Abstract
Epstein–Barr virus (EBV) associated gastric cancer, accounting for ~ 9% of all gastric cancers, has unique pathologic, genomic, and clinical features and is linked to a better prognosis. Therefore, we aim to develop and validate a robust deep learning-based classifier to detect EBV positivity for both biopsy and surgical resection specimens in an accurate and reliable manner. We propose a two-stage stain normalization-based robust artificial intelligence classifier for EBV-gastric cancer positivity detection (EBV-TRACER). The results were aggregated to compute three prediction scores including EBV positive cancer-to-tissue ratio, EBV positive cancer-to-tumor ratio, and EBV positive cancer size. To assess the classification performance, we measured the area under the receiver operating curve for detecting EBV status. We also conducted nuclei segmentation and classification to quantitatively analyze and visualize the relationship between the EBV status and lymphocyte features. Our analysis included 2684 gastric specimens with diagnostic clinical reports for gastric cancer collected from January 1, 2011 to December 31, 2023. In the internal validation cohorts, EBV-TRACER yielded AUCs ranging from 0.6596 (95% CI: 0.5359–0.7697) to 0.8414 (95% CI: 0.7267–0.9263) using the three scores. In the external validation cohort, AUCs of 0.7644 (95% CI: 0.6829–0.8399), 0.7652 (95% CI: 0.6850–0.8398), and 0.7221 (95% CI: 0.6134–0.8178) were obtained for the three scores, respectively. Overall, EBV-TRACER significantly outperforms models without stain normalization and those using conventional stain normalization. It could serve as a promising tool for improving the efficiency and accuracy of decision making in gastric cancer diagnosis, supporting more effective treatment planning and strategy development.
Similar content being viewed by others
Introduction
Gastric cancer is a prevalent disease worldwide, reported as the 5th most common cancer and 4th leading cause of death in 20201. Traditionally, gastric cancer was histologically classified into several sub-types such as papillary, tubular, mucinous, and poorly cohesive carcinoma sub-types2. Such histological classification was clinically ineffective in diagnosing and treating patients with gastric cancer. Based on the genomic and proteomic analysis, gastric cancer can be classified into four major molecular subtypes, including Epstein–Barr virus (EBV) associated subtype, microsatellite instable (MSI) subtype, genomically stable (GS) subtype, and chromosomal instability (CIN) subtype, which are related to extreme DNA hypermethylation, elevated mutation rates and hypermethylation, less distinctive genomic alterations, and marked aneuploidy and frequent focal amplification of receptor tyrosine kinases, respectively3. Such molecular sub-types appear to improve patient stratification for advanced treatment strategies, and thus better outcomes4.
EBV associated gastric cancer (EBV-GC) accounts for ~ 9% of all gastric cancers and is often found in young men and in the proximal location of the stomach with a moderate to poor degree of differentiation. EBV-GC involves distinct pathologic, genomic, and clinical characteristics such as a lower incidence of lymph node metastasis, a higher level of programmed death-ligand 1 (PD-L1) expression, and a better prognosis5. Although it has distinct histological features, including adenocarcinoma with infiltrating lymphocytes, lymphoepithelioma-like carcinoma with dense infiltration of lymphocytes (carcinoma with lymphoid stroma or medullary carcinoma), and an irregularly anastomosing tubule- or cord-like arrangement of tumor cells (lace pattern)6, definitive diagnosis still relies on detecting EBV-encoded small RNA (EBER) in gastric tissues using in situ hybridization (ISH), designated as EBER-ISH. EBV-GC is also considered as a biomarker for immune checkpoint inhibitor therapy7, making EBER-ISH test advantageous for confirming EBV-GC positivity. However, due to its high cost and low incidence, it is not routinely performed in clinics. Therefore, there is a need for alternative diagnostic methods to identify EBV-GC susceptibility using routine hematoxylin and eosin (H&E) stained slides.
In recent years, several artificial intelligence (AI) or deep learning-based algorithms have shown to be effective in predicting and analyzing molecular sub-types such as MSI8,9, generic alterations10, and patient outcomes11 in gastric cancer using H&E stained slides. A few research efforts have been made to develop AI models for the prediction of EBV-GC12,13,14,15,16. Though successful, these previous works face two major challenges. First, their generalization ability is uncertain. Most of these works used H&E stained whole-slide images (WSIs) digitized by a single slide scanner. In such cases, it is well known that variations in staining, caused by sample preparation, staining procedures, scanning devices, and etc., can lead to substantial performance degradation. A multi-center cohort study with multiple digital slide scanners demonstrated that the performance of the EBV-GC prediction significantly varies between centers, with an area under the receiver operating curve (AUC) ranging from 0.672 to 0.85915. Second, previous works primarily focused on WSIs from surgical resection specimens, not from biopsy specimens. The ability to predict EBV-GC in biopsy specimens is beneficial to patients since it enables early confirmation of EBV-GC and offers additional treatment options such as endoscopic submucosal dissection. In our previous study, we showed that an AI model can predict EBV-GC in biopsy specimens with high accuracy16; however, it has not yet been validated on the slide images from surgical resection specimens. Hence, no existing work identifies EBV-GC in both biopsy specimens and surgical resection specimens in an accurate and reliable fashion.
Herein, we propose a two-stage stain normalization-based robust AI classifier for EBV-GC detection (EBV-TRACER) that can predict EBV-GC positivity in H&E stained slides (Fig. 1). Employing multiple independent cohorts, we show that EBV-TRACER can identify the positive H&E slides for EBV-GC for both biopsy and surgical resection specimens with consistent accuracy and reliability, and thus the results are not sensitive to variations across different slides and cohorts.
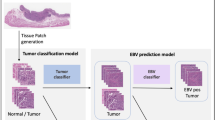
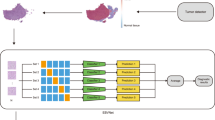
Overall pipeline of EBV-TRACER. A A slide is divided into a number of disjoint image patches, and each image patch is assigned a class label by using the corresponding EBER-ISH. These labeled patches are used to train an EBV classification model. The proposed two-stage stain normalization method conducts. B slide-level stain normalization using thumbnails of slide images and then. C patch-level stain normalization using the stained normalized thumbnail image and the original patch images. D The trained EBV classification model is applied to normalized slide images to compute three scores for slide-level classification. E A trained nuclei segmentation and classification model is used to identify lymphocytes and other cells in each patch image.
Materials and methods
Study design
In this retrospective study, we developed a robust AI framework, including a convolutional neural network and a two-stage stain normalization mechanism, that facilitates an automated and reliable prediction of EBV-GC positivity in both gastric biopsy and resection specimens. To evaluate the effectiveness of the proposed method, we collected gastric specimens from two different sources that are categorized into three cohorts, including an internal development cohort, an internal validation cohort, and an external validation cohort. The internal development and validation cohorts were obtained from Kangbuk Samsung Hospital (KSH). The external validation cohort was acquired from The Cancer Genome Atlas (TCGA) project. The internal development cohort was used to adjust the convolutional neural network to fit to the task of predicting EBV status. The internal and external validation cohorts were utilized to test the accuracy and robustness of the proposed framework. These cohorts involve gastric slides obtained from both biopsy and gastrectomy specimens and digitized using differing digital scanners.
This study was approved by the regional institutional review board of Kangbuk Samsung Hospital (IRB No. 2024-11-035-002) and was performed according to the ethical stands of the Declaration of Helsinki, as revised in 200817. This study is retrospective in nature, and the requirement for informed consent was waived by the Regional Institutional Review Board of Kangbuk Samsung Hospital in accordance with relevant guidelines and regulations.
Data collection and preparation
This study includes 2408 gastric specimens with diagnostic clinical reports for gastric cancer. Table 1 illustrates the details of this study cohorts. The internal development and validation cohorts were obtained and de-identified by KSH. The internal development cohort (KSH-DEV) includes 732 gastrectomy specimens, which were used to generate 16 tissue microarray (TMA) blocks with a total of 708 tissue cores and 24 WSIs. The internal validation cohort involves four gastric datasets. Two datasets were obtained from 285 gastric biopsy specimens. These specimens were scanned using two digital slide scanners, including Aperio AT2 (Leica Biosystem) and Aperio GT450 (Leica Biosystem) scanner, which resulted in KSH-GAT and KSH-GGT, respectively. KSH-DEV and KSH-GAT were previously utilized to develop an AI model for predicting EBV status in gastric biopsy specimens16. KSH-GAT was a part of the dataset with 286 specimens. Here, KSH-GAT contains 285 out of 286 specimens, for which we have both scans. The third dataset (KSH-GCB) includes 181 gastric biopsy slides. The last dataset (KSH-STO) involves 854 WSIs (758 gastrectomy specimens and 96 dissection specimens). All the slides were stained with hematoxylin and eosin (H&E) and reviewed by experienced pathologists (K. Kim and B. Song.). The external validation cohort comprises 347 gastric cancer slides sourced from TCGA, particularly focusing on stomach adenocarcinoma (TCGA-STAD). A variety of Aperio scanners were employed to digitize the slides.
Two-stage stain normalization
Stain normalization is an essential preprocessing step in digital pathology, aiming to reduce color distribution variability across histological slides stained with the same reagents. However, a single slide image often exceeds the capacity of standard computing environments. Consequently, previous methods such as the one proposed by Macenko et al.18 typically target small regions (or patches) for normalization, which can lead to a loss of intrinsic color distribution information, as they fail to capture the comprehensive heterogeneity of the slide. To overcome these challenges, our method involves a comprehensive analysis of color distribution across the entire slide, subsequently applying this information to individual regions.
We accomplished stain normalization of slide images in two stages: (1) slide-level stain normalization and (2) patch-level stain normalization. In the first stage, we used thumbnails of the slide images from the training dataset to fit a stain normalizer, i.e., Macenko stain normalizer, which learns the target color distribution of the training dataset. We then normalized the thumbnail of the input slide image to match the color distribution of the training dataset. In the second stage, we normalized the original input slide image (source) using the stained normalized thumbnail of the same slide image (target). For each image patch in the source slide image, we located the corresponding target patch from the target thumbnail, learned the color distribution of the target patch using Macenko stain normalizer, and normalized the image patch. Repeating the same process across all patches in the slide, we completed the two-stage stain normalization.
Image pre-processing
Following previous works16, we split each slide image and generated image patches using a sliding window method that moves a rectangular window of size 1024 × 1024 pixels with a stride of 512 pixels throughout the slide image. To avoid generating background image patches, we identified background areas by using an average pixel value (> 230) and excluded them for further analyses. The image patches were resized by half, corresponding to the spatial size of 512 × 512 pixels, and then used for training and validation purposes.
AI model for EBV-GC detection EBV-TRACER
We proposed EBV-GC positivity detection as a two-stage classification problem, in which a slide image is split into a number of image patches, the image patches are individually and independently classified by an AI model into three tissue types, and the patch-level classification results are summarized and merged together to produce the final decision, i.e., EBV-GC positive or EBV-GC negative. In a prior study16, multiple architectures were evaluated for patch-level EBV-GC classification, including ResNet, DeiT (a vision transformer), and EfficientNet. EfficientNet was shown to be the best among them. Following this study, EBV-TRACER is built based EfficientNet19 for EBV-GC classification.
EfficinetNet was trained on KSH-DEV to classify image patches into three types, including benign, EBV-GC positive, and EBV-GC negative. The trained EfficientNet was applied to each slide image, classifying all the image patches generated from the slide image. Since the sliding window method generates overlapping patches, we aggregated the classification results of these overlapping patches by computing the average probabilities for the three classes. Then, the class label with the highest probability was assigned to each pixel in the slide image. This process resulted in a tissue type prediction map for the entire slide image. Using this prediction map, we measured slide-level prediction scores for EBV-GC positivity detection using three criteria: (1) EBV+/Tissue: the ratio of image patches that are predicted as EBV-GC positive to the total number of image patches; (2) EBV+/Tumor: the ratio of image patches that are predicted as EBV-GC positive within the entire tumor region, i.e., both EBV-GC positive and EBV-GC negative image patches; (3) EBV + Size (mm2): the total area of image patches predicted as EBV-GC positive within the entire slide. For each criterion, we computed the AUC to evaluate model performance. The identical, trained EfficientNet was applied to both internal and external validation cohorts. We also note that both the stain normalization method was applied to each slide before generating image patches, if used.
Lymphocyte segmentation and quantification
EBV-GC positivity, especially for early gastric carcinoma, has been related to tumor infiltrating lymphocytes6. To quantitatively measure and analyze the relationship between EBV-GC and lymphocytes, we conducted lymphocyte segmentation by using Hover-Net20, which is considered the state-of-the-art nuclei segmentation and classification model. We trained Hover-Net on a public dataset (GLySAC)21, which contains 30,875 labeled nuclei of three categories such as lymphocytes, epithelium, and miscellaneous. In this study, we group epithelium and miscellaneous as one group. The trained Hover-Net was applied to the entire image patches from a slide image, producing a nuclei map per slide. Using the nuclei map, we quantified each slide by computing the ratios of the number and area of lymphocytes to the total number and area of all cells within a slide, respectively.
EBV-GC detection using combined EBV-GC positivity and lymphocyte features
To assess the added value of lymphocyte features, we combined three slide-level EBV-GC scores (EBV+/Tissue, EBV+/Tumor, and EBV + Size) with the ratios of lymphocyte number and area and conducted EBV-GC prediction. Specifically, given a five-dimensional feature vector (EBV+/Tissue, EBV+/Tumor, EBV + Size, ratio of lymphocyte number, and ratio of lymphocyte area), we adopted a multi-layer perceptron (MLP) classifier consisting of two hidden layers with 64 units each, ReLU activation functions, a dropout layer with a rate of 0.2, and an output layer with a sigmoid function to perform binary classification of EBV status. The model was trained using the Adam optimizer with a learning rate of 5 × 10⁻⁴ and cosine annealing learning rate scheduler over 30 epochs. To mitigate class imbalance, weighted sampling was applied based on label distribution in the training data. We conducted three separate experiments, each using a different normalization strategy, no normalization, conventional stain normalization, and the proposed two-stage stain normalization. For each normalization setting, we used the KSH-STO cohort for training, KSH-GGT and KSH-GCB for validation, and TCGA-STAD as the external test set. The same MLP architecture and training protocol were applied across all normalization conditions to enable fair comparison.
Statistical analysis
We separately applied EBV-TRACER to each cohort and measured the slide-level classification performance using AUC. For each AUC, 95% confidence interval (CI) was calculated and reported using bootstrap resampling with 2,000 repetitions following the implementation in scikit-learn. Specifically, we resampled the test set with replacement and calculated the AUC for each sample, discarding iterations in which only one class was present. The 95% confidence intervals were then estimated by computing the 2.5th and 97.5th percentiles of the bootstrapped AUC scores. For classification metrics such as accuracy, sensitivity, and specificity, we used the point estimates derived from the confusion matrix.
Results
Comparison with conventional method
The results of EBV-TRACER for the slide-level EBV-GC prediction are shown in Table 2, demonstrating that EBV-TRACER can predict EBV-GC positivity in an accurate and robust manner. Using the tumor ratio criterion (EBV+/Tumor), EBV-TRACER achieved AUCs ranged from 0.7443 (95% CI: 0.6218–0.8485; KSH-GCB) to 0.8073 (95% CI: 0.6883–0.9183; KSH-GAT) for the internal validation cohorts. It obtained an AUC of 0.7644 (95% CI: 0.6829–0.8399) for the external validation cohort (TCGA-STAD). Using other two criteria, the results were slightly varied; for instance, with the tissue ratio criterion (EBV+/Tissue), EBV-TRACER obtained AUCs for the internal validation cohorts ranging from 0.7439 (95% CI: 0.6145–0.8496; KSH-GCB) to 0.8414 (95% CI: 0.7267–0.9263; KSH-GGT) and an AUC of 0.7652 (95% CI: 0.6850–0.8398) for the external validation cohort. Similarly, in the internal validation cohorts, the size criterion (EBV + Size) had AUCs ranging from 0.6596 (95% CI: 0.5359–0.7697) in KSH-GCB to 0.7784 (95% CI: 0.6430–0.8911) in KSH-GGT and, in the external validation cohort, it had 0.7221 AUC (95% CI: 0.6134–0.8178). In a head-to-head comparison, the tissue criterion obtained the best AUC of 0.8387 (95% CI: 0.7013–0.9492) for KSH-GAT and 0.8414 (95% CI: 0.7267–0.9263) for KSH-GGT. For KSH-GCB, KSH-STO, and TCGA-STAD, the tumor ratio criterion obtained the best AUCs, suggesting its robustness to detect EBV-GC positivity.
The results of EBV-TRACER were substantially superior to the conventional approaches with and without stain normalization (Table 2). Without stain normalization, the model attained the best performance in KSH-GAT with AUCs ranging from 0.7964 (95% CI: 0.6681–0.8979) to 0.8727 (95% CI: 0.7499-0.9600) across three criteria; however, in the other internal and external validation cohorts, the results were substantially lower than those by EBV-TRACER. For example, without stain normalization, AUCs varied between 0.4514 (95% CI: 0.3318–0.5744; KSH-GCB using the size criterion) and 0.8026 (95% CI: 0.6749–0.8989; KSH-GGT using the tissue ratio criterion) in the other internal validation cohorts, whereas EBV-TRACER obtained AUCs from 0.6596 (95% CI: 0.5359–0.7697; KSH-GCB using the size criterion) to 0.8414 (95% CI: 0.7267–0.9263; KSH-GGT using the tissue criterion). In the external validation cohort, the model without stain normalization achieved 0.7295 AUC (95% CI: 0.6407–0.8164), which is considerably lower than the highest AUC of 0.7652 (95% CI: 0.6850–0.8398) obtained by EBV-TRACER. Moreover, while conventional stain normalization generally improved performance compared to the ones without stain normalization, these improvements remained lower than those achieved by EBV-TRACER, with the exception for KSH-GGT (0.8130 AUC, 95% CI: 0.7330–0.8832) and TCGA-STAD (0.7870 AUC, 95% CI: 0.6945–0.8757) using the tumor ratio criterion.
AUC values varied across cohorts, ranging from 0.6596 to 0.8414 in internal cohorts and from 0.7221 to 0.7652 in the external cohort, TCGA-STAD. Nonetheless, EBV-TRACER consistently enhanced performance compared to conventional and non-normalized settings. In contrast, the other two methods showed greater variability: No stain normalization ranged from 0.4514 to 0.8727 in internal cohorts and from 0.6849 to 0.7295 in the external cohort; Conventional stain normalization ranged from 0.5567 to 0.8402 in internal cohorts and from 0.6828 to 0.7870 in the external cohort.
For a fine-grained analysis of the prediction results, patch-level EBV-GC classification was compared in KSH-GAT and KSH-GGT (Table 3), which include identical slides but scanned using different scanners. Without stain normalization, the model obtained 0.7884 specificity, 0.9690 sensitivity, and 0.8694 F1 in KSH-GAT. However, its performance significantly decreased to 0.3694 specificity, 0.9969 sensitivity, and 0.5391 F1 in KSH-GGT, explaining the performance drop in the slide-level classification. Applying conventional stain normalization to KSH-GGT, specificity and F1 increased by 0.3608 and 0.2448, respectively, but sensitivity decreased by 0.1508. With the two-stage stain normalization (EBV-TRACER), specificity and F1 further improved by 0.0667 and 0.0813, respectively, over conventional stain normalization. Sensitivity also improved by 0.1003, but remained 0.0505 lower than the model without stain normalization. It is worth noting that EBV-TRACER was the only model to achieve comparable specificity, sensitivity, and F1 values in both KSH-GGT and KSH-GAT, diminishing the effect of different slide scanners.
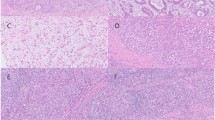
Figure 2 visualizes the prediction results by EBV-TRACER and other methods. For both biopsy slides (Fig. 2A and B) and WSIs (Fig. 2C), the prediction maps by EBV-TRACER closely matched to the ground truth maps. In contrast, the other two methods (no stain normalization and conventional stain normalization) tended to over-predict EBV-GC. In Fig. 2B and C, the close-up regions contain cells with a lace-like pattern and abundant lymphocytes, which are typical characteristics of EBV-GC. All models correctly predicted them as EBV-GC positive; however, other areas, including non-tumor regions, were also predicted as EBV-GC positive by the other two methods. As for Fig. 2A, the close-up region shows few lymphocytes and lacks lace-like patterns in tumor cell arrangement, which is suggestive of EBV-GC negativity. This region was correctly classified as EBV-GC negative by EBV-TRACER only.
Visualization of the predictive results by EBV-TRACER and other methods. Patch-level classification results for benign, EBV-GC negative, and EBV-GC positive are displayed as color maps per slide for ground truth, no stain normalization, conventional stain normalization, and EBV-TRACER. The lymphocyte number ratio is presented as a heatmap. For each slide, quantitative measures are provided, including the ratios of EBV-GC positive areas to total tumor and tissue areas and the size of EBV-GC positive areas, and close-up views of the corresponding rectangular regions. In the lower-left corner of each slide, a black scale bar represents 2 mm.
Comparison with deep learning-based stain normalization methods
The performance of EBV-TRACER was further compared against two deep learning-based stain normalization methods, including StainGAN22 and StainNet23, on the external validation cohort, TCGA-STAD. StainGAN was trained using unpaired slides from KSH-DEV and TCGA-STAD, and then used to normalize the entire TCGA-STAD cohort. Subsequently, StainNet was trained in a supervised manner using the original TCGA-STAD slides as inputs and the stained normalized slides processed by StainGAN as pseudo-ground truth. All normalized and original slides were processed using a pre-trained EfficientNet-based EBV classifier, enabling a fair performance comparison across different stain normalization methods.
The classification results are summarized in Table 4. Using the EBV+/tumor ratio criterion, EBV-TRACER achieved an AUC of 0.7644 (95% CI: 0.6829–0.8399), which was slightly lower than that of StainGAN (0.7987, 95% CI: 0.7192–0.8819), but comparable to StainNet (0.7725, 95% CI: 0.6925–0.8443). For the EBV+/tissue ratio criterion, EBV-TRACER achieved the highest AUC of 0.7652 (95% CI: 0.6850–0.8398), outperforming both StainGAN (0.7613, 95% CI: 0.6767–0.8380) and StainNet (0.7370, 95% CI: 0.6671–0.8125). With respect to the EBV + area criterion, EBV-TRACER reached an AUC of 0.7221 (95% CI: 0.6134–0.8178), which was higher than that of StainGAN (0.6928, 95% CI: 0.5881–0.7933) and comparable to StainNet (0.7224, 95% CI: 0.5985–0.8340). In addition to the quantitative results, Fig. 3 visually compares the normalized patch images from TCGA-STAD, produced by various methods including no stain normalization, conventional stain normalization, StainGAN, StainNet, and two-stage stain normalization (EBV-TRACER). While conventional normalization often results in color imbalance and loss of contrast, the proposed two-stage stain normalization method maintains realistic staining and structural fidelity. It is worth noting that the proposed two-stage stain normalization method does not require additional training on target slides, whereas the other two deep learning-based methods (StainGAN and StainNet) depend on further training for optimal performance.
Investigation of lymphocyte ratios and their relationship with EBV-GC classification
To quantitatively investigate the relationship between EBV-GC positivity and lymphocyte distribution, the ratio of the number and area of lymphocytes were computed and compared between EBV-GC positive and EBV-negative slides as shown in Table 5. Overall, EBV-GC positive slides exhibited a larger number and area of lymphocytes than EBV-GC negative slides regardless of cohort type or source. In the internal validation cohorts, lymphocyte number and area ratios increased by 0.0080 to 0.0778 and from 0.0129 to 0.0845, respectively. In the external validation cohort, these ratios increased by 0.0751 and 0.0755, respectively. These results confirm the previous findings on the relationship between EBV-GC and tumor infiltrating lymphocytes. Moreover, we utilized the ratios of lymphocyte number and area to perform slide-level EBV-GC classification (Table 5). For the internal validation cohorts, the lymphocyte number ratio yielded AUCs ranging from 0.5021 (95% CI: 0.3387–0.6557; KSH-GAT) to 0.7039 (95% CI: 0.5906–0.8130; KSH-STO), whereas the lymphocyte area ratio achieved AUCs ranging from 0.5214 (95% CI: 0.3610–0.6745; KSH-GAT) to 0.7103 (95% CI: 0.5993–0.8185; KSH-STO). In the external validation cohort, the two scores obtained 0.6411 AUC (95% CI: 0.5033–0.7627) and 0.6474 AUC (95% CI: 0.5130–0.7632), respectively. These results were substantially inferior to those obtained by EBV-TRACER, indicating the limited utility of lymphocytes and the superior ability of EBV-TRACER in EBV-GC diagnosis. A visual analysis (Fig. 2) further revealed that lymphocyte ratios alone are not specific enough to accurately identify EBV-GC regions. Higher lymphocyte ratios were generally observed in the tumor areas with EBV-GC as well as in non-tumor areas, corresponding to the lower classification performance.
Furthermore, we evaluated the combined effect of the three slide-level EBV-GC scores (EBV+/Tumor, EBV+/Tissue, and EBV + Size) and two lymphocyte ratios on EBV-GC positivity detection. Table 6 illustrates the classification results on TCGA-STAD with three different normalization methods. The results clearly demonstrate that the combination of these features produces superior performance compared to using either lymphocyte ratios or slide-level EBV-GC scores. Among the stain normalization methods, EBV-TRACER generally outperformed the other methods.
Discussion
EBV-TRACER is a two-stage AI model that enables the detection of EBV-GC positive slides in both biopsy and surgical resection specimens. Using multiple internal and external validation cohorts, it can predict the positivity of EBV-GC from standard H&E stained slides in an accurate and robust fashion, holding potential for improving treatment planning and patient outcome. Specifically, EBV-TRACER can assist pathologists in identifying EBV-GC without the need for extra sample preparation and examinations such as EBER-ISH, and thus reduce the workload of pathologists and the cost associated with gastric cancer diagnosis. Importantly, our method does not require re-training or fine-tuning the classification model to adapt to new data. Instead, we correct stain variability through the proposed two-stage normalization process alone, enabling practical application to unseen datasets without additional annotation or model modification.
AI models have been extensively applied to pathology image analysis. Several works exist for analyzing and diagnosing gastric cancer and its subtypes, including intestinal-type, diffuse-type, and EBV-GC16,24. Though successful, one of the urgent and unmet needs for these AI models is robustness against variability in slide images, mainly due to difference in staining quality. Without adequate adjustment, both our study and previous study have observed substantial performance degradation in classifying EBV-GC16. However, EBV-TRACER demonstrated consistent performance across different cohort types and specimen sources. Although conventional stain normalization method and/or AI-based methods18,23 can be applied to mitigate variability in slides images, these are either ineffective or require additional training and validation procedures. The effectiveness of conventional stain normalization has proved limited, and AI-based methods have not been extensively validated for gastric cancer and its subtypes. While conventional stain normalization methods focus on local patch-based adjustment, our approach first aligns global stain distribution using thumbnail-level normalization, followed by fine-grained patch-level correction. This two-stage process was particularly effective in reducing domain shifts between scanners, as demonstrated in scanner-controlled biopsy experiments. These results highlight the benefit of our method over conventional techniques, even without expanding the training dataset.
Due to the lack of publicly available EBV-labeled multi-center datasets beyond TCGA-STAD, we constructed diverse internal validation cohorts using multiple scanners, tissue types, and time periods to simulate real-world variability. This design choice aimed to provide a more robust evaluation of model performance across heterogeneous staining conditions. TCGA-STAD, the external validation cohort, has been widely employed in various GC studies. In13, a deep learning model was developed using TCGA-STAD alone to predict EBV status, achieving 0.85 AUC. This model was further used to analyze the associate between EBV status, the spatial pattern of lymphocyte infiltration, and patient prognosis. In14, TCGA-STAD served as the training dataset to develop an EBV-GC classifier. The classifier was tested on an external validation dataset, achieving 0.88 AUC. Another study12 sought to combine the predictions by a deep learning model and a human expert for detecting EBV-GC positivity, providing a performance gain on TCGA-STAD and another external validation dataset. Regardless of the usage of human expertise, lower performance was obtained on TCGA-STAD than the other dataset, which was acquired using the same scanner with the training dataset. These studies lacked adequate validation, particularly with respect to staining variability, which limits their clinical applicability. One multicenter cohort study on EBV-GC15 included eight cohorts, including TCGA-STAD, obtained from seven countries using different scanners. This study utilized TCGA-STAD and three cohorts a training dataset and the other four cohorts as external validation datasets. Within the training dataset, it achieved an AUC of 0.810; however, AUCs substantially varied across the validation datasets, with scores of 0.836, 0.672, 0.859, and 0.676. These findings highlight that the inclusion of multiple datasets with different scanners is insufficient to guarantee consistent performance in clinical settings.
Our study has several limitations. First, only one external validation cohort (TCGA-STAD) was employed. Due to the limited availability of public datasets, we were unable to include additional external cohorts. Although TCGA-STAD includes specimens from multiple institutes, EBV-TRACER needs to be validated on additional external cohorts to ensure its accuracy and robustness in clinical settings. Second, EBV-TRACER independently and individually used three criteria to conduct slide-level classification and its performance varied across validation cohorts. We may further improve and stabilize its diagnostic performance by combining these three criteria and optimizing the final decision rule. Third, EBV-TRACER relies on patch-level classification, which is accomplished by EfficieNetB0. Other AI models can be adopted to enhance the patch-level classification performance, potentially leading to improved slide-level classification for EBV-GC. Fourth, no biopsy samples were included in KSH-DEV. Including biopsy samples during training or developing a model specifically tailored to biopsy samples may further enhance the EBV-GC classification performance. Last, EBV-TRACER was shown to be effective in predicting the positivity of EBV-GC from slide images. However, its clinical implications have not been investigated. A follow-up study is desirable to understand and analyze the effects of EBV-TRACER on GC diagnosis.
Conclusion
EBV-TRACER, a robust deep learning-based classifier, effectively detects EBV positivity in gastric cancer specimens with high accuracy, leveraging a two-stage stain normalization approach. The model demonstrated strong performance in both internal and external validation cohorts, consistently outperforming conventional methods. Its ability to analyze biopsy and surgical resection specimens while providing detailed quantitative insights enhances diagnostic precision. EBV-TRACER holds significant potential to improve clinical decision-making and optimize treatment strategies for gastric cancer patients.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-a Cancer J. Clin. 71 (3), 209–249 (2021).
Nagtegaal, I. D. et al. The 2019 WHO classification of tumours of the digestive system. Histopathology 76 (2), 182–188 (2020).
Bass, A. J. et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513 (7517), 202–209 (2014).
Sohn, B. H. et al. Clinical significance of four molecular subtypes of gastric cancer identified by the cancer genome atlas project. Clin. Cancer Res. 23 (15), 4441–4449 (2017).
Lima, A. et al. PD-L1 expression in EBV associated gastric cancer: a systematic review and meta-analysis. Discover Oncol., 13(1). (2022).
Shinozaki-Ushiku, A., Kunita, A. & Fukayama, M. Update on Epstein-Barr virus and gastric cancer. Int. J. Oncol. 46 (4), 1421–1434 (2015).
Högner, A. & Moehler, M. Immunotherapy in gastric cancer. Curr. Oncol. 29 (3), 1559–1574 (2022).
Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in Gastrointestinal cancer. Nat. Med. 25 (7), 1054– (2019).
Hinata, M. & Ushiku, T. Detecting immunotherapy-sensitive subtype in gastric cancer using histologic image-based deep learning. Sci. Rep., 11(1). (2021).
Jang, H. J. et al. Prediction of genetic alterations from gastric cancer histopathology images using a fully automated deep learning approach. World J. Gastroenterol., 27(44). (2021).
Huang, B. L. et al. Accurate diagnosis and prognosis prediction of gastric cancer using deep learning on digital pathological images: A retrospective multicentre study. Ebiomedicine, 73. (2021).
Zheng, X. Y. et al. A deep learning model and human-machine fusion for prediction of EBV-associated gastric cancer from histopathology. Nat. Commun., 13(1). (2022).
Zhang, B. Y. et al. Deep Learning Predicts EBV Status in Gastric Cancer Based on Spatial Patterns of Lymphocyte Infiltration. Cancers, 13(23). (2021).
Jeong, Y. et al. Deep learning model to predict Epstein-Barr virus associated gastric cancer in histology. Sci. Rep., 12(1). (2022).
Muti, H. S., Heij, L. R. & Keller, G. Development and validation of deep learning classifiers to detect Epstein-Barr virus and microsatellite instability status in gastric cancer: a retrospective multicentre cohort study (vol 3, pg e654,). Lancet Digital Health, 2021. 3(10): pp. E622-E622.). Lancet Digital Health, 2021. 3(10): pp. E622-E622. (2021).
Le Vuong, T. T. et al. Prediction of Epstein-Barr virus status in gastric cancer biopsy specimens using a deep learning algorithm. Jama Netw. Open., 5(10). (2022).
Assoc, W. M. World medical association declaration of Helsinki ethical principles for medical research involving human subjects. Jama-Journal Am. Med. Association. 310 (20), 2191–2194 (2013).
Macenko, M. et al. A Method for Normalizing Histology Slides for Quantitative Analysis. Ieee International Symposium on Biomedical Imaging: From Nano to Macro, Vols 1 and 2, 2009: p. 1107-+. (2009).
Tan, M. X. & Le, Q. V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. International Conference on Machine Learning, Vol 97, 97. (2019).
Graham, S. et al. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image. Anal., 58. (2019).
Doan, T. N. N. et al. SONNET: A Self-Guided ordinal regression neural network for segmentation and classification of nuclei in Large-Scale Multi-Tissue histology images. Ieee J. Biomedical Health Inf. 26 (7), 3218–3228 (2022).
Shaban, M. T. et al. Staingan: Stain Style Transfer for Digital Histological Images. Ieee 16th International Symposium on Biomedical Imaging (Isbi 2019), 2019: pp. 953–956. (2019).
Kang, H. T. et al. StainNet: A Fast and Robust Stain Normalization Network8 (Frontiers in Medicine, 2021).
Ji, K. X. et al. Construction and interpretation of machine learning-based prognostic models for survival prediction among intestinal-type and diffuse-type gastric cancer patients. World J. Surg. Oncol., 22(1). (2024).
Funding
This study was supported by a grant of National Research Foundation of Korea (NRF) (No. 2021R1A2C2014557, No. RS-2025-00558322, and No. RS-2024-00397293) and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI21C1137).
Author information
Authors and Affiliations
Contributions
Jin Tae Kwak and Kyungeun Kim conceived and designed the study. Jin Tae Kwak and Keunho Byeon conducted data analysis and interpretation. Boram Song, Seung Wan Chae, and Kyungeun Kim curated and organized datasets and annotations. Keunho Byeon executed experiments and drafted the manuscript. All authors read and reviewed the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Byeon, K., Song, B., Chae, S. et al. A robust artificial intelligence system for predicting EBV status in gastric cancer biopsy and resection specimens. Sci Rep 15, 35100 (2025). https://doi.org/10.1038/s41598-025-18836-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-18836-8