Abstract
Autism Spectrum Disorder (ASD) is a neurodevelopmental condition characterized by impaired social interaction, and restricted and repetitive patterns of behavior. ASD presents as a clinical spectrum, with variable levels of severity and multiple co-occurring conditions. The etiology of ASD may involve hundreds of genes and there is evidence that neurotransmitter and synaptic (NS) pathways are implicated. Proton Magnetic Resonance Spectroscopy (1H-MRS) has made it possible to study the concentration of brain neurometabolites and compare their levels in the brains of ASD and control individuals. We integrated genetic variants in NS genes with 1H-MRS analysis, and identified 12 predicted damaging variants (PDVs) in 12 NS genes in 10 ASD individuals, most mapping to genes involved in Gamma-aminobutyric acid (GABA) and glutamate pathways. Total creatine (tCr) and total N-acetyl aspartate (tNAA), markers of bioenergetics and neuronal metabolism, respectively, were lower in ASD patients with genetic alterations in NS genes compared to a control group without ASD. We conclude that PDVs in NS genes that are important for the regulation of glutamate or involved in GABAergic functions are associated with neurometabolic alterations, and that dysfunction in glutamatergic and/or GABAergic pathways may be implicated as these pathways are linked to the metabolic measures altered in cases.
Similar content being viewed by others
Introduction
Autism Spectrum Disorder (ASD) is a neurodevelopmental condition characterized by social and communication deficits and repetitive behavioral patterns1. ASD is clinically and genetically very heterogeneous, presenting as a clinical spectrum of symptoms that are further complicated by multiple co-occurring conditions. The prevalent hypothesis proposes that a combination of genetic alterations with environmental exposure lead to ASD2. While hundreds of genes have been associated to ASD3, less than 40% of patients are diagnosed with genetic syndromes or have chromosomic alterations, single gene disorders, or gene variants and metabolic disturbances with mitochondrial dysfunction that can explain the disease etiology4.
Genes associated with ASD suggest the involvement of important physiological processes such as metabolism, chromatin remodeling, translation and synapse5,6,7,8. Experimental animal models of ASD provide evidence for compromised neurotransmission, synapse formation and stabilization, and synaptic plasticity mechanisms9,10,11,12. For instance, excitatory/inhibitory imbalance is often observed in ASD and thought to be due to abnormal glutamatergic excitatory and GABAergic inhibitory neurotransmission in several brain regions, affecting information processing and regulation of behavior13. Excitatory/inhibitory imbalance could potentially explain social and cognitive deficits associated with ASD14,15. Such imbalances may arise from alterations during the early stages of neural circuit development or involved in synaptic homeostasis, since several of the genes identified in linkage and association studies encode proteins involved in these processes16,17.
Several studies implicate synaptic genes encoding pre- and post-synaptic proteins in non-syndromic ASD, including synaptic adhesion molecules such as neuroligins, neurexins, cadherins and contactins18,19,20,21, synaptic scaffold proteins like the ProSAPs/SHANKs multidomain proteins22, ion channels like calcium channels23 and N-methyl-D-aspartate (NMDA) or Gamma-aminobutyric acid (GABA) neurotransmitter receptors24,25. Postmortem studies have discovered structural and functional alterations in glutamatergic and GABAergic pathways in ASD26,27. In addition, mutations in pre-synaptic genes such as RIMS3 and post-synaptic genes such as IL1RAPL1 and SYNGAP1, involved in either synaptic vesicle organization or synapse formation, were also associated with ASD28,29. While the biological validation of the involvement of the hundreds of genes that have been implicated in the disease is not trivial, because brain tissue is mostly available postmortem, a window to brain function and neurochemistry is offered by neuroimaging studies of in vivo metabolism. Conversely, identifying risk genes underlying structural, functional, and neurochemical alterations observed in the brain of ASD patients is important to provide a deeper understanding of the neurobiology of ASD. Proton Magnetic Resonance Spectroscopy (1H-MRS) has made it possible to study the cerebral concentration of key biochemical substances in clinical and non-clinical populations in a non-invasive way30. Specifically, 1H-MRS has revealed widespread neurochemical alterations in ASD, with a range of molecules and brain regions being implicated, but the picture is complex31. For instance, studies investigating glutamate and GABA have reported increased, decreased or unchanged levels of these metabolites in several brain regions32. However, these studies did not take individuals’ genetic background into account, which could help to further clarify how synaptic dysregulation contributes to ASD neurobiology. Given that prior studies have found changes in tNAA15,33, including a recent meta-analysis34 it is important to understand how this metabolite links to Glutamate and GABA, from a neurometabolic point of view. NAA is present almost exclusively in the central nervous system, and largely only in neurons. There is an intimate connection between glutamate and NAA, and other neurometabolites, based for example on precursor-product relationships35. Glutamate and aspartate levels can change in reverse ways. For example, a shift in the aspartate aminotransferase reaction leads to an increase in aspartate formation with a reduction of its glutamate precursor. Moreover, both glutamate and GABA metabolism are linked through intermediates of the tricarboxylic acid cycle, which explains changes in neurometabolic coupling with changes in the diet36 or in disorders such as diabetes37.
In spite of these methodological opportunities, the integration of genetic studies with brain imaging is still lagging behind, even though it can offer extremely important information to guide the development of novel therapeutic approaches38. In this study, we integrate genetic analysis and 1H-MRS to improve the understanding of how synaptic dysregulation contributes to ASD neurobiology. This approach may guide biomarker development for patient stratification, leading to more effective pharmacological therapies.
Methods
Sample population and clinical assessment of patients
In this study we analyzed 16 ASD cases, selected because we had comprehensive clinical information and genetic data from genomic scans as well as 1H-MRS information, and a control group of 14 subjects. The patients were recruited at the Autism Clinic from the Paediatric Hospital at the University Hospital in Coimbra, Portugal. All patients were previously diagnosed and assessed in the clinical setting at least twice a year by a team of neurodevelopmental pediatricians and psychologists with extensive experience in ASD assessment and diagnosis. Thus, a comprehensive clinical characterization was used to ensure that the patients met the inclusion and exclusion criteria by the time of the 1H-MRS session. Inclusion criteria for patients were: (i) age below 18 years old, (ii) full-scale intelligence quotient (FSIQ) above 70 as measured by the Portuguese versions of the Wechsler Intelligence Scales39,40, so to exclude subjects with intellectual disability, (iii) ASD diagnostic confirmed by positive results on the direct parental or caregiver interview performed by an experienced pediatrician, namely the Autism Diagnostic Interview - Revised (ADI-R)41, and proband direct assessment with Autism Diagnostic Observation Schedule (ADOS) performed by an experienced psychologist42.
Informed consent was obtained from the parents/guardians of the participants. Children and adolescents also gave oral informed consent.
The study was approved by the ethics committee of the Faculty of Medicine of the University of Coimbra and the Ethics Committee for Health from the Instituto Nacional de Saúde Doutor Ricardo Jorge, and was conducted in accordance with the declaration of Helsinki.
Genetic data analysed
Genetic data included Copy Number Variants (CNVs) and Single Nucleotide Variants (SNVs), obtained in the context of two large ASD international consortia datasets: (1) the Autism Genome Project (AGP) used SNP arrays to carry out a Genome Wide Association Study (GWAS) with CNV detection, for 2611 families with one affected subject and the parents, from North America and Europe, including our team43,44,45; (2) the Autism Sequencing Consortium (ASC) performed whole exome sequencing (WES) for SNV detection in 10,008 individuals with ASD, and shared the output for research purposes46.
Identification of predicted damaging variants in NS genes
We analyzed exomic data from the ASC. Quality control filters applied using the parameters defined in bcftools (https://samtools.github.io/bcftools/) were: Variant Quality Score VSQLOD= >-1.5; Read Depth DP > 8; Genotype Quality GQ > 20; Allelic Depth AD > 0.2 and Missingness < 10%. Predicted damaging SNVs in affected individuals were selected in genes from a well curated list of neurotransmission and synaptic candidate genes (NS genes) developed in a previous study47 and that resulted from the application of filters for these processes in different databases that combine information from several biological processes and the genes involved in those mechanisms, namely Gene Ontology (GO)48, KEGG pathway database49, Reactome50, SynaptomeDB51 and SynSysNet52. Variants with predicted damaging impact in protein function and/or structure were identified using the Ensemble Variant Effect Predictor (VEP) tool53. VEP determines the effect of a variant in genes, transcripts and protein sequences, as well as regulatory regions, and incorporates SIFT54 and PolyPhen-2 algorithms55 to score for changes to protein sequence. SIFT predicts whether an amino acid substitution affects protein function based on sequence homology and the physical properties of amino acids. PolyPhen-2 is a tool which predicts possible impact of an amino acid substitution on the structure and function of a human protein using physical and comparative considerations. We selected loss of function (LoF) variants (variants of frameshift, stop gain, start lost, splice acceptor and splice donor) and missense variants classified as deleterious by SIFT, and/or as possibly or probably damaging by Polyphen-2, according to the Ensembl VEP. We also selected variants classified as pathogenic or likely pathogenic by InterVar56. InterVar is a bioinformatics software tool for clinical interpretation of genetic variants by the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) 2015 guideline57. Finally, we filtered out the variants that were present in the large control datasets included in the Genome Aggregation Database (gnomAD)58; http://gnomad.broadinstitute.org/) with minor allele frequency (MAF) ≥ 5%. The gnomAD datasets were developed to provide large-scale data of genetic variation present in several populations, and can be used to filter out common benign/neutral variants and identify rare variants with clinical meaning based on the frequency in the human population. The dataset used (gnomAD v2.1.1) incorporates the information from 60146 controls sequenced as part of various disease-specific and population genetic studies. We further inspected the AGP dataset to identify NS genes targeted by CNVs in individuals with ASD. We only selected CNVs that were detected by more than one algorithm, as implemented in44.
Characterization of predicted damaging variants in NS genes and associated biological pathways
We enriched in biological pathways the list of genes identified in the SNVs and CNVs analysis previously done, using the KEGG pathway database49,59 and the Reactome database50. GeneCards (https://www.genecards.org/) and PubMed (https://pubmed.ncbi.nlm.nih.gov/) were used to identify gene functions and the key roles of genes in the human brain. Gene expression in human brain regions was obtained from the Human Protein Atlas brain section (https://www.proteinatlas.org/humanproteome/brain). Additionally, we used the ClinVar database60 to inspect the classification of variants identified in this dataset based on supporting evidence, ClinVar classifies variants as pathogenic or benign, or as Variants with Uncertain Significance (VUS) whenever a variant does not fulfill criteria for pathogenic or benign, or the evidence for benign and pathogenic is conflicting. It is important to take into consideration the difference between implicating a variant as pathogenic (causative) for a disease, and a variant that may be predicted to be damaging to the encoded protein, and not necessarily being implicated in a disease, since both classifications are independent57.
We also examined if the genes with the variants were identified in the SFARI list as ASD candidate genes, and the strength of the evidence associating these genes to ASD (https://gene.sfari.org/database/gene-scoring/). SFARI is an ASD dedicated database for the autism research community, containing up-to-date information on genes associated with ASD that incorporates a gene scoring module. This module establishes a gene rank according to the strength of the evidence that associates a given gene to the disease61. The strongest candidate genes (SFARI categories 1 and 2) come from well-defined evidence on human genetic studies. Category 1 considers rigorous statistical comparisons between cases and controls yielding genome-wide statistical significance with independent replication to be the strongest possible evidence for a gene, and in category 2 these criteria are slightly relaxed. Category 1 includes genes that have been clearly implicated in ASD, by the presence of at least 3 de novo likely-gene-disrupting mutations being reported in the literature, and are typically returned to the participants. Some of the genes in this category meet the most rigorous threshold of genome-wide significance (p < 5 × 10− 8); all at least meet a threshold of false discovery rate of < 0.1. Category 2 includes genes with 2 reported de novo likely-gene-disrupting mutations, and genes uniquely implicated by a genome-wide association study, either reaching genome-wide significance or, if not, consistently replicated and accompanied by evidence that the risk variant has a functional effect. Genes predisposing to ASD in the context of a syndromic disorder (e.g. Fragile X Syndrome) are placed in category (S) and, if there is additional evidence implicating them in ASD, will have a number in front of the S according to the strength of that evidence (1 and 2 are the strongest categories).
Proton magnetic resonance spectroscopy (1H-MRS) data acquisition and analysis
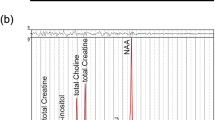
1H-MRS acquisition, data processing and quality check were done in the context of a previous study15. 1H-MRS data was acquired in a 3T Siemens TimTrio scanner (Erlangen, Germany) using 12-channel birdcage head coil, without participant sedation following the procedure described in15. Briefly, a T1-weighted Magnetization Prepared Rapid Acquisition Gradient-Echo (MPRAGE) sequence was acquired for each participant with 1 × 1 × 1 mm3 voxel size, repetition time (TR) = 2.3 s, echo time (TE) = 2.98 ms, flip angle (FA) = 9°, field of view (FOV) = 256 × 256, 160 slices, and used for spectroscopy voxel positioning in the medial prefrontal cortex/anterior cingulate cortex. A single-voxel with 3 × 3 × 3 cm was acquired with the MEGA-PRESS method62,63. Acquisition parameters were: repetition time 1.5 s, echo time 68 ms, 392 averages (196 ON, 196 OFF), 1024 data points, bandwidth 2000 Hz with automatic shimming and chemical shift selective (CHESS) weak water suppression with a bandwidth of 35 Hz, resulting in an acquisition time of approximately 10 min. No saturation bands were used. LCModel version 6.3–1D (Stephen Provencher Inc., Oakville, Canada15 was used to quantify total N-acetyl aspartate (tNAA) (n-acetylaspartate + n-acetylaspartilglutamate), gamma aminobutyric acid + macromolecules (GABA+) and glutamate + glutamine (Glx) from the ON-OFF difference spectrum, and total creatine from the OFF spectrum, which were corrected for the amount of cerebrospinal fluid (CSF-corrected).
Metabolites were quantified in three groups: (1) controls without ASD (n = 14); (2) subjects with ASD without predicted damaging variants (PDVs) in NS genes (n = 6); (3) subjects with ASD with PDVs in NS genes (n = 10). Statistical analysis of spectroscopy data was done with IBM SPSS statistics version 23 (IBM Corporation, IL, USA). Data distributions were assessed for normality (Shapiro–Wilk test). Normally distributed variables were analyzed using one-way ANOVA (Tukey correction for multiple comparisons was applied), while non-normally distributed variables were analyzed with the Kruskal-Wallis test, with metabolite (or ratio) as dependent variable and group as independent variable to test for differences across the 3 groups. An additional trend analysis was performed using the Jonckheere-Terpstra. Significance level was p = 0.05.
A post hoc power analysis was carried out with G*Power 3.1.9 software64,65 to evaluate the statistical power of the significant between groups results.
Results
Population characterization
In the present study, we analysed 16 patients and 14 control participants. The three groups defined were matched for age and performance intelligence quotient (Table 1). The two ASD groups were matched for full-scale intelligence and verbal quotients, but were different from the control group (post hoc Tukey p < 0.002) (Table 1).
Identification and characterization of predicted damaging variants in NS genes in the cases analysed
We identified 12 PDVs in NS genes in 10 of the cases analyzed, namely 10 SNVs and 2 CNVs (Table 2, and Supplementary Table 1 for a complete description of variants). Two patients had 2 variants in different genes, while all others had only 1 (Supplementary Table 1). Table 2 shows a summary of the characterization of variants. Most of the variants identified in the cases analysed are in genes involved in GABA and glutamate pathways. Additionally, 1 variant was found in the gene CHRND involved in the Acetylcholine pathway. The majority of the SNVs are classified by VEP as missense variants, except one stop-gain variant, and the two CNVs are deletions (Table 2 and Supplementary Table 1). Additionally, we checked if any of these variants are reported in the ClinVar database to inspect the classification of variants based on additional supporting evidence. ClinVar classifies variants as pathogenic or benign, or VUS whenever a variant does not fulfill criteria for pathogenic or benign or the evidence for benign and pathogenic is conflicting. Of 6 variants reported in the ClinVar database, 5 are VUS and 1 is classified as pathogenic. This pathogenic variant is found in the gene ALDH5A1 that encodes an enzyme that catalyzes the dehydrogenation of gamma-aminobutyraldehyde to GABA, and is related with Succinic semialdehyde dehydrogenase deficiency, which is characterized by infantile-onset hypotonia, developmental delay, cognitive impairment, expressive language deficit, and mild ataxia (Table 2 and Supplementary Table 1).
We also analyzed whether NS genes affected by PDVs in these 10 cases are part of the SFARI gene list of ASD candidates. Of 5 genes with PDVs involved in GABA metabolism (VPS18, ALDH5A1, GAD1, ALDH9A1, SYNE1), 1 has a SFARI score of 1 S (ALDH5A1) and another has a score of 2 (SYNE1), and of 6 genes with PDVs involved in the glutamate pathway (CREBBP, AKAP9, EPB41L2, RTN3, CACNA1C, PANX1), 2 have a SFARI score of 1 S (CREBBP and CACNA1C) and another has a score of 2 (AKAP9) (Table 2 and Supplementary Table 1). Categories 1 and 2 are the strongest SFARI categories with well-defined evidence on human genetic studies. An “S” is added to the numeric score when the gene also predisposes to ASD in the context of a syndromic disorder.
Magnetic resonance spectroscopy
One-way ANOVA revealed significant differences for tNAA levels (F(2,27) = 3.734, p = 0.037, η² = 0.217, 95% CI [0.000, 0.420] ). Post hoc Tukey test for multiple comparisons showed significant differences between the control group without ASD and the ASD group with genetic alterations in NS genes, such that tNAA levels were lower in the ASD group with such genetic alterations (p = 0.038, 95% CI: [0.002, 0.00005], Cohen’s d = 0.114). The post hoc power analysis based on the tNAA effect size indicated a power of 0.68 at α = 0.05. The tCr levels showed a near threshold level difference (Kruskal-Wallis: H(2) = 5.583, p = 0.061). A trend pattern of decreased levels for all metabolites across the 3 groups was observed (Fig. 1), such that metabolite levels decreased from control group > ASD without genetic alterations > ASD with genetic alterations in the ASD group with PDVs identified, which was significant for tCr (J = 86.00, p = 0.030) and tNAA levels (J = 73.00, p = 0.007) (Fig. 1). The effect size of this significant trend was calculated as Z/√N (rank bi-serial correlation), and was r = 0.40 (95% CI [0.05, 0.66]) and r = 0.49 (95% CI [0.16, 0.72]), respectively, which is considered a moderate effect size. There were no statistical differences between groups or significant trends for GABA + and Glx analysis.
Metabolites quantified in the medial prefrontal cortex/anterior cingulate cortex of ASD individuals and controls without ASD. Circles: controls without ASD (ctrl, n = 14); Squares: ASD individuals without predicted damaging variants in NS genes (no, n = 6); Triangles: ASD individuals with predicted damaging variants in NS genes (yes, n = 10); (a) tCr: total creatine (creatine + phosphocreatine); (b) tNAA: total N-acetyl aspartate (n-acetylaspartate + n-acetylaspartilglutamate); (c) GABA+: gamma aminobutyric acid + macromolecules; (d) Glx: glutamate + glutamine. Graphs depict individual values, mean ± 95% confidence interval. * P Significance of the trend analysis using the the Jonckheere-Terpstra test.
Discussion
In this study, we investigated how synaptic dysregulation contributes to ASD integrating measures of brain metabolites through 1H-MRS analysis with data on genetic variation. In line with the trend analysis, the levels of tCr and tNAA were significantly lower in the ASD group with PDVs in NS genes when compared with the control group. While the post hoc power analysis based on the tNAA effect size already indicated a power of 0.68 at α = 0.05, this actually underestimates the power of the performed trend analysis, which detected a significant monotonic trend of moderate effect size using the Jonckheere-Terpstra test. This method is more powerful than a one-way ANOVA when evaluating ordered differences, even with non-parametric data, and accordingly also revealed a significant trend for the tCr levels, further supporting the main conclusions of the study. Nonetheless, future research with larger cohorts and independent replication, potentially through multicenter collaborations, will be useful to validate and extend the current results.
The results are in agreement with the notion that tNAA is one of the most consistently altered neurometabolites in ASD as we observed before15, and was confirmed recently33,34. Its metabolism is tightly linked to the regulation of Glutamate and GABA levels. We further observed that the NS genes with PDVs in 10 of the individuals analysed encode proteins implicated either in GABA or glutamate functions.
The genes VPS18, SYNE1, ALDH9A1, ALDH5A1 and GAD1 are implicated in GABA pathways. The gene VPS18 is involved in dendrite development of Purkinje cells, which are a class of GABAergic inhibitory neurons located in the cerebellum66. The SYNE1 gene is also expressed in the Purkinje cells and is important for movement coordination67. The gene ALDH9A1 encodes an enzyme that catalyzes the dehydrogenation of gamma-aminobutyraldehyde to GABA68,69. The gene ALDH5A1 also encodes an aldehyde dehydrogenase family member that catalyzes one step in the degradation of GABA. Diseases associated with ALDH5A1 include Succinic Semialdehyde Dehydrogenase Deficiency (SSADHD), which is an autosomal-recessively inherited disorder characterized by global developmental delay, hypotonia or Epilepsy70. Interestingly, guanidino-species involved in the production of creatine are altered in SSADHD71 which may provide clues for the participation of ALDH5A1 in some of the alterations in tCr levels observed. The GAD1 gene is responsible for encoding an isoform of the enzyme glutamic acid decarboxylase (GAD1) that converts glutamate to GABA72.
The genes PANX1, CACNA1C, RTN3, EPB41L2, AKAP9 and CREBBP are involved in glutamatergic function. The PANX1 gene encodes a protein that forms plasma membrane channels that play a role in the release of ATP and glutamate in neurons and astrocytes73,74. The gene CACNA1C is involved in glutamate release and in the regulation of calcium concentration in the intracellular space75. Similarly, studies show that the RTN3 gene has been implicated in early phases of the glutamate secretory pathway76,77. The protein encoded by the gene EPB41L2 interacts with other proteins to regulate the insertion of AMPA receptors (AMPARs) which play a critical role in glutamatergic neurotransmission. The gene AKAP9 is also involved in glutamatergic signaling and is part of a protein network implicated in ASD78,79. The gene CREBBP also participates in the modulation of the glutamatergic neurotransmission as is involved in long-term potentiation mechanisms in glutamatergic synapses80. Most variants indentified in this study don´t have a clear outcome. Functional validation is needed to understand their impact in protein function to consolidate our results and, eventually, identify drug targets.
Our results are in accordance with other studies that reported reduced levels of NAA, glutamate, glutamine and GABA in children with ASD31,34. 1H-MRS studies measuring metabolite concentrations in participants with ASD have shown inconsistent findings, particularly for glutamate, GABA, NAA, and creatine; however, some patterns are evident. Lower GABA and NAA concentrations are frequently observed, suggesting that altered brain metabolism, particularly regarding neuronal integrity and excitation/inhibition balance, may be implicated in the pathophysiology of ASD34. Creatine levels can fluctuate with age and brain region. Some studies have found higher Cr levels in adults with ASD compared to children, with specific increases in the temporal lobe and decreases in the occipital lobe31. Age-related differences in metabolite concentrations are observed in ASD, with some metabolites showing distinct patterns in children and adults. The inconsistency of findings can also be attributed to differences in the MRS methodology used (e.g., voxel size, echo time, repetition time), as well as variations in sample demographics (e.g., age, sex, severity of ASD symptoms)31. Different methods for data analysis, including metabolite ratios calculations and data normalization, can also contribute to the variability on results. Understanding the specific metabolite alterations in ASD may help developing targeted therapies that address the underlying neurobiological mechanisms.
Multiple lines of evidence indicate that the dysfunction of glutamate receptors such as NMDARs is associated with ASD81,82. NMDAR is an ionotropic glutamate receptor (iGluR) that is activated by the neurotransmitter glutamate83 and mediates excitatory synaptic neurotransmission, which is important for synaptic plasticity, learning and memory. Earlier studies demonstrated that NMDARs are fundamental during neuronal development and synapse maturation processes84. Pharmacological enhancement or suppression of NMDAR function attenuate social withdrawal and repetitive behaviors81. On the other hand, GABA is an important inhibitory neurotransmitter in the central nervous system and there is evidence for an association between mutations affecting GABA function and ASD, including studies that reported reduced expression of GABAergic genes and GABA-related proteins in postmortem brain samples from individuals with ASD85. One of the most commonly reported chromosomal loci where CNVs observed in ASD in the 15q11-q13 chromosomal region86,87 that contains a number of genes encoding subunits of GABA receptors. GABA-A and GABA-B receptors alterations were identified in ASD brains88,89,90,91. Additionally, postmortem studies and experiments with animal models support GABAergic dysfunction playing a role in ASD85,92. It is estimated that Epilepsy co-occurs in 30% of ASD patients, probably in consequence of an excitation and inhibition imbalance. There are studies that estimated that subclinical epileptiform activity may occur in up to 85% of children with ASD93,94. Altogether, several lines of evidence support the imbalance of excitation to inhibition ratio proposal in ASD, possibly caused by alterations in GABA and glutamate pathways95.
The present study shows that having PDVs in NS genes might have a marked impact in brain neurochemistry. Specifically, we observed significant alterations in tCr (bioenergetics) and tNAA (neuronal metabolism and neuromodulation) levels when compared to the levels measured in the control group without ASD. We showed that almost all the variants identified involved glutamate and GABA pathways and there is previous evidence of the interaction between NMDARs and GABA with N-acetylaspartate (NAA) and creatine (Cr). Creatine modulates GABAergic and glutamatergic pathways96, and creatine treatment was shown to promote the differentiation of GABAergic neuronal precursors in cultured fetal rat spinal cord97 and GABAergic cells in cultured striatal tissue98. In addition, previous works suggest that creatine may also modulate brain NMDARs99,100 and exert positive cognitive effects through this action101. Other compounds may also act on brain NMDARs on oligodendrocyte function, and may interfere in the myelination of axons. Demyelination is associated to several disorders such as leucodystrophies and there is evidence that this can reflect an action of NAA on oligodendrocyte NMDA receptors102. The myelination of axons by oligodendrocytes is essential for normal brain activity103, and failures in this process results in compromised mental and physical activity. The activation of glutamate receptors on oligodendrocytes or their precursor’s cells can contribute to the loss of myelin102. Glutamate release was shown to kill precursor and immature oligodendrocytes104,105, and the activation of oligodendrocytes NMDARs by glutamate causes demyelination and loss of axonal action potential propagation106,107. However, the role of NMDA receptors in oligodendrocytes has been controversial. Previous studies showed that transient NMDA-receptor activation induces a Ca2+ -dependent efflux of NAA in rat hippocampus. There is evidence suggesting that Ca2+ -influx via the NMDAR modulates the efflux of NAA from neurons and may contribute to pathologies108. More recently, it was shown that NMDARs also have a role in glucose uptake regulation in response to axonal glutamate release, modulating the cooperation between oligodendrocytes and axons109. A role for NMDA receptor signaling in oligodendrocyte differentiation and promoting myelination has been suggested25,110. Increasing evidence suggests that myelination is a dynamic process that may be disrupted in the ASD brain111. NAA catabolism in oligodendrocytes is a precursor to fatty acids involved in the myelination of neuronal axons112. A pilot study in children with ASD suggested that alterations in axons and myelin of the corona radiata appear to be associated with the clinical severity of ASD111,113. Importantly, the 1H-MRS tNAA signal is a combination of NAA and n-acetylaspartilglutamate (NAAG) which may contribute to 10–25% of the total signal captured114. NAAG is thought to play a role in neuroprotection and synaptic plasticity due to modulatory properties of neurotransmission involving glutamate, GABA and other monoamines115. Thus, while no Glx or GABA + differences were directly observed, these neurotransmitters are tightly linked to brain bioenergetics and metabolism measures that were altered, as explained above. Importantly, 1H-MRS captures the bulk levels of metabolites and is not able to discriminate between the metabolic and neurotransmitter pools which are in constant flow112. Hence, we cannot exclude that synaptic glutamate and GABA levels are altered in the ASD participants with genetic alterations. Future studies disentangling the neurotransmitter and neurometabolic pools, for example using carbon-labeled spectroscopic approaches116, could provide further insight.
The interconnection between the observed metabolite alterations and ASD symptoms is still under investigation. NAA is a marker of neuronal integrity and its levels can be affected by several factors, including mitochondrial dysfunction117. Instead of directly aiming to increase NAA levels, research have focused on addressing underlying neurological and metabolic factors that may contribute to ASD symptoms and potentially improve overall neuronal health118,119. Potential treatments and interventions may include nutritional approaches that may help improve mitochondrial function, which is implicated in some cases of ASD. Dietary guanidinoacetic acid (GAA) has been shown to increase brain creatine levels. Studies have demonstrated that supplementing with GAA can lead to a notable increase in creatine levels in several brain regions, which may improve energy production and utilization within the brain120. Experiments with Dodecyl creatine ester (DCE), a prodrug of creatine, are very promising in increasing brain creatine concentrations and improving cognitive function in mice121.
In conclusion, the present study shows that the ASD cases analysed which have significant alterations in tNAA and tCr levels also have PDVs in genes that are important for processes associated with the regulation of glutamate and GABAergic functions. According with evidence resulting from previous studies, it is possible that the implications of the 1H-MRS alterations detected in the patients of the present study are associated with glutamatergic and/or GABAergic pathways dysregulation.
Data availability
Sequence data that support the findings of this study was downloaded from the database of Genotypes and Phenotypes (dbGaP) web site, under dbGaP accession phs000298.v1.p1; phs000298.v2.p2; phs000298.v3.p2; phs000298.v4.p3. Version 1–3: The data sets were deposited by the ARRA Autism Sequencing Collaborative, an ARRA-funded research initiative. The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
References
A. P. Association, Diagnostic and statistical manual of mental disorders : DSM-5. American Psychiatric Association, 2013. Acedido: 26 de fevereiro de 2025. [Online]. Disponível em: https://thuvienso.hoasen.edu.vn/handle/123456789/9853
Santos, J. X. et al. A role for gene-environment interactions in autism spectrum disorder is supported by variants in genes regulating the effects of exposure to xenobiotics. Front. Neurosci. 16, 862315. https://doi.org/10.3389/fnins.2022.862315 (2022).
Ramaswami, G. & Geschwind, D. H. Genetics of autism spectrum disorder. Handb. Clin. Neurol. 147, 321–329. https://doi.org/10.1016/B978-0-444-63233-3.00021-X (2018).
Genovese, A. & Butler, M. G. Clinical assessment, genetics, and treatment approaches in autism spectrum disorder (ASD). Int. J. Mol. Sci. 21(13), 4726. https://doi.org/10.3390/ijms21134726 (2020).
Synaptic, transcriptional and chromatin genes disrupted in autism | Nature. Acedido: 26 de fevereiro de 2025. [Online]. Disponível em: https://www.nature.com/articles/nature13772
Lord, C. et al. Autism spectrum disorder. Nat. Rev. Dis. Primer 6(1), 5. https://doi.org/10.1038/s41572-019-0138-4 (2020).
Hoffmann, A. & Spengler, D. Chromatin remodeler CHD8 in autism and brain development. J. Clin. Med. 10(2), 366. https://doi.org/10.3390/jcm10020366 (2021).
Bonsi, P., De Jaco, A., Fasano, L. & Gubellini, P. Postsynaptic autism spectrum disorder genes and synaptic dysfunction. Neurobiol. Dis. 162, 105564. https://doi.org/10.1016/j.nbd.2021.105564 (2022).
Giovedí, S., Corradi, A., Fassio, A. & Benfenati, F. Involvement of synaptic genes in the pathogenesis of autism spectrum disorders: The case of synapsins. Front. Pediatr. 2, 94. https://doi.org/10.3389/fped.2014.00094 (2014).
Kuo, H.-Y. & Liu, F.-C. Molecular pathology and pharmacological treatment of autism spectrum disorder-like phenotypes using rodent models. Front. Cell. Neurosci. 12, 422. https://doi.org/10.3389/fncel.2018.00422 (2018).
Sgritta, M. et al. Impaired synaptic plasticity in an animal model of autism exhibiting early hippocampal GABAergic-BDNF/TrkB signaling alterations. iScience 26(1), 105728. https://doi.org/10.1016/j.isci.2022.105728 (2023).
Urrutia-Ruiz, C. et al. Deletion of the autism-associated protein SHANK3 abolishes structural synaptic plasticity after brain trauma. Int. J. Mol. Sci. 23(11), 6081. https://doi.org/10.3390/ijms23116081 (2022).
Uzunova, G., Pallanti, S. & Hollander, E. Excitatory/inhibitory imbalance in autism spectrum disorders: Implications for interventions and therapeutics. World J. Biol. Psychiatry Off. J. World Fed. Soc. Biol. Psychiatry 17(3), 174–186. https://doi.org/10.3109/15622975.2015.1085597 (2016).
Gao, R. & Penzes, P. Common mechanisms of excitatory and inhibitory imbalance in schizophrenia and autism spectrum disorders. Curr. Mol. Med. 15(2), 146–167. https://doi.org/10.2174/1566524015666150303003028 (2015).
Carvalho Pereira, A., Violante, I. R., Mouga, S., Oliveira, G. & Castelo-Branco, M. Medial frontal lobe neurochemistry in autism spectrum disorder is marked by reduced N-acetylaspartate and unchanged gamma-aminobutyric acid and glutamate + glutamine levels. J. Autism Dev. Disord. 48(5), 1467–1482. https://doi.org/10.1007/s10803-017-3406-8 (2018).
Toro, R. et al. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. TIG 26(8), 363–372. https://doi.org/10.1016/j.tig.2010.05.007 (2010).
Penzes, P., Buonanno, A., Passafaro, M., Sala, C. & Sweet, R. A. Developmental vulnerability of synapses and circuits associated with neuropsychiatric disorders. J. Neurochem. 126(2), 165–182. https://doi.org/10.1111/jnc.12261 (2013).
Jamain, S. et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 34(1), 27–29. https://doi.org/10.1038/ng1136 (2003).
Zoghbi, H. Y. Postnatal neurodevelopmental disorders: Meeting at the synapse?. Science 302(5646), 826–830. https://doi.org/10.1126/science.1089071 (2003).
Südhof, T. C. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455(7215), 903–911. https://doi.org/10.1038/nature07456 (2008).
Betancur, C., Sakurai, T. & Buxbaum, J. D. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 32(7), 402–412. https://doi.org/10.1016/j.tins.2009.04.003 (2009).
Jiang, Y.-H. & Ehlers, M. D. Modeling autism by SHANK gene mutations in mice. Neuron 78(1), 8–27. https://doi.org/10.1016/j.neuron.2013.03.016 (2013).
Schmunk, G. & Gargus, J. J. Channelopathy pathogenesis in autism spectrum disorders. Front. Genet. 4, 222. https://doi.org/10.3389/fgene.2013.00222 (2013).
Shao, Y. et al. Fine mapping of autistic disorder to chromosome 15q11-q13 by use of phenotypic subtypes. Am. J. Hum. Genet. 72(3), 539–548. https://doi.org/10.1086/367846 (2003).
Li, X., Zou, H. & Brown, W. T. Genes associated with autism spectrum disorder. Brain Res. Bull. 88(6), 543–552. https://doi.org/10.1016/j.brainresbull.2012.05.017 (2012).
Hutsler, J. J. & Zhang, H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 1309, 83–94. https://doi.org/10.1016/j.brainres.2009.09.120 (2010).
Chattopadhyaya, B. & Cristo, G. D. GABAergic circuit dysfunctions in neurodevelopmental disorders. Front. Psychiatry 3, 51. https://doi.org/10.3389/fpsyt.2012.00051 (2012).
Kumar, R. A. et al. A de novo 1p34.2 microdeletion identifies the synaptic vesicle gene RIMS3 as a novel candidate for autism. J. Med. Genet. 47(2), 81–90. https://doi.org/10.1136/jmg.2008.065821 (2010).
Piton, A. et al. Mutations in the calcium-related gene IL1RAPL1 are associated with autism. Hum. Mol. Genet. 17(24), 3965–3974. https://doi.org/10.1093/hmg/ddn300 (2008).
Soares, D. P. & Law, M. Magnetic resonance spectroscopy of the brain: Review of metabolites and clinical applications. Clin. Radiol. 64(1), 12–21. https://doi.org/10.1016/j.crad.2008.07.002 (2009).
Ford, T. C. & Crewther, D. P. A comprehensive review of the (1)H-MRS metabolite spectrum in autism spectrum disorder. Front. Mol. Neurosci. 9, 14. https://doi.org/10.3389/fnmol.2016.00014 (2016).
Ajram, L. A. et al. The contribution of [1H] magnetic resonance spectroscopy to the study of excitation-inhibition in autism. Prog. Neuropsychopharmacol. Biol. Psychiatry 89, 236–244. https://doi.org/10.1016/j.pnpbp.2018.09.010 (2019).
Dionísio, A. et al. Neurochemical differences in core regions of the autistic brain: A multivoxel 1H-MRS study in children. Sci. Rep. 14(1), 2374. https://doi.org/10.1038/s41598-024-52279-x (2024).
Du, Y. et al. Neurometabolite levels in the brains of patients with autism spectrum disorders: A meta-analysis of proton magnetic resonance spectroscopy studies (N = 1501). Mol. Psychiatry 28(7), 3092–3103. https://doi.org/10.1038/s41380-023-02079-y (2023).
Shen, J., Shenkar, D., An, L. & Tomar, J. S. Local and interregional neurochemical associations measured by magnetic resonance spectroscopy for studying brain functions and psychiatric disorders. Front. Psychiatry https://doi.org/10.3389/fpsyt.2020.00802 (2020).
Ribeiro, M. et al. High-fat diet induces a neurometabolic state characterized by changes in glutamate and N-acetylaspartate pools associated with early glucose intolerance: An in vivo multimodal MRI study. J. Magn. Reson. Imaging JMRI https://doi.org/10.1002/jmri.25942 (2018).
d’Almeida, O. C. et al. The neurometabolic profiles of GABA and Glutamate as revealed by proton magnetic resonance spectroscopy in type 1 and type 2 diabetes. PLoS ONE 15(10), e0240907. https://doi.org/10.1371/journal.pone.0240907 (2020).
Hashem, S. et al. Genetics of structural and functional brain changes in autism spectrum disorder. Transl. Psychiatry 10(1), 1–17. https://doi.org/10.1038/s41398-020-00921-3 (2020).
de Coimbra, U. Escala de Inteligência de Wechsler para Crianças – Terceira Edição (WISC-III). uc.pt. Acedido: 26 de fevereiro de 2025. [Online]. Disponível em: https://www.uc.pt/fpce/psyassessmentlab/testes/inteligencia/wisc-iii/.
de Coimbra, U. Escala de Inteligência de Wechsler para Adultos – Terceira edição (WAIS-III). uc.pt. Acedido: 26 de fevereiro de 2025. [Online]. Disponível em: https://www.uc.pt/fpce/psyassessmentlab/testes/inteligencia/wais-iii/.
Rutter, M., LeCouteur, A. & Lord, C. Autism diagnostic interview-revised. 12 de dezembro de https://doi.org/10.1037/t18128-000 (2016).
Lord, C. et al. The autism diagnostic observation schedule-generic: A standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 30(3), 205–223 (2000).
Autism Genome Project Consortium et al., Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 39 (3), 319–328. https://doi.org/10.1038/ng1985 (2007).
Pinto, D. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466(7304), 368–372. https://doi.org/10.1038/nature09146 (2010).
Anney, R. et al. Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum. Mol. Genet. 21(21), 4781–4792. https://doi.org/10.1093/hmg/dds301 (2012).
Buxbaum, J. D. et al. The autism sequencing consortium: Large-scale, high-throughput sequencing in autism spectrum disorders. Neuron 76(6), 1052–1056. https://doi.org/10.1016/j.neuron.2012.12.008 (2012).
Vilela, J. et al. Identification of neurotransmission and synaptic biological processes disrupted in autism spectrum disorder using interaction networks and community detection analysis. Biomedicines 11(11), 2971. https://doi.org/10.3390/biomedicines11112971 (2023).
Carbon, S. et al. AmiGO: Online access to ontology and annotation data. Bioinform. Oxf. Engl. 25(2), 288–289. https://doi.org/10.1093/bioinformatics/btn615 (2009).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1), 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Jassal, B. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 48(D1), D498–D503. https://doi.org/10.1093/nar/gkz1031 (2020).
Pirooznia, M. et al. SynaptomeDB: An ontology-based knowledgebase for synaptic genes. Bioinformatics 28(6), 897–899. https://doi.org/10.1093/bioinformatics/bts040 (2012).
von Eichborn, J. et al. SynSysNet: Integration of experimental data on synaptic protein–protein interactions with drug-target relations. Nucleic Acids Res. 41(D1), D834–D840. https://doi.org/10.1093/nar/gks1040 (2013).
McLaren, W. et al. The ensembl variant effect predictor. Genome Biol. 17(1), 122. https://doi.org/10.1186/s13059-016-0974-4 (2016).
Ng, P. C. & Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31(13), 3812–3814. https://doi.org/10.1093/nar/gkg509 (2003).
Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 7, 20. https://doi.org/10.1002/0471142905.hg0720s76 (2013).
Li, Q. & Wang, K. InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet. 100(2), 267–280. https://doi.org/10.1016/j.ajhg.2017.01.004 (2017).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 17(5), 405–424. https://doi.org/10.1038/gim.2015.30 (2015).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581(7809), 434–443. https://doi.org/10.1038/s41586-020-2308-7 (2020).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. Publ. Protein Soc. 28(11), 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Landrum, M. J. et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46(D1), D1062–D1067. https://doi.org/10.1093/nar/gkx1153 (2018).
Banerjee-Basu, S. & Packer, A. SFARI Gene: An evolving database for the autism research community. Dis. Model. Mech. 3(3–4), 133–135. https://doi.org/10.1242/dmm.005439 (2010).
Edden, R. A. E., Pomper, M. G. & Barker, P. B. In vivo differentiation of N-acetyl aspartyl glutamate from N-acetyl aspartate at 3 Tesla. Magn. Reson. Med. 57(6), 977–982. https://doi.org/10.1002/mrm.21234 (2007).
Mescher, M., Merkle, H., Kirsch, J., Garwood, M. & Gruetter, R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 11(6), 266–272 (1998).
Faul, F., Erdfelder, E., Lang, A.-G. & Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 39(2), 175–191. https://doi.org/10.3758/bf03193146 (2007).
Faul, F., Erdfelder, E., Buchner, A. & Lang, A.-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods 41(4), 1149–1160. https://doi.org/10.3758/BRM.41.4.1149 (2009).
Peng, C. et al. Vps18 deficiency inhibits dendritogenesis in Purkinje cells by blocking the lysosomal degradation of Lysyl Oxidase. Biochem. Biophys. Res. Commun. 423(4), 715–720. https://doi.org/10.1016/j.bbrc.2012.06.021 (2012).
Gros-Louis, F. et al. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat. Genet. 39(1), 80–85. https://doi.org/10.1038/ng1927 (2007).
Končitíková, R. et al. Kinetic and structural analysis of human ALDH9A1. Biosci. Rep. 39(4), BSR20190558. https://doi.org/10.1042/BSR20190558 (2019).
McPherson, J. D., Wasmuth, J. J., Kurys, G. & Pietruszko, R. Human aldehyde dehydrogenase: Chromosomal assignment of the gene for the isozyme that metabolizes γ-aminobutyraldehyde. Hum. Genet. 93(2), 211–212. https://doi.org/10.1007/BF00210615 (1994).
Vogel, K. R. et al. Thirty years beyond discovery—clinical trials in succinic semialdehyde dehydrogenase deficiency, a disorder of GABA metabolism. J. Inherit. Metab. Dis. 36(3), 401–410. https://doi.org/10.1007/s10545-012-9499-5 (2013).
Brown, M., Turgeon, C., Rinaldo, P., Roullet, J.-B. & Gibson, K. M. Temporal metabolomics in dried bloodspots suggests multipathway disruptions in aldh5a1-/- mice, a model of succinic semialdehyde dehydrogenase deficiency. Mol. Genet. Metab. 128(4), 397–408. https://doi.org/10.1016/j.ymgme.2019.10.003 (2019).
Geller, B. et al. Linkage disequilibrium of the brain-derived neurotrophic factor Val66Met polymorphism in children with a prepubertal and early adolescent bipolar disorder phenotype. Am. J. Psychiatry 161(9), 1698–1700. https://doi.org/10.1176/appi.ajp.161.9.1698 (2004).
Ma, W., Hui, H., Pelegrin, P. & Surprenant, A. Pharmacological characterization of pannexin-1 currents expressed in mammalian cells. J. Pharmacol. Exp. Ther. 328(2), 409–418. https://doi.org/10.1124/jpet.108.146365 (2009).
Seo, J. H., Dalal, M. S., Calderon, F. & Contreras, J. E. Myeloid Pannexin-1 mediates acute leukocyte infiltration and leads to worse outcomes after brain trauma. J. Neuroinflam. 17(1), 245. https://doi.org/10.1186/s12974-020-01917-y (2020).
Scotti-Muzzi, E., Chile, T., Vallada, H., Otaduy, M. C. G. & Soeiro-de-Souza, M. G. Association between CACNA1C gene rs100737 polymorphism and glutamatergic neurometabolites in bipolar disorder. Eur. Neuropsychopharmacol. J. Eur. Coll. Neuropsychopharmacol. 59, 26–35. https://doi.org/10.1016/j.euroneuro.2022.04.001 (2022).
Wakana, Y. et al. Reticulon 3 is involved in membrane trafficking between the endoplasmic reticulum and Golgi. Biochem. Biophys. Res. Commun. 334(4), 1198–1205. https://doi.org/10.1016/j.bbrc.2005.07.012 (2005).
Chiurchiù, V., Maccarrone, M. & Orlacchio, A. The role of reticulons in neurodegenerative diseases. Neuromol. Med. 16(1), 3–15. https://doi.org/10.1007/s12017-013-8271-9 (2014).
Poelmans, G., Franke, B., Pauls, D. L., Glennon, J. C. & Buitelaar, J. K. AKAPs integrate genetic findings for autism spectrum disorders. Transl. Psychiatry 3(6), e270–e270. https://doi.org/10.1038/tp.2013.48 (2013).
Kantamneni, S. Cross-talk and regulation between glutamate and GABAB receptors. Front. Cell. Neurosci. 9, 135. https://doi.org/10.3389/fncel.2015.00135 (2015).
Hu, S. C., Chrivia, J. & Ghosh, A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron 22(4), 799–808. https://doi.org/10.1016/s0896-6273(00)80738-2 (1999).
Lee, E.-J., Choi, S. Y. & Kim, E. NMDA receptor dysfunction in autism spectrum disorders. Curr. Opin. Pharmacol. 20, 8–13. https://doi.org/10.1016/j.coph.2014.10.007 (2015).
Lim, C.-S. et al. Dysfunction of NMDA receptors in neuronal models of an autism spectrum disorder patient with a DSCAM mutation and in Dscam-knockout mice. Mol. Psychiatry 26(12), 7538–7549. https://doi.org/10.1038/s41380-021-01216-9 (2021).
Traynelis, S. F. et al. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol. Rev. 62(3), 405–496. https://doi.org/10.1124/pr.109.002451 (2010).
Adesnik, H., Li, G., During, M. J., Pleasure, S. J. & Nicoll, R. A. NMDA receptors inhibit synapse unsilencing during brain development. Proc. Natl. Acad. Sci. U. S. A. 105(14), 5597–5602. https://doi.org/10.1073/pnas.0800946105 (2008).
Pizzarelli, R. & Cherubini, E. Alterations of GABAergic signaling in autism spectrum disorders. Neural Plast. 2011, 297153. https://doi.org/10.1155/2011/297153 (2011).
Wang, L. et al. Association study and mutation sequencing of genes on chromosome 15q11-q13 identified GABRG3 as a susceptibility gene for autism in Chinese Han population. Transl. Psychiatry 8(1), 152. https://doi.org/10.1038/s41398-018-0197-4 (2018).
Tamada, K. et al. Genetic dissection identifies Necdin as a driver gene in a mouse model of paternal 15q duplications. Nat. Commun. 12(1), 4056. https://doi.org/10.1038/s41467-021-24359-3 (2021).
Collins, A. L. et al. Investigation of autism and GABA receptor subunit genes in multiple ethnic groups. Neurogenetics 7(3), 167–174. https://doi.org/10.1007/s10048-006-0045-1 (2006).
Fatemi, S. H., Folsom, T. D., Reutiman, T. J. & Thuras, P. D. Expression of GABA(B) receptors is altered in brains of subjects with autism. Cerebellum Lond. Engl. 8(1), 64–69. https://doi.org/10.1007/s12311-008-0075-3 (2009).
Oblak, A. L., Gibbs, T. T. & Blatt, G. J. Decreased GABA(B) receptors in the cingulate cortex and fusiform gyrus in autism. J. Neurochem. 114(5), 1414–1423. https://doi.org/10.1111/j.1471-4159.2010.06858.x (2010).
Oblak, A. L., Gibbs, T. T. & Blatt, G. J. Reduced GABAA receptors and benzodiazepine binding sites in the posterior cingulate cortex and fusiform gyrus in autism. Brain Res. 1380, 218–228. https://doi.org/10.1016/j.brainres.2010.09.021 (2011).
Zhao, H. et al. GABAergic system dysfunction in autism spectrum disorders. Front. Cell Dev. Biol. 9, 781327. https://doi.org/10.3389/fcell.2021.781327 (2021).
Gillberg, C. & Billstedt, E. Autism and Asperger syndrome: Coexistence with other clinical disorders. Acta Psychiatr. Scand. 102(5), 321–330. https://doi.org/10.1034/j.1600-0447.2000.102005321.x (2000).
Yasuhara, A. Correlation between EEG abnormalities and symptoms of autism spectrum disorder (ASD). Brain Dev. 32(10), 791–798. https://doi.org/10.1016/j.braindev.2010.08.010 (2010).
Rubenstein, J. L. R. & Merzenich, M. M. Model of autism: Increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2(5), 255–267 (2003).
Joncquel-Chevalier Curt, M. et al. Creatine biosynthesis and transport in health and disease. Biochimie 119, 146–165. https://doi.org/10.1016/j.biochi.2015.10.022 (2015).
Creatine treatment promotes differentiation of GABA-ergic neuronal precursors in cultured fetal rat spinal cord - Pesquisa Google.. Acedido: 18 de março de 2025. [Online]. Disponível em: https://www.google.com/search?q=Creatine+treatment+promotes+differentiation+of+GABA-ergic+neuronal+precursors+in+cultured+fetal+rat+spinal+cord&rlz=1C1GCEA_enPT1092PT1092&oq=Creatine+treatment+promotes+differentiation+of+GABA-ergic+neuronal+precursors+in+cultured+fetal+rat+spinal+cord&gs_lcrp=EgZjaHJvbWUyBggAEEUYOdIBCTE4NTRqMGoxNagCCLACAfEFL-AY8nDpuB0&sourceid=chrome&ie=UTF-8
Ducray, A. et al. Creatine and neurotrophin-4/5 promote survival of nitric oxide synthase-expressing interneurons in striatal cultures. Neurosci. Lett. 395(1), 57–62. https://doi.org/10.1016/j.neulet.2005.10.051 (2006).
Oliveira, M. S. et al. The involvement of the polyamines binding sites at the NMDA receptor in creatine-induced spatial learning enhancement. Behav. Brain Res. 187(1), 200–204. https://doi.org/10.1016/j.bbr.2007.09.004 (2008).
Royes, L. F. F. et al. Neuromodulatory effect of creatine on extracellular action potentials in rat hippocampus: Role of NMDA receptors. Neurochem. Int. 53(1–2), 33–37. https://doi.org/10.1016/j.neuint.2008.04.008 (2008).
Brosnan, J. T. & Brosnan, M. E. Creatine: Endogenous metabolite, dietary, and therapeutic supplement. Annu. Rev. Nutr. 27, 241–261. https://doi.org/10.1146/annurev.nutr.27.061406.093621 (2007).
Kolodziejczyk, K., Hamilton, N. B., Wade, A., Káradóttir, R. & Attwell, D. The effect of N-acetyl-aspartyl-glutamate and N-acetyl-aspartate on white matter oligodendrocytes. Brain J. Neurol. 132(Pt 6), 1496–1508. https://doi.org/10.1093/brain/awp087 (2009).
Fields, R. D. White matter in learning, cognition and psychiatric disorders. Trends Neurosci. 31(7), 361–370. https://doi.org/10.1016/j.tins.2008.04.001 (2008).
Follett, P. L., Rosenberg, P. A., Volpe, J. J. & Jensen, F. E. NBQX attenuates excitotoxic injury in developing white matter. J. Neurosci. Off. J. Soc. Neurosci. 20(24), 9235–9241. https://doi.org/10.1523/JNEUROSCI.20-24-09235.2000 (2000).
Khwaja, O. & Volpe, J. J. Pathogenesis of cerebral white matter injury of prematurity. Arch. Dis. Child. Fetal Neonatal Ed. 93(2), F153–F161. https://doi.org/10.1136/adc.2006.108837 (2008).
Micu, I. et al. NMDA receptors mediate calcium accumulation in myelin during chemical ischaemia. Nature 439(7079), 988–992. https://doi.org/10.1038/nature04474 (2006).
Bakiri, Y., Hamilton, N. B., Káradóttir, R. & Attwell, D. Testing NMDA receptor block as a therapeutic strategy for reducing ischaemic damage to CNS white matter. Glia 56(2), 233–240. https://doi.org/10.1002/glia.20608 (2008).
Tranberg, M. et al. NMDA-receptor mediated efflux of N-acetylaspartate: Physiological and/or pathological importance?. Neurochem. Int. 45(8), 1195–1204. https://doi.org/10.1016/j.neuint.2004.06.005 (2004).
Saab, A. S. et al. Oligodendroglial NMDA receptors regulate glucose import and axonal energy metabolism. Neuron 91(1), 119–132. https://doi.org/10.1016/j.neuron.2016.05.016 (2016).
Lundgaard, I. et al. Neuregulin and BDNF induce a switch to NMDA receptor-dependent myelination by oligodendrocytes. PLoS Biol. 11(12), e1001743. https://doi.org/10.1371/journal.pbio.1001743 (2013).
Galvez-Contreras, A. Y., Zarate-Lopez, D., Torres-Chavez, A. L. & Gonzalez-Perez, O. Role of oligodendrocytes and myelin in the pathophysiology of autism spectrum disorder. Brain Sci. 10(12), 951. https://doi.org/10.3390/brainsci10120951 (2020).
Rae, C. D. A guide to the metabolic pathways and function of metabolites observed in human brain 1H magnetic resonance spectra. Neurochem. Res. 39(1), 1–36. https://doi.org/10.1007/s11064-013-1199-5 (2014).
Carmody, D. P. & Lewis, M. Regional white matter development in children with autism spectrum disorders. Dev. Psychobiol. 52(8), 755–763. https://doi.org/10.1002/dev.20471 (2010).
Pouwels, P. J. & Frahm, J. Differential distribution of NAA and NAAG in human brain as determined by quantitative localized proton MRS. NMR Biomed. 10(2), 73–78 (1997).
Neale, J. H., Bzdega, T. & Wroblewska, B. N-acetylaspartylglutamate: The most abundant peptide neurotransmitter in the mammalian central nervous system. J. Neurochem. 75(2), 443–452. https://doi.org/10.1046/j.1471-4159.2000.0750443.x (2000).
Mandal, P. K., Guha Roy, R., Samkaria, A., Maroon, J. C. & Arora, Y. In vivo 13C magnetic resonance spectroscopy for assessing brain biochemistry in health and disease. Neurochem. Res. 47(5), 1183–1201. https://doi.org/10.1007/s11064-022-03538-8 (2022).
Marotta, R. et al. The Neurochemistry of Autism. Brain Sci. 10(3), 163. https://doi.org/10.3390/brainsci10030163 (2020).
Zheng, H.-F., Wang, W.-Q., Li, X.-M., Rauw, G. & Baker, G. B. Body fluid levels of neuroactive amino acids in autism spectrum disorders: A review of the literature. Amino Acids 49(1), 57–65. https://doi.org/10.1007/s00726-016-2332-y (2017).
Frye, R. E. Mitochondrial dysfunction in autism spectrum disorder: Unique abnormalities and targeted treatments. Semin. Pediatr. Neurol. 35, 100829. https://doi.org/10.1016/j.spen.2020.100829 (2020).
Ostojic, S. M., Ostojic, J., Drid, P., Vranes, M. & Jovanov, E. P. Dietary guanidinoacetic acid increases brain creatine levels in healthy men. Nutr. Burbank Los Angel. Cty. Calif 33, 149–156. https://doi.org/10.1016/j.nut.2016.06.001 (2017).
Mabondzo, A., van de Kamp, J. & Mercimek-Andrews, E. S. Dodecyl creatine ester therapy: From promise to reality. Cell. Mol. Life Sci. CMLS 81(1), 186. https://doi.org/10.1007/s00018-024-05218-y (2024).
Acknowledgements
The authors would like to thank the database of Genotypes and Phenotypes (dbGaP) data, including the dbGaP accession phs000298.v4.p3 used in this study. The data/analyses presented in the current publication are based on the use of study data downloaded from the dbGaP web site, under dbGaP accession phs000298.v1.p1; phs000298.v2.p2; phs000298.v3.p2; phs000298.v4.p3. Version 1–3: The data set(s) were deposited by the ARRA Autism Sequencing Collaborative, an ARRA-funded research initiative. Support for the Autism Sequencing Collaborative was provided by the following grants: R01- MH089208, awarded to Mark Daly; R01-MH089175, awarded to Richard Gibbs; R01-MH089025, awarded to Joseph Buxbaum; R01-MH089004, awarded to Gerard Schellenberg; and R01-MH089482, awarded to James Sutcliffe. Version 4: This work was supported by National Institutes of Health (NIH) grants U01MH100233, U01MH100209, U01MH100229 and U01MH100239 to the Autism Sequencing Consortium. Sequencing at Broad Institute was supported by NIH grants R01MH089208 (M.J.D.) and sequencing by U54 HG003067 (S. Gabriel, E. Lander) and UM1HG008895 (Gabriel, Lander). We acknowledge the clinicians and organizations that contributed to samples used in this study. Finally, we are grateful to the many families whose participation made this study possible. Copy Number Variant analysis was applied to the dataset of the Autism Genome Project (AGP), funded by NIMH, HRB, MRC, Autism Speaks, Hilibrand Foundation, Genome Canada, OGI and CIHR. We acknowledge the families who participated in these projects.
Funding
This research was funded by Fundação para a Ciência e a Tecnologia (FCT), Portugal, Centre grants to BioISI (DOI: https://doi.org/10.54499/UIDB/04046/2020, UIDP/04046/2020, UID/04046/2025), PAC-POCI-01-0145-FEDER-016428 MEDPERSYST and the National Institute of Health Doutor Ricardo Jorge. J.V., A.R.M. and J.X.S. are recipients of a fellowship from the BioSys PhD program PD65-2012 (J.V. Ref: PD/BD/131390/2017; A.R.M. Ref: PD/BD/113773/2015; and J.X.S. Ref: PD/BD/114386/2016) from Fundação para a Ciência e a Tecnologia (FCT), (Portugal). SM was funded under Fundação para a Ciência e a Tecnologia (Portugal) CEEC Grant ref 2022.00474.CEECIND/CP1714/CT0030(https://doi.org/10.54499/2022.00474.CEECIND/CP1714/CT0030).
Author information
Authors and Affiliations
Contributions
JV, AMV, ACP, IRV, MC-B and GO conceived and designed the study. JV, ARM, ACP, and SM collected the data, and JV, HM, ACP and IRV analyzed data under supervision from AMV, MC-B and GO. GO, AMV and MC-B provided resources to support data collection and analysis. ACP and JV created figures and tables. JV, ACP AMV, MC-B wrote the manuscript with support from GO, HM, JXS, ARM, CR, IRV and SM. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vilela, J., Pereira, A.C., Violante, I.R. et al. Neurometabolic profiles of autism spectrum disorder patients with genetic variants in specific neurotransmission and synaptic genes. Sci Rep 15, 36318 (2025). https://doi.org/10.1038/s41598-025-20090-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-20090-x