LEDs on potato shoot tip recovery has been observed in our previous study where an identical RB light treatment promoted the post-cryopreservation regeneration of five potato cultivars, resulting in double regeneration percentages compared to CW17. A prolonged incubation in darkness may alter shoot recovery, and therefore, in general, a transfer to light is beneficial for the recovery of high-quality plants14. On the other hand, red light has been shown to strongly inhibit potato sprout elongation50. Chen et al. (2018) found that combined red-blue light, specifically at a red-to-blue ratio of 3:1, improved overall growth and microtuberization of in vitro potato plantlets compared to white, red, or blue light alone51. Similarly, Chen et al. (2021) observed that monochromatic red light promoted stem elongation, weaker roots, reduced photosynthetic activity, but increased carbohydrate accumulation on potato plantlets grown in vitro. In contrast, monochromatic blue light caused dwarfing, stronger roots, increased protein accumulation, and lower chlorophyll content without reduced photosynthetic activity. Red-blue light, on the other hand, produced the most balanced and favorable growth effects52.
In other plant species, the combination of red and blue light has been shown to affect, e.g., growth and medicinal component accumulation of Dendrobium denneanum53, root biomass and flavonoid content of Scutellaria baicalensis54. However, the effect of modified light conditions on cryopreservation success is still not well studied. Optimization of the light spectral properties to decrease light-related stress and to gain a maximum benefit through specific wavelengths that drive photomorphogenesis could greatly benefit the cryopreservation efficiency of plant germplasm. In the current study, two genes which were specifically strongly upregulated in RB + encoded bHLH1 transcription factors (PGSC0003DMG401007298 and PGSC0003DMG400009058). These genes represent interesting candidates responsible for controlling responses of shoot tips to light spectra during post-cryopreservation recovery. According to a BLAST search in NCBI (National Center for Biotechnology Information), the former gene encodes a homolog of UPBEAT1 that is known to regulate the expression of certain peroxidases55. Therefore, it is possible that UPBEAT1 is contributing to ROS homeostasis after cryopreservation. However, to verify this hypothesis, gene functional studies would be needed in the future. Previously, it has been shown that transcription factor WRKY22, which affects osmotic stress, is required during cryo-stress acclimation in Arabidopsis shoot tips22.
Cryopreservation effect
Overall, cryopreservation had an extensive effect on the gene expression of potato, as 5,767 DEGs common to all cryopreserved samples were up- or down-regulated in comparison to non-cryopreserved shoot tips. Similarly, a high number of genes were differentially expressed in Arabidopsis seedlings cryopreserved by the droplet vitrification method in comparison to non-cryopreserved controls20. In the study by Ren et al.20, 10,049 genes were differentially expressed between non-cryopreserved controls and cryopreserved seedlings, which were recovered for 24 h post cryopreservation under controlled growth conditions (8/16-h light/dark cycle with 150 µmol m− 2s− 1 light). These studies illustrate the extensive and complex effect of cryopreservation on the transcriptome in both Arabidopsis and potato. Taking into account the differences in genome size between potato (844 Mb, 39,031 genes28, and Arabidopsis (135 Mbp, 27,655 genes, Ensembl Plants Database, https://plants.ensembl.org/Arabidopsis_thaliana/Info/Annotation), the proportion of differentially expressed genes was lower in potato compared to Arabidopsis. This can be due to the different experimental material, as in our study in vitro shoot tips were used, while Ren et al.20 used 60-h germinated seedlings. In addition, the cryopreservation protocol, recovery phase, and light conditions were different in the study by Ren et al.20 from the present one. The plant vitrification protocol used by Ren et al.20 consists of many steps (dehydration, cryoprotection with PVS2 or PVS3, cooling in liquid nitrogen, rewarming, and removal of cryoprotectant) and aims to the removal of all freezable water from the cell solutes so that they can freeze without the formation of water crystals. The steps in the vitrification protocol are known to induce several abiotic stress responses, and therefore, to prepare the plant to better cope with cooling to ultra-low temperatures.
In Arabidopsis seedlings, several abiotic stress responses were activated, resulting in the up-regulation of heat- and cold-stress-responsive genes during osmo- and cryoprotection, while the cooling-rewarming phase caused the most drastic changes20. In the study by Gross et al.21, cryoprotectant treatment during the vitrification protocol caused the main transcriptional changes in Arabidopsis shoot tips excised from 7-day-old seedlings. In that study, the changes in gene expression caused by cryoprotection exceeded those induced by the liquid nitrogen cooling phase21. Both studies reflect the cryoprotectant-related stress responses in the vitrification protocol and highlight the benefit of using shoot tips of young in vitro plants.
The DMSO-droplet method used in this study was selected for its prior application in related studies investigating light quality effects on cryopreservation17,25. This method involves fewer steps than vitrification-based protocols, requiring only cryoprotection with 10% DMSO, cooling, rewarming, and cryoprotectant removal. However, despite these operational advantages, the DMSO-droplet method generally results in lower regeneration percentages compared to droplet-vitrification methods based on PVS2 or PVS312,56. It has been shown that, with DMSO-droplet method some freezable water remains in the shoot tips which may affect post-cryopreservation recovery47 and participate in the extensive cryopreservation effect on transcript abundance as observed in the present study. The superior performance of vitrification-based methods has led institutions such as the International Potato Center (CIP) in Peru and the Leibniz Institute for Plant Genetics and Crop Plant Research (IPK) in Germany to adopt PVS2 or PVS3 droplet-vitrification as their standard protocols12.
In our study, cryopreservation induced a cascade of stress and defense responses related to biotic stimuli, which may be related to the quantitative defense responses caused by the overall cryopreservation process. It is well-known that several key regulatory factors in signalling pathways respond to both biotic and abiotic stresses. Consequently, the extreme conditions of cryopreservation may have activated these shared pathways leading to the significant biotic stress related GO term enrichment58. However, previous cryopreservation-related studies have mainly reported abiotic stress factors such as cold, heat, water deprivation and oxidative stress20,21. Therefore alternatively, the defense responses observed in the current study may be induced by endophytic symbionts residing in potato shoot tips. Latent endophytic bacteria and fungi are commonly present in in vitro plant tissues with no sign of contamination until severe stress59,60. Endophyte appearance is a common post-cryopreservation drawback of many clonally propagated species12,25,61,62,63. In the present study, no signs of contamination were observed, but responses to biotic stimuli were activated in all cryopreservation treatments. In some light conditions, biotic responses were more pronounced, but the effect was inconsistent. For example, the GO terms ‘response to biotic stimulus’, ‘defense response’, ‘response to external biotic stimulus’, and ‘response to other organism’ were enriched in all cryopreserved samples, but in CW + and D+, the effect was more pronounced. Moreover, ‘response to chitin’ and ‘chitin metabolic process’, ‘chitin catabolic process’ were enriched only in CW + and D + but not in RB+. This could be due to the non-homogenous distribution of endophytes in donor plants, causing variability in endophyte occurrence in different batches of samples, as has been observed during mint cryopreservation62. Overall, in the present study, the analysis of light quality effect during post-cryopreservation recovery indicates that light conditions play a role in defense responses. However, since the different light spectra were not tested on non-cryopreserved controls and the study included only a single cultivar, the interpretation and generalization of the results are limited.
In RB+, the ‘response to biotic stimulus’, ‘defense response to bacterium’ and ‘defense response to fungus’ were down-regulated in comparison to CW+. Light conditions are known to regulate plant defense mechanisms, but also to influence bacterial and fungal physiological responses. For example, red light has been shown to inhibit mycelial growth of the symbiotic fungus, Trichoderma atroviridae, while normal growth of the fungus was observed under blue and white light64. Similarly, for Colletotrichum acutatum, a fungal pathogen, higher fungal growth was observed under blue and white wavelengths as well as under dark compared to red light65. Moreover, white and red light inhibit the mobility, while blue light can increase the infection capacity of the tomato pathogen Pseudomonas syringae66. Therefore, post-cryopreservation light quality may affect the responses of endophytic organisms residing in plant tissues.
In addition to procedural and environmental variables, genotypic differences likely contribute to the observed variation in transcriptomic responses following cryopreservation. The regeneration of ‘Agrie Dzeltenie’ in previous studies was 26% and 21% under CW treatment17,25. Therefore, the transcriptomic responses observed in this study may, in part, reflect genotype-specific reactions of ‘Agrie Dzeltenie’, and future studies involving multiple cultivars are necessary to determine the broader applicability of these findings. While this study focuses on a single cultivar using the DMSO-droplet method, future investigations could benefit from comparing the effect of light spectral quality across a broader range of genotypes, other vegetatively propagated crops and cryopreservation protocols. Specifically, applying light treatments with different light quality and intensity combinations, in conjunction with high-performance vitrification methods could help optimize recovery for recalcitrant cultivars. Additionally, understanding the transcriptional responses of plant material preserved with DMSO-based (e.g., PVS2) versus DMSO-free (e.g., PVS3) methods may provide further insights into the stress responses and recovery mechanisms, which could be influenced by both the chemical composition of the cryoprotectants and the post-thaw light environment.
Conclusions
Potato cryopreservation has progressed markedly over the past decades and is now widely used for long-term preservation of both improved cultivars and landraces in major genebanks worldwide. For example, the International Potato Center (CIP), which houses more than 4100 accessions from seven cultivated species have been successfully cryopreserved with minimum recovery rate over 20%. Through continuous protocol refinement, CIP has recently raised the average full-plant recovery rate from 58 to 73%. Despite these advances, some accessions remain recalcitrant and exhibit poor recovery. Therefore, further improvements to the protocols are still needed. Many large genebanks have cryopreserved numerous accessions decades ago, prior to the latest protocols, highlighting the importance of post-cryopreservation enhancements. In particular, optimizing post-thaw conditions—such as oxidative stress management, growth regulator application, and environmental parameters—could further improve the recovery of cryobanked material and help achieve higher post-cryopreservation success rates4,11.
This study provides the first comprehensive transcriptome analysis of cryopreserved potato, thereby revealing unique transcriptional changes related to cryopreservation of vegetatively propagated species. Our findings show that cryopreservation triggers complex effects on gene expression, including activation of defense responses to both biotic stimuli and oxidative stress. Specifically, we observed that varying light spectral quality—particularly red–blue (RB) illumination—significantly influences transcript accumulation during shoot tip recovery.
In previous work, we have shown that RB light is beneficial for the regeneration of potato after cryopreservation. In line with this, the shoot tips recovering in RB + had fewer stress-related transcripts in comparison to CW + treatment in the current study. In addition, we identified transcription factors specific to RB + treatment that represent interesting candidates for controlling responses of potato shoot tips to light spectra during post-cryopreservation recovery. Interestingly, among all cryopreserved samples, enriched GO terms included various defense and stress responses to biotic stimuli, such as ‘response to other organism’, which may relate to endophytes commonly detected in clonally propagated materials.
The obtained knowledge can be used to improve the preservation of plant genetic resources for food and agriculture. Therefore, optimization of light spectral properties in order to decrease light-induced stress, and on the other hand, to gain the maximum benefit through specific wavelengths that drive photomorphogenesis, may greatly benefit the cryopreservation efficiency of plant germplasm. These findings may help to further understand the critical steps needed for successful cryopreservation of clonal crops and therefore improve the cryopreservation efficiency of plant germplasm in the future.
Data availability
The RNA-seq data generated in this study is publicly available from GenBank’s GEO ( https://www.ncbi.nlm.nih.gov ) under accession number GSE262492. All other datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- ADK:
-
Adenosine kinase
- APRT:
-
Adenine phosphoribosyltransferase
- BP:
-
Biological processes
- CW:
-
Cool white fluorescent tubes
- D:
-
Dark
- DEG:
-
Differentially expressed genes
- GA3 :
-
Gibberellic acid
- GO:
-
Gene Ontology
- IAA:
-
Indole acetic acid
- LED:
-
Light emitting diodes
- LN:
-
Liquid nitrogen
- MS:
-
Murashige and Skoog
- PCA:
-
Principal component analysis
- qPCR:
-
Quantitative real-time PCR
- RB:
-
90% red and 10% of blue light
- ROS:
-
Reactive oxygen species
- RT:
-
Room temperature
References
Niino, T. & Arizaga, M. V. Cryopreservation for preservation of potato genetic resources. Breed Sci. 65, 41–52. https://doi.org/10.1270/jsbbs.65.41 (2015).
Pence, V.C. Evaluating costs for the in vitro propagation and preservation of endangered plants. Vitro Cell. Dev. Biol. Plant. 47, 176–187. https://doi.org/10.1007/s11627-010-9323-6 (2011).
Panta, A. et al. Improved cryopreservation method for the long-term conservation of the world potato germplasm collection. Plant. Cell. Tissue Organ. Cult. 120, 117–125. https://doi.org/10.1007/s11240-014-0585-2 (2015).
Vollmer, R. et al. The potato cryobank at the international potato center (CIP): a model for long term conservation of clonal plant genetic resources collections of the future. CryoLetters 37, 318–329 (2016).
Mix-Wagner, G., Schumacher, H. M. & Cross, R. J. Recovery of potato apices after several years of storage in liquid nitrogen. CryoLetters 24, 33–41 (2003).
Kim, H. H. et al. Cryopreservation of potato cultivated varieties and wild species: Critical factors in droplet vitrification. CryoLetters 27, 223–234 (2006).
Vollmer, R., Villagaray, R., Castro, M., Anglin, N. L. & Ellis, D. Cryopreserved potato shoot tips showed genotype-specific response to sucrose concentration in rewarming solution (RS). Plant. Cell. Tissue Organ. Cult. 136, 353–363. https://doi.org/10.1007/s11240-018-1520-8 (2019).
Folgado, R. et al. Changes in sugar content and proteome of potato in response to cold and dehydration stress and their implications for cryopreservation. J. Proteom. 98, 99–111. https://doi.org/10.1016/j.jprot.2013.11.027 (2014).
Kaczmarczyk, A., Rutten, T., Melzer, M. & E.R.J. Keller. Ultrastructural changes associated with cryopreservation of potato (Solanum tuberosum L.) shoot tips. CryoLetters 29, 145–156 (2008).
Bettoni, J. C., Bonnart, R. & Volk, G. M. Challenges in implementing plant shoot tip cryopreservation technologies. Plant. Cell. Tissue Organ. Cult. 144, 21–34. https://doi.org/10.1007/s11240-020-01846-x (2021).
Popova, E., Kulichenko, I. & Haeng-Hoon, K. Critical role of regrowth conditions in post-cryopreservation of in vitro plant germplasm. Biol 12, 542. https://doi.org/10.3390/biology12040542 (2023).
Köpnick, C. Changes of soluble sugars and ATP content during DMSO droplet freezing and PVS3 droplet vitrification of potato shoot tips. Cryobiol 85, 79–86. https://doi.org/10.1016/j.cryobiol.2018.09.005 (2018).
Seabrook, J. E. A. Light effects on the growth and morphogenesis of potato (Solanum tuberosum) in vitro: A review. Amer J Potato Res. 82, 353–367. https://doi.org/10.1007/BF02871966 (2005).
Benson, E. E., Harding, K. & Smith, K. Variation in recovery of cryopreserved shoot-tips of Solanum tuberosum exposed to different pre- and post-freeze light regimes. CryoLetters 10, 323–344 (1989).
Gonzalez-Arnao, M. T. & Engelmann, F. Cryopreservation of plant germplasm using the encapsulation-dehydration technique: review and case study on sugarcane. CryoLetters 27, 155–168 (2006).
Zhao, Y., Wu, Y., Chang, Y. & Reed, B. M. Cryopreservation of fruit and ornamental trees. In Plant Cryopreservation: A Practical Guide (ed. Reed, B.M.) 387–420 (Springer Science and Business Media, 2008); https://doi.org/10.1007/978-0-387-72276-4
Edesi, J., Kotkas, K., Pirttilä, A. M. & Häggman, H. Does light spectral quality affect survival and regeneration of potato (Solanum tuberosum L.) shoot tips after cryopreservation? Plant. Cell. Tissue Organ. Cult. 119, 599–607. https://doi.org/10.1007/s11240-014-0559-4 (2014).
Harding, K., Benson, E. E. & Smith, H. The effects of pre-freeze in vitro culture period on the recovery of cryopreserved shoot-tips of solanum tuberosum. CryoLetters 12, 17–22 (1991).
Bukhov, N. G., Popova, E. V. & Popov, A. S. Photochemical activities of two photosystems in Bratonia Orchid protocorms cryopreserved by vitrification method. Russ. J. Plant. Physiol.. 53, 793–799. https://doi.org/10.1134/S1021443706060100 (2006).
Ren, L. et al. Transcriptomic profiling revealed the regulatory mechanism of Arabidopsis seedlings response to oxidative stress from cryopreservation. Plant. Cell. Rep. 34, 2161–2178. https://doi.org/10.1007/s00299-015-1859-9 (2015).
Gross, B. L., Henk, A. D., Bonnart, R. & Volk, G. M. Changes in transcript expression patterns as a result of cryoprotectant treatment and liquid nitrogen exposure in Arabidopsis shoot tips. Plant. Cell. Rep. 36, 459–470. https://doi.org/10.1007/s00299-016-2095-7 (2017).
Stock, J. et al. The transcription factor WRKY22 is required during cryo-stress acclimation in Arabidopsis shoot tips. J Exp Bot. 71, 4993–5009. https://doi.org/10.1093/jxb/eraa224 (2020).
Quijada-Rivera, M. et al. Transcriptome assessment in red Globe grapevine zygotic embryos during the cooling and warming phase of the cryopreservation procedure. Cryobiology 110, 56–68. https://doi.org/10.1016/j.cryobiol.2022.12.016 (2023).
Htwe, C. S. S., Subramani, R., Pathania, P. & Agrawal, A. Transcriptome profiling during sequential stages of cryopreservation in banana (Musa AAA cv Borjahaji) shoot meristem. Plants 12, 1165; (2023). https://doi.org/10.3390/plants12051165
Edesi, J., Pirttilä, A. M. & Häggman, H. Modified light spectral conditions prior to cryopreservation alter growth characteristics and cryopreservation success of potato (Solanum tuberosum L.) shoot tips in vitro. Plant. Cell. Tissue Organ. Cult. 128, 409–421. https://doi.org/10.1007/s11240-016-1119-x (2017).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 15, 473–497 (1962).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. (2010). https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
The Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. https://doi.org/10.1038/nature10158 (2011).
Sharma, S. K. et al. Construction of reference chromosome-scale pseudomolecules for potato: integrating the potato genome with genetic and physical maps. G3-Genes Genom Genet. 3, 2031–2047. https://doi.org/10.1534/g3.113.007153 (2013).
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: An efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. https://doi.org/10.1093/bioinformatics/btt656 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated Estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106. https://doi.org/10.1186/gb-2010-11-10-r106 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 57, 289–300 (1995).
Yao, F., Coquery, J. & Le Cao, K. A. Independent principal component analysis for biologically meaningful dimension reduction of large biological data sets. BMC Bioinform. 13, 24. https://doi.org/10.1186/1471-2105-13-24 (2012).
Jin, J. et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. https://doi.org/10.1093/nar/gkw982 (2017).
Kolde, R. Pheatmap: pretty heatmaps. R package version 1.0.12, 790 ; (2018). https://cran.r-project.org/web/packages/pheatmap/index.html
Maere, S., Heymans, K. & Kuiper, M. BiNGO: A cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinform 21, 3448–3449. https://doi.org/10.1093/bioinformatics/bti551 (2005).
Bustin, S. A. et al. The MIQE guidelines: Minimum information for publication of quantitative Real-Time PCR experiments. Clin. Chem. 55, 611–622. https://doi.org/10.1373/clinchem.2008.112797 (2009).
Kaczmarczyk, A., Rokka, V. M. & Keller, E. R. J. Potato shoot tip cryopreservation. A review. Potato Res. 54, 45–79. https://doi.org/10.1007/s11540-010-9169-7 (2011).
Domingues, N., Matos, A., da Silva, J. & Cartaxana, P. Response of the diatom Phaeodactylum tricornutum to photooxidative stress resulting from high light exposure. PLoS ONE. 7, e38162. https://doi.org/10.1371/journal.pone.0038162 (2012).
Takahashi, S. et al. The solar action spectrum of photosystem II damage. Plant. Physiol. 153, 988–993. https://doi.org/10.1104/pp.110.155747 (2010).
Lee, G., Roseman, A., Saibil, H. & Vierling, E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 16, 659–671. https://doi.org/10.1093/emboj/16.3.659 (1997).
Wang, W., Vinocur, B. & Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 218, 1–14. https://doi.org/10.1007/s00425-003-1105-5 (2003).
Berrocal-Lobo, M. et al. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant. Physiol. 128, 951–961. https://doi.org/10.1104/pp.010685 (2002).
Iqbal, A. & Khan, R. Snakins: Antimicrobial potential and prospects of genetic engineering for enhanced disease resistance in plants. Mol. Biol. Rep. 50, 8683–8690. https://doi.org/10.1007/s11033-023-08734-5 (2023).
Bernier, F. & Berna, A. Germins and germin-like proteins: Plant do-all proteins. But what do they do exactly? Plant. Physiol. Biochem. 39, 545–554. https://doi.org/10.1016/S0981-9428(01)01285-2 (2001).
Rudenko, N. et al. Carbonic anhydrases in photosynthetic cells of higher plants. Biochem (Moscow). 80, 74–684. https://doi.org/10.1134/S0006297915060048 (2015).
DiMario, R. et al. Plant carbonic anhydrases: Structures, locations, evolution, and physiological roles. Mol. Plant. 10, 30–46. https://doi.org/10.1016/j.molp.2016.09.001 (2017).
Rudenko, N. et al. Role of plant carbonic anhydrases under stress conditions. Plant. Stress Physiol. https://doi.org/10.5772/intechopen.91971 (2020). IntechOpen.
Mølmann, J. A. B. & Johansen, T. J. Sprout growth Inhibition and photomorphogenic development of potato seed tubers (Solanum tuberosum L.) under different LED light colours. Potato Res. 63, 199–215. https://doi.org/10.1007/s11540-019-09435-y (2020).
Chen, L. et al. Effects of red and blue leds on in vitro growth and microtuberization of potato single-node cuttings. Front. Agr Sci. Eng. 5, 197–205. https://doi.org/10.15302/J-FASE-2018224 (2018).
Chen, L. et al. Transcriptome analysis reveals effects of red and blue light-emitting diodes (LEDs) on the growth, chlorophyll fluorescence and endogenous plant hormones of potato (Solanum tuberosum L.) plantlets cultured in vitro. J. Integr. Agric. 20, 2914–2931. https://doi.org/10.1016/S2095-3119(20)63393-7 (2021).
Fan, Y. et al. Transcriptome analysis reveals the effects of red and blue light on the physiological and medicinal components of Dendrobium denneanum. Ind. Crop Prod. 180, 114798. https://doi.org/10.1016/j.indcrop.2022.114798 (2022).
Zhang, T. et al. Transcriptome and targeted metabolome analysis revealed the effects of combined red and blue light on the growth and secondary metabolism of scutellaria baicalensis Georgi. Ind. Crop Prod. 188, 115598. https://doi.org/10.1016/j.indcrop.2022.115598 (2022).
Tsukagoshi, H., Busch, W. & Benfey, P. N. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616. https://doi.org/10.1016/j.cell.2010.10.020 (2010).
Kryszczuk, A., Keller, J., Grübe, M. & Zimnoch-Guzowska, E. Cryopreservation of potato (Solanum tuberosum L.) shoot tips using vitrification and droplet method. J. Food Agric. Environ. 4, 196–200 (2006).
Keller, E. R. J. et al. Experience in large-scale cryopreservation and links to applied research for safe storage of plant germplasm. Acta Hortic. 1113, 239–250. https://doi.org/10.17660/ActaHortic.2016.1113.36 (2016).
Fujita, M., Fujita, Y. & Noutoshi, Y. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin. Plant. Biol 9, 436–442 ; https://doi.org/10.1016/j.pbi.2006.05.014
Podolich, O., Laschevskyy, V., Ovcharenko, L., Kozyrovska, N. & Pirttilä, A. M. Methylobacterium sp. resides in unculturable state in potato tissues in vitro and becomes culturable after induction by Pseudomonas fluorescens IMGB163. J. Appl. Microbiol. 106, 728–737. https://doi.org/10.1111/j.1365-2672.2008.03951.x (2009).
Podolich, O., Ardanov, P., Zaets, I., Pirttilä, A. M. & Kozyrovska, N. Reviving of the endophytic bacterial community as a putative mechanism of plant resistance. Plant. Soil. 388, 367–377. https://doi.org/10.1007/s11104-014-2235-1 (2015).
Keller, E. R. J., Senula, A., Zanke, C., Grübe, M. & Kaczmarczyk, A. Cryopreservation and in vitro culture - state of the Art as conservation strategy for genebanks. Acta Hortic. 918, 99–111. https://doi.org/10.17660/ActaHortic.2011.918.11 (2011).
Senula, A. & Keller, E. R. J. Cryopreservation of mint - routine application in a genebank, experience and problems. Acta Hortic. 908, 467–476. https://doi.org/10.17660/ActaHortic.2011.908.60 (2011).
Jenderek, M. M. & Reed, B. M. Cryopreserved storage of clonal germplasm in the USDA National plant germplasm system. Vitro Cell. Dev. Biol. – Plant. 53, 299–308. https://doi.org/10.1007/s11627-017-9828-3 (2017).
Casas-Flores, S., Rios-Momberg, M., Bibbins, M., Ponce-Noyola, P. & Herrera-Estrella, A. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiol 150, 3561–3569. https://doi.org/10.1099/mic.0.27346-0 (2004).
Yu, S., Ramkumar, G. & Lee, Y. H. Light quality influences the virulence and physiological responses of Colletotrichum acutatum causing anthracnose in pepper plants. J. Appl. Microbiol. 115, 509–516. https://doi.org/10.1111/jam.12252 (2013).
Wu, L., McGrane, R. S. & Beattie, G. A. Light regulation of swarming motility in Pseudomonas syringae integrates signaling pathways mediated by a bacteriophytochrome and a LOV protein. mBio 4 https://doi.org/10.1128/mBio.00334-13 (2013). 10.1128/mbio.00334 – 13.
Acknowledgements
The authors would like to thank Dr. Leila Eshraghi for the initial bioinformatic analyses, Matti Rauman for the light treatment setup and spectral measurements, and laboratory technicians Tarja Törmänen and Taina Uusitalo for their help with the laboratory work. The RNA-Sequencing was done at the Finnish Functional Genomics Centre of the Turku Centre for Biotechnology.
Funding
This work was funded by the national scholarship programme Kristjan Jaak (to J.E.), which is funded and managed by the Archimedes foundation in collaboration with the Estonian Ministry of Education and Research. Additional funding to J.E. was provided by the Finnish Cultural Foundation, North Ostrobothnia Regional Fund and Oulun läänin talousseuran maataloussäätiö.
Author information
Authors and Affiliations
Contributions
J.E., A.M.P. and H.H. designed the study. J.E. performed the cryopreservation experiments and the RNAseq analyses. S.J.-L. and H.M.S. performed the qPCR analyses. J.E. wrote the manuscript which was then reviewed by all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Edesi, J., Jokipii-Lukkari, S., Salo, H.M. et al. Light spectrum modulates stress and defense gene expression in potato (Solanum tuberosum L.) shoot tips during post-cryopreservation recovery. Sci Rep 15, 34570 (2025). https://doi.org/10.1038/s41598-025-18012-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-18012-y