Abstract
Prostate cancer is characterized by an immunosuppressive tumour environment. This work combines Raman spectroscopy with group-and-bases-restricted non-negative matrix factorization (GBR-NMF) and machine learning to assemble models of immune cell densities within the needle-core biopsies of patients undergoing high-dose-rate brachytherapy (HDR-BT). Raman spectral acquisition, as well as immunohistochemistry staining of CD68\(^+\), CD3\(^+\), and \(\hbox {CD8}^+\) cells, was completed for biopsies collected before and 2 weeks following the first fraction of HDR-BT. Regression techniques, constructed using GBR-NMF scores, that produced the most accurate predictions of immune cell density by metrics of root mean-squared error (RMSE) and R\(^{2}\) were the gradient-boosted trees model of \(\hbox {CD68}^{+}\hbox {+ CD3}^{+}\) density (RMSE: 163 counts\(\hbox {/mm}^2\), \(\hbox {R}^{2}\): 0.65) and the elastic net model of \(\hbox {CD68}^{+}\)/ \(\hbox {CD8}^{+}\) (RMSE: 0.25, \(\hbox {R}^{2}\): 0.82). The accuracy of these models, herein defined as the fraction of patient predictions within \(\pm 1\) standard deviation of their measured values was 11/16 and 12/16, for CD68\(^{+}+\) CD3\(^{+}\) and CD68\(^{+}\)/ CD8\(^{+}\) models, respectively. To further delineate which metabolites were most important in the CD68\(^{+}\)/ CD8\(^+\) model, this ratio was further predicted in stromal and epithelial tissues within the biopsies, and resulting models utilized the GBR-NMF scores of glutathione, collagen, palmitic acid, and the pre- or post-HDR-BT label to produce an optimal performance level according to RMSE and R\(^2\). In summary, this study illustrates a novel methodology in which supervised machine learning techniques are used to model immune cells, which are prognostic indicators of disease progression.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) is the most frequent cancer diagnosis made in men and the third leading cause of cancer-related deaths in Canada1. When cancer is suspected, one of the most common methods of diagnosis is tissue biopsy2, and the gold standard of assessment for the grading and prognosis of PCa is done through histological assessment of Gleason score3,4. Standards for treatment include surgery and radiotherapy of localized disease, followed by active monitoring post-radiotherapy for the potential of biochemical recurrence2,5. Notably, fractionated high-dose-rate brachytherapy (HDR-BT), in which high doses are delivered using a high dose-rate radioactive source directly to the prostate glands through implanted catheters, has been successful as a monotherapy for localized PCa6,7.
Tumour-infiltrating immune cells are linked to tumorigenesis, as cancer-associated inflammation is a contributor to genomic instability, induction of cancer cell proliferation, stimulation of angiogenesis, and, eventually, cancer metastasis8,9. Localized PCa is characterized by an immunosuppressed phenotype whereby tumour-associated immune cells, such as macrophages and lymphocytes, are present at the tumour margin and within benign tissue but only sparsely present within the tumour itself10,11,12. Additionally, there is evidence to suggest immune cells, such as macrophages and T cells, are selectively enriched or depleted in cancerous prostate epithelium relative to surrounding normal tissue, and that these changes are distinct from those seen between benign and malignant prostate stroma13. The immune infiltration landscape within benign prostate tissue is furthermore heterogeneous among individuals, and although it remains evident that inflammation influences PCa development, the precise immune profile that determines clinical outcome remains unresolved as varying associations with prognosis have been found13,14,15.
A candidate thread of investigation to determine an underlying basis for individual-to-individual variation in immune infiltration is the interrogation of subcellular elements and their relative quantities. Metabolites which are integral to cellular processes are being investigated to ascertain their effects on antitumour activity and to derive new treatments and nutritional strategies for cancer patients16,17,18. Notable examples of metabolites found to alter cancer progression include methionine19, cysteine20,21, glutathione22,23,24, arginine25, and asparagine26. Several of these metabolites, such as methionine, cysteine, and glutathione are vital for T cell activation and proliferation27,28,29,30,31. Similarly, these metabolites have also been found to alter cellular responses to oxidative stress and DNA damage, factors which are crucial in radiation response32,33,34,35.
Tumour-infiltrating immune cells have been investigated for their predictive ability to delineate radiation treatment response in patients receiving radiation therapy36. The quantity of macrophages (CD68\(^+\)), cytotoxic cells (CD8\(^+\)), and lymphocytes (CD3\(^+\)) measured in biopsy cores have been associated with tumour aggression and biochemical recurrence14,37,38,39. In general, lower concentrations of CD68\(^+\) cells and higher concentrations of CD8\(^+\)CD3\(^+\) cells have been indicative of a more favourable prognosis40,41,42, although there remains variation in the strength of the association between these cells and the ability to accurately predict treatment outcome. Machine learning techniques have been employed to stratify patients based on immune cell densities and classify their treatment response43,44,45,46. For example, small to intermediate cohorts of patients are currently being used to identify associations between pre-treatment immune cell quantities in biopsies and post-treatment outcomes such as biochemical recurrence40,41. Here, we propose modelling the immune landscape directly prior to and after the first fraction of HDR-BT treatment utilizing the underlying biochemical signature of a patient’s tissue.
Raman spectroscopy (RS) is a label-free, non-invasive optical technique which results in a unique spectrum that is consistent with the composition of biochemicals present in the sample under investigation47. With the use of machine learning techniques, Raman spectra of biological materials can be deconstructed into multiple biochemical constituents, providing information on their relative abundance48. As an application tool for radiation therapy, RS has been used to study cellular response to ionizing radiation and the consequent effects on cellular metabolism49,50. Investigations of RS utility in radiation response monitoring have revealed its capacity to monitor in vivo biochemical changes and supplement understanding of the tumour microenvironment51. Furthermore, RS is currently being utilized as a predictive tool for radiation response in cell lines and clinical samples in conjunction with supervised machine learning techniques52,53,54. There remains significant potential for the use of RS as a predictive tool in the clinical setting, both for monitoring patient response to therapy and delineating effective treatment groups based on individual biochemical status. Herein, we propose the use of RS for the investigation of biochemical changes in biopsies and the prediction of immune infiltrates, with the overarching aim of elucidating the underlying biochemicals that are most relevant to the influx of immune cells in cancer.
In this study, we used the combination of RS with group- and basis-restricted non-negative matrix factorization (GBR-NMF)55 and supervised machine learning to build regression models of immune cell counts in biopsies of intermediate-risk prostate adenocarcinoma prior to and after the first fraction of HDR-BT. Using multiple immune cell densities involved in antitumor activity, as well as the spectral library of 36 biochemicals previously established by our group53,56,57, the ratio of CD68\(^+\)/ CD8\(^+\) and the sum of CD68\(^{+}+\) CD3\(^{+}\) cells were the most successfully predicted by metrics of relative root mean square error and accuracy. The major advantage of this methodology, and more generally, the goal of the development of a predictive tool for immune activity in biopsies from cancer patients, is the ability to uncover underlying relationships between the biochemical constituents of a patient’s sample and their unique intertumoral immune changes resulting from radiation treatment. The differentiation of those metabolites that pilot antitumoral immune activity and treatment response are necessary components for the discovery of new therapeutic targets and the building of individualized treatment regimens for cancer patients.
Results
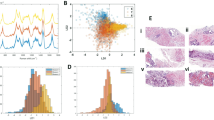
A A representative image of IHC multiplex staining showing a needle-core biopsy (i) pre-HDR-BT and (ii) post-first-fraction of HDR-BT from the same individual, highlighting an influx of CD68\(^+\) cells following radiation. Blue boxes are utilized to show the regions of each biopsy in which immune cells were counted. B Densities of immune cells expressing surface markers (i) CD68\(^+\), (ii) CD3\(^+\)CD8\(^-\), (iii) CD3\(^-\)CD8\(^+\) and (iv) CD3\(^+\)CD8\(^+\) across all regions and biopsies, separated by whether the biopsy was collected prior to or after the first fraction of HDR-BT. The thick horizontal bar represents the mean and the thin bar represents the median. C GBR-NMF score distributions of metabolites which significantly differed (\(\hbox {p} < 0.05\)) in pre- and post-first-fraction prostate biopsies. D Heatmaps displaying Spearman correlations between average individual immune parameters and average patient GBR-NMF metabolites in (i) both pre- and post-first-fraction data (n = 16), (ii) only pre-radiation data (n=8) and (iii) only post-first-fraction data (n=8). P-values were computed with the Wilcoxon sign-ranked matched pair test. ns = no significance, * \(\hbox {p} < 0.05\), ** \(\hbox {p} < 0.01\), *** \(\hbox {p} < 0.001\).
Immune cell density and group and basis restricted non-negative matrix factorization (GBR-NMF) score changes with radiation treatment
The major advantage of using a semi-supervised NMF method is the ability to track pre- and post-radiation changes in the biochemical composition of patients’ tumour biopsies. A summary of the RS and immune data obtained, as well as the tissue area sampled, from biopsy slices are presented in Table 1 for each of the eight individuals included in this study. For individual #4, a second pre-HDR-BT biopsy yielded no viable spectra following spectral pre-processing, and therefore this biopsy was not included in further analysis.
As shown in Fig. 1A and B, across all types of tissues, biopsies collected following the first fraction of HDR-BT contained significantly higher CD68\(^+\) density and significantly lower CD3\(^-\)CD8\(^+\) and CD8\(^+\)CD3\(^+\) densities, with no change observed in CD3\(^+\)CD8\(^-\) density. Patient biopsies contained, on average, a 2.5-fold increase in CD68\(^+\) density and a 2-fold decrease in CD8\(^+\)CD3\(^+\) density following the first 13.5 Gy fraction of HDR-BT. A similar trend of elevated CD68\(^+\) density and depleted CD3\(^+\)CD8\(^+\) density in post-radiation biopsies was observed in cell counts obtained from the stromal and epithelial tissue compartments (Figure S1). The mean pre-radiation cell density for cells expressing CD68\(^+\), CD3\(^+\) and CD8\(^+\) were 38 counts/mm\(^2\) (95% CI: 16 counts/mm\(^2\)– 59 counts/mm\(^2\)), 626 counts/mm\(^2\) (95% CI: 354 counts/mm\(^2\)– 897 counts/mm\(^2\)) and 60 counts/mm\(^2\) (95% CI: 28 counts/mm\(^2\)– 91 counts/mm\(^2\)), respectively. The mean post-radiation cell density for cells expressing CD68\(^+\), CD3\(^+\) and CD8\(^+\) were 93 counts/mm\(^2\) ( 95% CI: 38 counts/mm\(^2\)– 149 counts/mm\(^2\)), 476 counts/mm\(^2\) (95% CI: 321 counts/mm\(^2\)– 632 counts/mm\(^2\)) and 40 counts/mm\(^2\) (95% CI: 19 counts/mm\(^2\)– 62 counts/mm\(^2\)).
Additionally, an average GBR-NMF score for each metabolite of known spectra was obtained from each region of the RS slices to perform an analysis of the pre- and post-radiation changes in the distribution of metabolites across all individuals. A list of the 36 biochemical bases for which GBR-NMF scores were obtained is provided in Table S1. A two-tailed Wilcoxon rank-sum test was performed for all region-averaged GBR-NMF metabolite scores; glutathione scores significantly increased following radiation, and phosphatidylcholine scores significantly decreased following radiation (Fig. 1C). Examples of patient spectra, before their subsequent decomposition into GBR-NMF scores, are shown in Figure S2 for both pre- and post-first-fraction biopsies.
Figure 1D displays notable correlations (\(p<0.05\)) between average immune density values and average GBR-NMF scores (i) both pre- and post-radiation, (ii) pre-radiation only and (iii) post-radiation only. DNA, glycogen and phosphatidylserine scores were positively correlated with T cell density (\(p<0.05\)) and cysteine scores were negatively correlated with T cell density in biopsies prior to radiation (\(p<0.05\)). Following a fraction of HDR-BT, citric acid and glutathione scores were highly negatively correlated with CD8\(^+\) T cell cell density (\(p=0.0009\) and \(p=0.002\), respectively), and methionine and phosphatidylserine scores were positively correlated with macrophage density (CD68\(^+\); \(p=0.004\) and \(p=0.007\), respectively). Across all biopsies pre- and post-radiation, glutathione and glycerol scores were negatively correlated with CD8\(^+\) T cell density (\(p=0.001\)) and combined macrophage and T cell density (CD68\(^{+}+\) CD3\(^{+}\); \(p=0.002\)), respectively.
A A global root-mean-squared-error from the predictions made for all 8 patients, for several machine learning techniques including the median guess model. Models utilized median GBR-NMF scores from biopsies, and prediction RMSEs are provided for the following cell densities: (i) CD68\(^+\), (ii) CD8\(^+\), (iii) CD3\(^+\), (iv) CD68\(^+\) + CD3\(^+\), and the ratio (v) CD68\(^+\)/ CD8\(^+\). B Predictions of CD68\(^+\) + CD3\(^+\) for each patient produced by gradient-boosted trees regression using the average GBR-NMF scores, plotted with an average ± standard deviation alongside the features with the highest importance. C Predictions of CD68\(^+\)/ CD8\(^+\) for each patient produced by elastic net regression using the median GBR-NMF scores, plotted with an average ± standard deviation alongside the features with the highest absolute coefficients. For each patient’s immune cell density predictions, models were optimized based on the training set with that patient’s data left out. The elastic net technique was optimized by grid search of \(\alpha\) and L1 ratio resulting in lowest RMSE, and random forest, gradient-boosted trees and extra trees methods were optimized by iteratively removing features based on importance ranking until the feature list resulted in the lowest RMSE for a patient’s predictions. Top features shown here were ranked by (i) feature importance when utilized in the extra trees model and (ii) highest absolute coefficient value when utilized in the elastic net model. RMSE = root mean squared error; SD = standard deviation.
Regression model performance and optimization
Figure 2A displays RMSE values for linear regression, elastic net, random forest, gradient-boosted trees, and extra trees methods when predicting values of CD68\(^{+}\), CD8\(^{+}\) and CD3\(^{+}\) density. Additionally, the error in prediction results obtained for CD68\(^{+}+\) CD3\(^{+}\) density and CD68\(^{+}\)/ CD8\(^{+}\) ratio are displayed in Fig. 2A (iv) and (v), as these models resulted in the lowest RMSE (163 counts/mm\(^2\) and 0.25, respectively), highest R\(^2\) (0.65 and 0.82, respectively) and the highest accuracy in predictions falling within \(\pm 1\) standard deviation of measured data of all the immune parameters modeled (11/16 and 12/16, respectively). Moreover, the relative RMSE (prediction RMSE / range of measured immune quantity \(\times\) 100%) for the prediction of CD68\(^+\)+ CD3\(^+\) was 11%, and the relative RMSE for the prediction of CD68\(^+\)/ CD8\(^+\) was 6% across immune predictions for all patients.
To add context to the varying model performances shown, a median guess model was included that predicted the median value of the training set data (i.e., immune cell density) for each patient. Successful prediction models were visually distinct from unsuccessful models, as both outperformed the median guess model (Fig. 2A) and displayed predictions which followed the trend of the patient-averaged immune cell data (Fig. 2B). The top ranked features used in the gradient-boosted trees model of CD68\(^{+}+\) CD3\(^{+}\) and the elastic net model of CD68\(^{+}\)/ CD8\(^{+}\) are displayed alongside a plot of the predictions produced by these models in Fig. 2B and C. The (i) feature importance metric was utilized to rank the features displayed for the gradient-boosted tree model, and (ii) the absolute magnitude of the coefficients was used to rank the features displayed for the elastic net model with the lowest errors, defined by RMSE and R\(^{2}\). Glycerol, citric acid and DNA scores were ranked as the top 3 most important features for prediction of CD68\(^{+}+\) CD3\(^+\) cell density, and the treatment status with regards to the first fraction of HDR-BT (pre/post), followed by collagen, palmitic acid and glutathione scores were ranked as the most important features for prediction of the CD68\(^{+}\)/ CD8\(^{+}\) ratio. As a complement to 2B–C, the RMSE for both the training and cross-validation testing sets is provided for differing modelling techniques in the supplementary file (Figure S4). Furthermore, the images in 2B–C (ii) display truncated lists of the ranked features utilized in each model, comprising a total of 13 and 27 features of the original 38 feature list, for the gradient-boosted CD68\(^{+}+\) CD3\(^{+}\) and elastic net CD68\(^+\)/ CD8\(^+\) models. These complete lists are displayed in the supplementary file (Figure S5).
Furthermore, the ratio of CD68\(^+\)/ CD8\(^+\) was found to be highly indicative of treatment status. A majority (7/8) of pre-radiation biopsies had a CD68\(^+\)/ CD8\(^+\) ratio \(< 0.75\) and a majority (7/8) of biopsies collected following the first fraction of HDR-BT had a CD68\(^+\)/ CD8\(^+\) ratio \(> 0.75\). This demarcation is congruent with the aforementioned result that CD68\(^+\) and CD8\(^+\) densities differed between pre- and post-radiation samples (Fig. 1A).
Plots depicting predicted vs. measured values for CD68\(^+\)/ CD8\(^+\) ratio modelled by A random forest, B gradient boosted trees, C extra trees and D elastic net models prior to (i) and after (ii) feature selection. The red diagonal represents the trend line along which predictions would perfectly match the measured CD68\(^+\)/ CD8\(^+\) values. RMSE = root mean squared error.
An alternate representation of model performance is displayed in Fig. 3 to highlight model performance in predictions of CD68\(^{+}\)/ CD8\(^{+}\) using median GBR-NMF scores from each patient biopsy. The red diagonal line represents perfect predictions, wherein predictions result in an RMSE of zero. The R\(^{2}\) score is displayed to represent the proportion of the variation in the sampled CD68\(^{+}\)/ CD8\(^{+}\) values that is explained by each model: (A) random forest, (B) gradient-boosted trees, (C) extra trees, and (D) elastic net, respectively. Model performance is displayed for the instance of using (i) all 38 features and (ii) a shortened list of features, following feature selection. Predictions for biopsies collected prior to, and after, the first fraction of HDR-BT are included. Consequently, for 8 patients, each model produces 16 predictions. Therefore, improvements in the model predictions occur due to a meaningful condensing of the list of features included in the models.
Prediction performance across models utilizing different GBR-NMF score datasets
To capture the effect of the distribution of GBR-NMF scores observed in patient biopsies on model predictions, multiple features of the scores were utilized in prediction, including the average, median, variance, interquartile range, maximum, kurtosis and skew of the GBR-NMF scores across patient biopsies, as illustrated in Fig. 4. The most successful prediction models of CD68\(^+\)/ CD8\(^+\) ratio and CD68\(^{+}+\) CD3\(^{+}\) density used the median and average GBR-NMF scores from the biopsies, respectively, to obtain predictions with the lowest global RMSE (highlighted by the deepest green in Fig. 4). The feature of the GBR-NMF scores which allowed for the best prediction performance differed between cell types, for example, the model producing the lowest global RMSE in prediction of CD68\(^+\) density utilized the patients’ maximum GBR-NMF scores as its inputs. However, despite this variation in the dataset which produced the highest performance, it was observed that the models which were successful predictors of immune parameters (i.e., those which produced the smallest RMSE and greatest R\(^{2}\)), utilized either the mean or the median GBR-NMF scores for immune cell desnity modelling.
Heatmaps displaying global RMSE across all individuals for GBR-NMF datasets and machine learning techniques utilized in the prediction of A CD68\(^+\), B CD3\(^+\), C CD68\(^+\) + CD3\(^+\), D CD8\(^+\), and E CD68\(^+\)/ CD8\(^+\). The median guess model is included to contextualize prediction results and for comparison to the cumulative RMSE produced by a simple guess of the median in the training dataset for each individual.
Prediction of immune parameters in stromal and epithelial tissue
The degree of macrophage infiltration and cytotoxic T cell depletion following HDR-BT was found to differ between epithelial and stromal compartments (see Figure S1). Figure 5 illustrates the global prediction RMSE for models of (A) stromal CD68\(^+\)/ CD8\(^+\) ratio and (B) epithelial CD68\(^+\)/ CD8\(^+\) ratio. Moreover, the predictions for the model that produced the lowest RMSE (extra trees, utilizing median GBR-NMF scores) is displayed for both tissue compartments, along with the features used in the model. The extra tree model was most successful for predicting stromal and epithelial CD68\(^+\)/ CD8\(^+\); this model structure produced the lowest RMSE and highest R\(^2\) scores using only the top 3 features from the original 38 features. The median metabolite scores obtained from the GBR-NMF algorithm produced the lowest RMSE compared to utilizing other biopsy metrics, such as the average, maximum or variance of the scores. Furthermore, the optimized elastic net and extra trees models which were obtained following feature analysis agreed on which of the input features produced the most accurate model: glutathione and collagen scores, alongisde the treatment response label (pre/post) were utilized in prediction models of CD68\(^+\)/ CD8\(^+\). In particular, glutathione scores were used in the stromal immune model, collagen scores were used in the epithelial immune model, and the treatment status label (pre/post) was utilized across all tissue models of CD68\(^+\)/ CD8\(^+\).
Analysis of A stromal CD68\(^+\)/ CD8\(^+\) and B epithelial CD68\(^+\)/ CD8\(^+\) prediction, visualized by (i) a heatmap of global RMSE across all patients for GBR-NMF datasets and varying machine learning techniques, (ii) a plot of predictions and measured immune data using median GBR-NMF scores and optimized extra trees, and (iii) a plot of feature importances, ordered from highest importance to lowest importance. RMSE root mean squared error; SD standard deviation.
Discussion
The Raman spectra of biopsies collected prior to, and following, the first fraction of HDR-BT from eight patients with intermediate risk prostate adenocarcinoma were decomposed using a GBR-NMF model55, producing relative scores indicative of the contribution of 36 metabolites known to be present in human tissue. The GBR-NMF scores of the metabolites were summarized as average, median, variance, interquartile range, maximum, kurtosis or skew measures, illustrating the distribution of select metabolites across an patient’s biopsy prior to, and following, the first fraction of HDR-BT. The metabolite scores were subsequently used to construct linear (simply, elastic net) and ensemble decision tree (random forest, gradient-boosted trees, extra trees) predictive models to sample the features of the metabolite distributions across all individuals which yielded superior predictions of CD68\(^{+}\), CD8\(^{+}\) and CD3\(^{+}\) immune cell densities. The GBR-NMF elastic net and GBR-NMF extra trees models allowed for the prediction of CD68\(^{+}\)/ CD8\(^{+}\) in patient biopsies across all tissue types, and within the subcategories of tissues within the biopsy (stromal, epithelial), respectively, using median metabolite scores and a pre/post radiation label for each patient biopsy as input features to the model. The GBR-NMF gradient-boosted trees model allowed for the prediction of CD68\(^{+}+\) CD3\(^{+}\) across all tissue types, using average metabolite scores for each patient biopsy. Predictive elastic net models were optimized by tuning \(\alpha\) and L1 ratio parameters and feature significance to the model was interpreted using the absolute coefficients in the linear equation constructed by the model. Predictive tree models (gradient-boosted trees, extra trees and random forest) were optimized by iterative elimination of features when ranked by importance. Hence, the GBR-NMF model in conjunction with machine learning methods allowed for the prediction of immune cell parameters and the identification of biochemicals which were important in the regression.
This study demonstrated the capability of RS as a predictive tool for monitoring radiation response within cancerous prostate biopsies by establishing a novel methodology which combines GBR-NMF with comparative machine learning, in turn producing successful regression models of select intra-biopsy immune markers. The ability to predict immune parameters is significant from a clinical viewpoint, as tumour-infiltrating immune cells have been thoroughly investigated for their prognostic potential in determining radiotherapy treatment outcomes58. Our analysis supported the literature evidence that macrophage density increases in tumour-surrounding tissue following radiotherapy to the prostate59,60. Similar to Kane et al., our data demonstrated a depletion by 50% in CD8\(^+\)CD3\(^+\) density in prostate biopsies 2 weeks following radiation to the prostate61.
Of the 36 biochemical bases for which GBR-NMF scores were obtained, including amino acids, antioxidants, fatty acids and phospholipids, only glutathione and phosphatidylcholine scores were found to significantly change in post-radiation biopsies relative to pre-radiation biopsies (\(p = 0.025\) and \(p = 0.030\), respectively). Post-HDR-BT glutathione scores displayed a right skew and phosphatidylcholine scores displayed a left skew relative to pre-HDR-BT scores (Fig. 1C). Higher post-radiation glutathione supports the conclusion by Mukha et al. that prostate cancer cells have a high demand for the glutathione precursor, glutamine62. Glutathione has also been found to provide greater radioresistance through the neutralization of reactive oxygen species, thereby fostering a more favourable environment for carcinogenesis23,63 and providing resistance to chemotherapy drugs64,65. Another GBR-NMF metabolite found to change following radiation was phosphatidylcholine. Likewise, Ferreira et al. found a persistent decrease in serum phosphatidylcholine in the weeks following pelvic intensity-modulated radiotherapy in a cohort of thirty-two high-risk PCa patients66. While glutathione and phosphatidylcholine scores differed post-radiation across all patient measurements, these radiation changes in these metabolites were not seen uniformly in data collected from each individual. Glutathione scores and phosphatidylcholine significantly increased and decreased, respectively, in 5/8 patients following radiation. Future work may be done to validate whether these changes, on an individual scale, can be linked to post-treatment outcomes.
Correlations between average GBR-NMF scores and average immune parameters revealed relationships between methionine scores in prostate biopsies and the presence of macrophages and T cells. Exogenous methionine is required during T-cell proliferation and differentiation to support protein synthesis and for the methylation reactions required during T-cell activation67. Methionine derived from apoptotic cells further supports tissue resolution and efferocytosis by macrophages68. Furthermore, correlations were found between scores of glutathione, cysteine and cholesterol and ratios of T cells and cytotoxic cells. These findings are supported by evidence that activated T cells control their intracellular concentrations of reactive oxygen species with endogenous antioxidants such as glutathione, whose synthesis is controlled by glutamate and cysteine concentrations69. Cholesterol is also required for the activation and proliferation of T cells, and Ma et al. describe an association between T cell function and exhaustion, a characteristic found in immunosuppressed prostate tumor environments, and cholesterol concentration within prostate tissue70.
A multitude of studies have stratified patients with PCa receiving radiotherapy into risk groups based on immune marker-based prognostic factors46. Herein, we demonstrate the first utilization of RS, an optical spectroscopy technique that has recently displayed improved accuracy in the diagnosis of prostate cancer71,72, with supervised machine learning techniques to develop a predictive model of intraprostatic immune infiltration.
Linear regression and decision tree ensemble methods were investigated in this study for their potential to predict immune parameters using RS data from prostate tissue biopsies. Overall, elastic net regression was found to outperform linear regression as an immune prediction model. Elastic net regression addresses the limitations of Lasso and Ridge methods to provide a regularization method that accomplishes feature selection and can utilize a higher number of features in its prediction than samples73,74, therefore making this method ideal for the typically small sample sizes seen in studies using clinical samples. We observed in our study that the extra trees and gradient-boosted trees regression methods consistently outperformed random forest regression by the metric of global RMSE across all individual immune predictions. The extra trees method is similar to random forests, but rather than utilizing a bootstrapped subset of the data for each tree, the algorithm creates a dataset for each tree with unique samples from the entire dataset, without replacement75. This reduces overfitting of the training set but may lead to a tradeoff of increased bias76. Gradient-boosted trees, on the other hand, combines multiple weaker decision trees to create a strong predictor by iteratively improving the model with consecutive trees that predict the residual error of the previous trees76. Gradient-boosted trees, although more computationally expensive, may lead to stronger predictions in regression tasks. Therefore, these features of the extra trees and gradient-boosted trees algorithms likely led to superior predictions over the random forest algorithm in our immune dataset of small sample size.
In the prediction of CD68\(^+\)/ CD8\(^+\) using elastic net regression, the radiation label alongside the GBR-NMF scores of palmitic acid, glutathione, and collagen were among the largest absolute coefficients in the model. In subsequent analyses of CD68\(^+\)/ CD8\(^+\) in stroma and epithelial tissue segments, extra trees models confirmed the importance of these metabolites, as glutathione, collagen and radiation label remained the highest ranked features by importance. Optimized regression models were constructed using only a few of the original 37 features; in particular, stromal CD68\(^+\)/ CD8\(^+\) ratio was predicted using the radiation label, glutathione and citric acid, while epithelial CD68\(^+\)/ CD8\(^+\) ratio was predicted using the radiation label, collagen and histidine. As the most abundant antioxidant that maintains cellular homeostasis, glutathione biosynthesis is required for inflammatory T cell responses69,77 and regulatory T cell suppressive function78. Subpopulations of innate immune cells, such as M1 macrophages, are characterized by high glycolysis and increased glutathione79. Li et al. found that the isoenzyme glutathione s-transferase pi was upregulated in the stroma of surgical specimens from patients who developed hormone-independent PCa80. Citrate has also been shown to induce pro- or anti-inflammatory macrophage polarization79 and has been found to significantly lower with the increase in reactive stromal tissue grading81. Additionally, glutathione scores and citric acid scores in patient biopsies analysed in this study were found to be highly negatively correlated (\(R < -0.9\)) with cytotoxic cells (Fig. 1D). For the prediction of CD68\(^+\)/ CD8\(^+\) in epithelial tissue, collagen and histidine were found to be the most important metabolites for predictive performance. There is evidence to support that collagen protein expression is up-regulated in human PCa and in also in PCa metastases82. Afik et al. also found collagen deposition and linearization to be positively associated with macrophage activity, and that denatured collagen is a chemoattractant for macrophages that mediate the promotion of cancer83. Histidine is influenced immunologically by the enzyme histidine decarboxylase, which converts histidine to histamine and is present on macrophages and T cells84,85. Histamine is a potent inflammatory molecule with high chemoattractant activity, and is associated with cellular maturation, activation, polarization, and effector functions, as well as the promotion of vascular and tissue changes86,87. Finally, although palmitic acid was found to be an important feature in the prediction of CD68\(^+\)/ CD8\(^+\) across all tissue types, it was not utilized in the modelling of this ratio in stromal or epithelial tissue subtypes; it is possible that palmitic acid’s high feature importance arose from its metabolic contribution to the tissue type labelled as”other” in the biopsies, and a more variable or heightened concentration of this saturated fatty acid could be a result of prostatic lipogenesis, a process vital in the growth and survival of PCa cells88. The prediction of CD68\(^+\)/ CD8\(^+\) in the tissue compartment labelled as”other” is displayed in Figure S3, and illustrates the high feature importance of palmitic acid.
Similar means and ranges of immune cell quantities as the ones displayed here (Fig. 1B) have been found in other studies which utilize IHC evaluation to obtain immune cell counts in prostate biopsy tissue40,60,61. Since the predictive modelling of immune parameters was demonstrated using regression techniques, errors were provided as global RMSEs across all individual predictions for each method. Furthermore, the relative RMSEs in the CD68\(^{+}+\) CD3\(^{+}\) and CD68\(^{+}\)/ CD8\(^{+}\) models which performed the best are approximately 13% and 6% of the overall range of these quantities measured in the patient biopsies. Therefore, the combination of RS and machine learning techniques may be a promising methodology for stratifying patients into prognostic groupings, in addition to providing information regarding the underlying biochemical composition of cancer tissue.
Machine learning analysis with RS data is well suited for studies that are posed to investigate the radiation response in the form of acute or late radiation toxicity, in which case the use of spectroscopy may be an additional form of tissue interrogation— along with the standard methods of histopathological staining— that provides metabolic information with a heightened resolution relative to cellular components and tissue substructures. In the broader scope, Raman spectroscopy may provide real-time, in vivo biological information during surgical application, with the field progressing toward instruments that may be used to probe for the presence and severity of malignancy directly in the surgical context89,90,91. Although the standard of practice for utilizing Raman spectroscopy to investigate radiation response and treatment outcomes has thus far been through the interrogation of tissue biopsies, recent studies have outlined the use of wet and dry blood plasma for detecting and diagnosing various cancers92,93. For example, Schiemer et al. describe the potential of both Attenuated Total Reflection-Fourier Transform Infrared (ATR-FtIR) and Raman spectroscopy techniques for the diagnosis of varying histological types of endometrial cancer94. Furthermore, comparisons between Raman spectra of plasma from patients with PCa and healthy volunteers reveal an increase in the bands associated with nucleic acids, potentially deriving from cell-free DNA within the blood95. These examples provide an avenue for building the utility of Raman spectroscopy for non-invasive cancer detection.
Finally, liquid biopsies may provide an attractive alternative to tissue biopsies for monitoring radiation treatment response due to their ability to provide a real-time view of the patient’s biomolecular composition using a non-invasive collection method. Using FITR spectroscopy and partial least squares discriminant analysis, Medipally et al. analysed spectra from 53 PCa patients receiving hormone and radiation therapies and achieved sensitivity and specificity rates from 80–99% when discriminating between pre- and post-radiation timepoints and severe acute or late toxicities96. RS has also demonstrated potential to distinguish the lymphocyte spectral signatures in PCa patients with minimal or severe late toxicity following dose escalation in IMRT up to 81 Gy97. Blood serum has been used to classify responders from non-responders following radiochemotherapy in rectal adenocarcinoma patients with a 73% accuracy98 and to distinguish between different phases of radiochemotherapy in cervical carcinoma with moderate results99. RS has further been used to explore radiation effects on whole peripheral blood as well as peripheral mononuclear cells and lymphocytes100. Components of peripheral blood, and in particular lymphocytes and circulating tumour DNA, have been thoroughly investigated for their relation to radiation treatment outcomes101,102,103.
The limitations of this study included the relatively small sample size of individuals (\(n = 8\)), as well as the drawback which accompanies working with small (\(\sim 0.1\) cm thick), frozen samples: the lack of an ability to maintain the structural similarity between adjacent slices of tissue due to folding, stretching, and uneven mounting in frozen media. Hence, while the slices used for RS and immunostaining from each biopsy were taken from adjacent slices of the same tissue, there is a lack of spatial correlation between regions of sampling in the RS slice and regions of sampling in the slice used for immunostaining. As such, there is no certainty that the stromal and epithelial compartments of the biopsies were equivalently sampled within the RS and IHC slices, and the area sampled in each region of the RS slice (120–160 \(\upmu\)m\(^2\)) was much smaller than the area of the adjacent slice sampled for cell counts (1–2 mm\(^2\)). Future work may be done to target the spatial distribution of immune cells within biopsies and whether these are associated with the relative distribution of the biochemicals measured and sampled with RS. Lastly, it was observed that the range of the immune data across regions of a biopsy varied between individuals, with some patients exhibiting a much larger variation in cell count across their sampled tissue; future work may investigate whether these variations in immune response were due to the sampling of tissue structures differing between patients or whether this variation bears any relation to individual radiation response.
The results reported here, including the predictive modelling methodology and the biochemicals associated with immune response, could be utilized in identifying key biological targets for monitoring radiation response. As RS was used to model immune cell densities, these results may further contribute to the development of a biological profile which can serve as a prognostic indicator for radiotherapy treatment outcome.
Methodology for determining the machine learning model with the lowest global RMSE while using a LOO cross-validation structure. Permutations of the 36 GBR-NMF summary scores and immune targets are computed and aggregated for each individual to produce a dataset to for immune prediction modelling. Model training is then completed for each training/testing split and the model which produces the smallest RMSE is utilized for further feature analysis. IQR interquartile range; RMSE root mean squared error.
Methods
Patient cohort
The eight patients included in this study were eligible for HDR-BT as a monotherapy for intermediate-risk prostate adenocarcinoma; inclusion criteria for this trial are specified in the clinical trial ID NCT03426748 (https://clinicaltrials.gov/study/NCT03426748). Informed consent for the use of bio-specimens in research was provided from all patients included in this study. Moreover, all biopsies discussed in this study were taken from patients subsequent to, and in addition to, those collected for the purpose of histopathological assessment of tumour changes due to radiotherapy treatment, as described by the aforementioned clinical trial.
Methods and procedures are in accordance with guidelines provided by Health Canada and Public Health Agency of Canada Research Ethics Board, and the study has been approved by the University of British Columbia and BC Cancer Research Ethics Board (REB# H17-02904). Prior to biopsy collection, each patient was given a multi-parametric MRI to identify the location of the dominant lesion; this location was then transferred by rigid registration to transrectal ultrasound (TRUS) such that it could be used for biopsy collection and dose escalation during therapy. The prescribed dose for the HDR-BT regimen was 27 Gy and was delivered in 2 fractions during procedures that were 2 weeks apart, as previously described56.
Biopsy collection
Under general anesthesia, biopsies were collected by radiation oncologists from the patients’ dominant lesion site(s) immediately preceding each fraction of HDR-BT: the first was taken prior to any radiation treatment, and a second was taken 2 weeks following the initial fraction (13.5 Gy) and directly prior to the delivery of the second fraction (13.5 Gy). The biopsies were collected transperineally using a needle-grid template and ultrasound guidance based on prior MRI and TRUS image registration. Once removed, the needle core biopsies (approximately \(0.1 \times 1.0\) cm) were placed in hypothermosol (Termo Fisher Scientifc, MA, USA) and transported to the University of British Columbia Okanagan.
Sample preparation for RS and IHC
Needle-core biopsies were laid flat and embedded in mounting medium (Tissue-Plus™ O.C.T. Compound, Fisher Scientific, MA, USA) and stored at \(-80^{\circ }\,\hbox {C}\). A 20 micron-thick slice of the embedded tissue was taken from each O.C.T. block using a rotary cryostat (HM 550; MICROM International GmbH, Walldorf, Germany) and placed on a magnesium fluoride slide. The O.C.T. blocks containing the remainder of the tissue biopsies were transported on dry ice to the Trev & Joyce Deeley Research Centre at BC Cancer Victoria for a triple IHC staining protocol of CD3, CD8 and CD68.
Triple IHC staining
CD3, CD8 and CD68 antigen retrieval was performed at \(110^{\circ }\,\hbox {C}\) for 15 min in a Biocare decloaking chamber using Diva decloaking solution (Biocare Medical, Concord, CA, USA) before loading the slides onto a Biocare Intellipath FLX Autostainer. The multi-colour immunofluorescence process involved polymers from Biocare.
The first round of staining involved performing endogenous peroxidase blocking with Peroxidazed 1 (Biocare) followed by non-specific blocking with Background Sniper (Biocare). Primary antibodies CD3 and CD3 (shown in Table 2) are manually diluted with Da Vinci Green (Biocare) before application to slides for 30 min at room temperature, followed by Mach2 Rabbit-HRP polymer (Biocare) for 30 min at room temperature. Ferrengi Blue (Biocare) and DAB (Biocare) chromogens was then applied for 10 min at room temperature; then the slides were incubated with periodic agitation in denaturing solution for 45 min at room temperature. The second round of staining followed suite similarly to the first, with CD68 diluted with Da Vinci Green (Biocare) before application to slides for 30 min at room temperature, followed by Rabbit-AP polymer (Biocare) for 30 min at room temperature. Warp Red (Biocare) and counter stain CAT hematoxylin (Biocare) were then applied for 7 and 5 min, respectively, at room temperature. Finally, the slides were removed from the Intellipath FLX, air dried, and coverslipped with Prolong Diamond Antifade mounting media (Thermo Fisher Scientific).
IHC image analysis
All slides were then scanned using a Vectra automated imaging system (Perkin Elmer, Waltham, MA, USA) at 4\(\times\), and 10 fields were captured for each slide at 20\(\times\). inForm image analysis software was used to process the images (Perkin Elmer, Waltham, MA, USA), running tissue segmentation algorithms to distinguish the cells present in the tissue stroma (stromal compartment) from the cells found within the tumour tissue (epithelial compartment) and generating cell phenotype counts for each slide, defined as the combination of markers that each cell expresses. The counts were inspected visually to ensure the results were reliable. The tissue segmented regions (epithelial or stromal) for each slide were converted from the number of pixels to the area of tissue (mm\(^2\)). The average cell counts for each phenotype were then normalised by the segmented area to obtain the cell density counts for each region, and these regions were averaged across the entirety of each individual’s biopsy for pre- and post-radiation samples.
To capture a range of potential intra-biopsy immune interactions, summations and divisions of the four immune cell markers counted with IHC (CD68\(^+\), CD3\(^+\)CD8\(^-\), CD\(^-\)CD8\(^+\) and CD3\(^+\)CD8\(^+\)) were computed for each biopsy region, and an average was computed for these metrics across each individual’s pre- and post-radiation biopsy tissues. The list of immune parameters investigated is summarized in Table S2. Significance values between the pre- and post-radiation immmune cell densities and ratios were calculated using a two-tailed Wilcoxon rank-sum test.
Spectral acquisition
Spectra were acquired using a Renishaw InVia Raman microscope (\(100 \times\) dry objective, 785 nm excitation laser, 30 s acquisition time and a power density of \(0.5 \text {mW}/\upmu \text {m}^3\)). Spectra were baseline corrected, normalised and smoothed using a Savitsky-Golay filtering algorithm. For each biopsy slice, 3 randomly selected regions along the core were chosen for analysis, and Raman spectra were acquired from a 90 to 120 micron squared area, using 15 micron step size (\(8 \times 8\), \(6 \times 8\) or \(6 \times 9\) grid). Spectra were acquired over the spectral range of 450-1800 cm-1. The number of biopsy cores obtained from each patient, and the spectra retained from each biopsy collected pre- and post-first-fraction of HDR-BT, are listed in Table 1.
Dimensionality reduction and supervised machine learning
As previously described55, the GBR-NMF algorithm was used to decompose the spectral matrix X into its matrix constituents WAS , where W is the weight matrix, A is the auxiliary matrix, and S is the matrix containing spectral bases. For each region of Raman data collected from a patient’s biopsy, the GBR-NMF algorithm was used to decompose the acquired spectra into a library of 36 known spectral bases and one unconstrained base. As illustrated in Fig. 6, several summary metrics (i.e., average, median, variance, inter-quartile range, maximum, kurtosis and skew) were computed for the GBR-NMF bases scores for each individual’s pre- and post-radiation tissue and these summary metrics were subsequently standardized using a z-score and utilized for predictive modelling of immune cell densities.
Datasets consisted of 38 total features for each patient biopsy, including 37 bases of metabolite scores and one radiation label, describing whether the biopsy was collected pre- or post-first-fraction of HDR-BT. These features were used to model average immune cell values comprising of the four cell markers measured for all patients. GBR-NMF in tandem with decision tree ensemble (random forest, extra trees, gradient-boosted trees) and linear (simple, elastic net) methods were used to model immune cell density parameters using a Leave-One-Out (LOO) cross-validation methodology. In the LOO methodology, one individual’s data was left out of the training set and then utilized to test the model by predicting that patient’s immune value, both pre- and post-first-fraction of HDR-BT. This resulted in 8 folds of cross-validation, one for each patient, with two predictions made in each fold for pre- and post-treatment immune values. Prediction errors were reported as a global RMSE across all cross-validation folds for each model structure, resulting in an RMSE for each type of model (simple linear, elastic net, random forest, gradient-boosted trees, and extra trees). A baseline median guess model was added for context on the various models’ performances; this guess model predicted the median value of the training set for each left-out patient, and this was repeated for all cross-validation folds. Therefore, the RMSE of the median guess model may be interpreted as the error that would result from an educated guess of each patient’s immune value, and models which have smaller RMSEs have, on average, made predictions which out-perform an educated guess.
Feature selection across models
For each decision tree ensemble method, feature selection using importance ranking was utilized to determine a limited feature list which improved the model’s performance. The top 3 to 15 features ranked by feature importance in the 38-feature model were iteratively implemented in the training and testing of random forest, extra trees, and gradient-boosted trees methods, using 100 estimators and a tree depth of 3, to ascertain the relevant features pertaining to the modelling of a particular type, or combination of, immune cells. Through this process, features ranked to have the lowest importance were not included in the next iteration of the decision tree model. Models with decreasing numbers of features were found to display recurring metabolites in their top 3 to 5 list of features when the metabolites were ranked by the importance metric. This process indicates the validity of this metric for keeping and discarding features from the overall feature list. Once feature selection was complete, the decision tree model that produced the lowest global RMSE across all cross-validation folds was stored.
In the elastic net method, a grid search was performed to obtain the \(\alpha\) and L1 ratio parameters that optimized the RMSE performance within the LOO cross-validation structure, thereby obtaining an optimal \(\alpha\) and L1 ratio for each of the models produced for the eight individuals included in this study. The elastic net model which used the optimal \(\alpha\) and L1 ratio was compared to other models using their global RMSE.
The feature selection steps outlined above included comparing the global RMSEs produced by each stored linear and decision tree-based model to the baseline median guess model, to determine the relative quality of their predictions with that of a model replicating an educated guess of the median of the training set.
Models that produced the smallest global RMSE utilizing the LOO cross-validation structure were then visualized with plots of predictions vs. actual data to ascertain whether these predictions were successful (as shown in Fig. 3). Immune targets that were successfully modelled were those in which the (1) stratification of model predictions was seen, such that the optimal model following feature selection performed better than the median guess model, and (2) the predictions followed the trendline in the actual immune data across all individuals. Finally, these successful parameters were plotted using their average and standard deviation on the same axes as the true measured immune parameter quantities to illustrate model performance and RMSE.
All data analysis was completed using Python v3.12. The packages and modules utilized for plotting, implementing machine learning techniques and feature selection, and statistical analysis were matplotlib and seaborn, scikit-learn, and statsmodels, respectively.
Data availibility
The code and data from this study have been uploaded to an OSF repository and are available upon request. Please contact the corresponding author, Andrew Jirasek (email: andrew.jirasek@ubc.ca).
References
Brenner, D. et al. Canadian cancer statistics 2023. Tech. Rep., Canadian Cancer Statistics Advisory Committee in collaboration with the Canadian Cancer Society, Statistics Canada and the Public Health Agency of Canada (2023).
Sekhoacha, M. et al. Prostate cancer review: Genetics, diagnosis, treatment options, and alternative approaches. Molecules https://doi.org/10.3390/molecules27175730 (2022).
Gleason, D. F. Histologic grading of prostate cancer: A perspective. Hum. Pathol. 23, 273–279 (1992).
Egevad, L., Granfors, T., Karlberg, L., Bergh, A. & Stattin, P. Prognostic value of the Gleason score in prostate cancer. BJU Int. 89, 538–542. https://doi.org/10.1046/j.1464-410X.2002.02669.x (2002).
Strouthos, I., Karagiannis, E., Zamboglou, N. & Ferentinos, K. High-dose-rate brachytherapy for prostate cancer: Rationale, current applications, and clinical outcome. Cancer Rep. 5, e1450. https://doi.org/10.1002/cnr2.1450 (2022).
Mendez, L. C. & Morton, G. C. High dose-rate brachytherapy in the treatment of prostate cancer. Transl. Androl. Urol. 7, 357–370. https://doi.org/10.21037/tau.2017.12.08 (2018).
Soatti, C. P. et al. High-dose-rate brachytherapy as monotherapy for localized prostate cancer using three different doses - 14 years of single-centre experience. J. Contemp. Brachyther. 12, 533–539. https://doi.org/10.5114/JCB.2020.101685 (2021).
Gonzalez, H., Hagerling, C. & Werb, Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. https://doi.org/10.1101/gad.314617 (2018).
Strasner, A. & Karin, M. Immune infiltration and prostate cancer. Front. Oncol. 5, 128. https://doi.org/10.3389/fonc.2015.00128 (2015).
Ebelt, K. et al. Prostate cancer lesions are surrounded by foxp3+, pd-1+ and b7–h1+ lymphocyte clusters. Eur. J. Cancer 45, 1664–1672. https://doi.org/10.1016/j.ejca.2009.02.015 (2009).
Fujii, T. et al. Immunohistochemical analysis of inflammatory cells in benign and precancerous lesions and carcinoma of the prostate. Pathobiology 80, 119–126. https://doi.org/10.1159/000342396 (2013).
Hussein, M. R. A., AL-Assiri, M. & Musalam, A. O. Phenotypic characterization of the infiltrating immune cells in normal prostate, benign nodular prostatic hyperplasia and prostatic adenocarcinoma. Exp. Mol. Pathol. 86, 108–113. https://doi.org/10.1016/j.yexmp.2008.11.010 (2009).
Andersen, L. B. et al. Immune cell analyses of the tumor microenvironment in prostate cancer highlight infiltrating regulatory t cells and macrophages as adverse prognostic factors. J. Pathol. 255, 155–165. https://doi.org/10.1002/path.5757 (2021).
Zhang, Q. et al. Tumor infiltrating m2 macrophages could predict biochemical recurrence of localized prostate cancer after radical prostatectomy. Exp. Cell Res. 384, 111588. https://doi.org/10.1016/j.yexcr.2019.111588 (2019).
Rådestad, E. et al. Characterization of infiltrating lymphocytes in human benign and malignant prostate tissue. Oncotarget 8, 60257–60269 (2017).
Ahmad, F., Cherukuri, M. K. & Choyke, P. L. Metabolic reprogramming in prostate cancer. Br. J. Cancer 125, 1185–1196. https://doi.org/10.1038/s41416-021-01435-5 (2021).
Ma, S., Ming, Y., Wu, J. & Cui, G. Cellular metabolism regulates the differentiation and function of t-cell subsets. Cell. Mol. Immunol. 21, 419–435. https://doi.org/10.1038/s41423-024-01148-8 (2024).
Cardoso, H. J. et al. Revisiting prostate cancer metabolism: From metabolites to disease and therapy. Med. Res. Rev. 41, 1499–1538. https://doi.org/10.1002/med.21766 (2021).
Ji, M., Xu, Q. & Li, X. Dietary methionine restriction in cancer development and antitumor immunity. Trends Endocrinol. Metabol. 35, 400–412. https://doi.org/10.1016/j.tem.2024.01.009 (2024).
Rudzińska, M. et al. The role of cysteine cathepsins in cancer progression and drug resistance. Int. J. Mol. Sci. 20, 3602. https://doi.org/10.3390/ijms20143602 (2019).
Srivastava, M. K., Sinha, P., Clements, V. K., Rodriguez, P. & Ostrand-Rosenberg, S. Myeloid-derived suppressor cells inhibit t-cell activation by depleting cystine and cysteine. Cancer Res. 70, 68–77. https://doi.org/10.1158/0008-5472.CAN-09-2587 (2010).
Diotallevi, M. et al. Glutathione fine-tunes the innate immune response toward antiviral pathways in a macrophage cell line independently of its antioxidant properties. Front. Immunol. 8, 289507. https://doi.org/10.3389/fimmu.2017.01239 (2017).
Kanwal, R. et al. Protection against oxidative dna damage and stress in human prostate by glutathione s-transferase p1. Mol. Carcinogenes 53, 8–18. https://doi.org/10.1002/mc.21939 (2014).
Kwon, D. H. et al. Protective effect of glutathione against oxidative stress-induced cytotoxicity in raw 264.7 macrophages through activating the nuclear factor erythroid 2-related factor-2/heme oxygenase-1 pathway. Antioxidants 8, 82 (2019).
Matos, A., Carvalho, M., Bicho, M. & Ribeiro, R. Arginine and arginases modulate metabolism, tumor microenvironment and prostate cancer progression. Nutrients 13, 4503. https://doi.org/10.3390/nu13124503 (2021).
Wang, X., Gong, W., Xiong, X., Jia, X. & Xu, J. Asparagine: A key metabolic junction in targeted tumor therapy. Pharmacol. Res. 206, 107292. https://doi.org/10.1016/j.phrs.2024.107292 (2024).
Pandit, M. et al. Methionine consumption by cancer cells drives a progressive upregulation of pd-1 expression in cd4 t cells. Nat. Commun. 14, 2593. https://doi.org/10.1038/s41467-023-38316-9 (2023).
Li, T. et al. Methionine deficiency facilitates antitumour immunity by altering m 6 a methylation of immune checkpoint transcripts. Gut 72, 501–511. https://doi.org/10.1136/gutjnl-2022-326928 (2023).
Mak, T. W. et al. Glutathione primes t cell metabolism for inflammation. Immunity 46, 675–689. https://doi.org/10.1016/j.immuni.2017.03.019 (2017).
Ma, S., Ming, Y., Wu, J. & Cui, G. Cellular metabolism regulates the differentiation and function of t-cell subsets. Cell. Mol. Immunol. 21, 419–435. https://doi.org/10.1038/s41423-024-01148-8 (2024).
Wang, W. & Zou, W. Amino acids and their transporters in t cell immunity and cancer therapy. Mol. Cell 80, 384–395. https://doi.org/10.1016/j.molcel.2020.09.006 (2020).
Bin, P., Huang, R. & Zhou, X. Oxidation resistance of the sulfur amino acids: Methionine and cysteine. BioMed Res. Int. 2017, 9584932. https://doi.org/10.1155/2017/9584932 (2017).
Yang, P. et al. Ionizing radiation upregulates glutamine metabolism and induces cell death via accumulation of reactive oxygen species. Oxidative Med. Cell. Longev. 2021, 5826932. https://doi.org/10.1155/2021/5826932 (2021).
Islam, M. M. et al. Impact of dietary ingredients on radioprotection and radiosensitization: a comprehensive review. Ann. Med. 56, 2396558. https://doi.org/10.1080/07853890.2024.2396558 (2024).
Miousse, I. R. et al. Methionine dietary supplementation potentiates ionizing radiation-induced gastrointestinal syndrome. Am. J. Physiol. Gastrointest. Liver Physiol. 318, 439–450. https://doi.org/10.1152/ajpgi.00351.2019.-Methionine (2020).
Cheng, S., Cheadle, E. J. & Illidge, T. M. Understanding the effects of radiotherapy on the tumour immune microenvironment to identify potential prognostic and predictive biomarkers of radiotherapy response. Cancers 12, 1–20. https://doi.org/10.3390/cancers12102835 (2020).
Nardone, V. et al. Tumor infiltrating t lymphocytes expressing foxp3, ccr7 or pd-1 predict the outcome of prostate cancer patients subjected to salvage radiotherapy after biochemical relapse. Cancer Biol. Ther. 17, 1213–1220. https://doi.org/10.1080/15384047.2016.1235666 (2016).
Yamamoto, Y. et al. Tumour and immune cell dynamics explain the psa bounce after prostate cancer brachytherapy. Br. J. Cancer 115, 195–202. https://doi.org/10.1038/bjc.2016.171 (2016).
Vilaça, M., Pinto, J. C., Magalhães, H., Reis, F. & Mesquita, A. Tumor-infiltrating lymphocytes in localized prostate cancer: Do they play an important role?. Cureus https://doi.org/10.7759/cureus.34007 (2023).
Yanai, Y. et al. Cd8-positive t cells and cd204-positive m2-like macrophages predict postoperative prognosis of very high-risk prostate cancer. Sci. Rep. 11, 22495. https://doi.org/10.1038/s41598-021-01900-4 (2021).
Yang, Y. et al. High intratumoral cd8+ t-cell infiltration is associated with improved survival in prostate cancer patients undergoing radical prostatectomy. Prostate 81, 20–28. https://doi.org/10.1002/pros.24068 (2021).
Yuri, P. et al. Increased tumor-associated macrophages in the prostate cancer microenvironment predicted patients’ survival and responses to androgen deprivation therapies in indonesian patients cohort. Prostate Int. 8, 62–69. https://doi.org/10.1016/j.prnil.2019.12.001 (2020).
Zeigler-Johnson, C., Morales, K. H., Lal, P. & Feldman, M. The relationship between obesity, prostate tumor infiltrating lymphocytes and macrophages, and biochemical failure. PLoS ONE 11, e0159109. https://doi.org/10.1371/journal.pone.0159109 (2016).
Zorko, N. A. et al. Natural killer cell infiltration in prostate cancers predict improved patient outcomes. Prostate Cancer Prostatic Dis. https://doi.org/10.1038/s41391-024-00797-0 (2024).
Zhou, Q. et al. B7 score and t cell infiltration stratify immune status in prostate cancer. J. Immunother. Cancer 9, e002455. https://doi.org/10.1136/jitc-2021-002455 (2021).
Apusiga, K. Immune cell infiltration-based prognosis in prostate cancer: a review of current knowledge. Bull. Natl. Res. Centre 47, 131. https://doi.org/10.1186/s42269-023-01106-w (2023).
Ember, K. J. I. et al. Raman spectroscopy and regenerative medicine: a review. npj Regen. Med. 2, 12. https://doi.org/10.1038/s41536-017-0014-3 (2017).
Allakhverdiev, E. S. et al. Raman spectroscopy and its modifications applied to biological and medical research. Cells 11, 386. https://doi.org/10.3390/cells11030386 (2022).
Huser, T. & Chan, J. Raman spectroscopy for physiological investigations of tissues and cells. Adv. Drug Deliv. Rev. 89, 57–70. https://doi.org/10.1016/j.addr.2015.06.011 (2015).
Jermyn, M. et al. A review of Raman spectroscopy advances with an emphasis on clinical translation challenges in oncology. Phys. Med. Biol. 61, R370–R400. https://doi.org/10.1088/0031-9155/61/23/R370 (2016).
Hill, I. E. et al. Understanding radiation response and cell cycle variation in brain tumour cells using Raman spectroscopy. Analyst 148, 2594–2608. https://doi.org/10.1039/d3an00121k (2023).
Roman, M., Wrobel, T. P., Panek, A., Paluszkiewicz, C. & Kwiatek, W. M. Physicochemical damage and early-stage biological response to x-ray radiation studied in prostate cancer cells by raman spectroscopy. J. Biophotonics 13, e202000252. https://doi.org/10.1002/jbio.202000252 (2020).
Deng, X. et al. Monitor ionizing radiation-induced cellular responses with Raman spectroscopy, non-negative matrix factorization, and non-negative least squares. Appl. Spectrosc. 74, 701–711. https://doi.org/10.1177/0003702820906221 (2020).
Tipatet, K. S. et al. Detection of acquired radioresistance in breast cancer cell lines using Raman spectroscopy and machine learning. Analyst 146, 3709–3716. https://doi.org/10.1039/d1an00387a (2021).
Shreeves, P., Andrews, J. L., Deng, X., Ali-Adeeb, R. & Jirasek, A. Nonnegative matrix factorization with group and basis restrictions. Stat. Biosci. 15, 608–632. https://doi.org/10.1007/s12561-023-09398-2 (2023).
Milligan, K. et al. Prediction of disease progression indicators in prostate cancer patients receiving hdr-brachytherapy using Raman spectroscopy and semi-supervised learning: a pilot study. Sci. Rep. 12, 15104. https://doi.org/10.1038/s41598-022-19446-4 (2022).
Milligan, K. et al. Raman spectroscopy and group and basis-restricted non negative matrix factorisation identifies radiation induced metabolic changes in human cancer cells. Sci. Rep. 11, 3853. https://doi.org/10.1038/s41598-021-83343-5 (2021).
Wang, C., Zhang, Y. & Gao, W. Q. The evolving role of immune cells in prostate cancer. Cancer Lett. 525, 9–21. https://doi.org/10.1016/j.canlet.2021.10.027 (2022).
Erlandsson, A. et al. Infiltrating immune cells in prostate cancer tissue after androgen deprivation and radiotherapy. Int. J. Immunopathol. Pharmacol. 37, 03946320231158025. https://doi.org/10.1177/03946320231158025 (2023).
Keam, S. P. et al. High dose-rate brachytherapy of localized prostate cancer converts tumors from cold to hot. J.r Immunother. Cancer 8, e000792. https://doi.org/10.1136/jitc-2020-000792 (2020).
Kane, N. et al. Significant changes in macrophage and cd8 t cell densities in primary prostate tumors 2 weeks after sbrt. Prostate Cancer Prostatic Dis. 26, 207–209. https://doi.org/10.1038/s41391-022-00498-6 (2023).
Mukha, A. et al. Gls-driven glutamine catabolism contributes to prostate cancer radiosensitivity by regulating the redox state, stemness and atg5-mediated autophagy. Theranostics 11, 7844–7868. https://doi.org/10.7150/THNO.58655 (2021).
Kumar, S. & Clair, D. S. Radioresistance in prostate cancer: Focus on the interplay between nf-\(\kappa\)b and sod. Antioxidants 10, 1925. https://doi.org/10.3390/antiox10121925 (2021).
Cheteh, E. H. et al. Human cancer-associated fibroblasts enhance glutathione levels and antagonize drug-induced prostate cancer cell death. Cell Death Dis. 8, e2848. https://doi.org/10.1038/CDDIS.2017.225 (2017).
Traverso, N. et al. Role of glutathione in cancer progression and chemoresistance. Oxidative Med. Cell. Longev. 7, 8. https://doi.org/10.1155/2013/972913 (2013).
Ferreira, M. R. et al. Impact of pelvic radiation therapy for prostate cancer on global metabolic profiles and microbiota-driven gastrointestinal late side effects: A longitudinal observational study. Int. J. Radiat. Oncol. Biol. Phys. 111, 1204–1213. https://doi.org/10.1016/j.ijrobp.2021.07.1713 (2021).
Zhao, T. & Lum, J. J. Methionine cycle-dependent regulation of t cells in cancer immunity. Front. Oncol. 12, 969563. https://doi.org/10.3389/fonc.2022.969563 (2022).
Ampomah, P. B. et al. Macrophages use apoptotic cell-derived methionine and dnmt3a during efferocytosis to promote tissue resolution. Nat. Metab. 4, 444–457. https://doi.org/10.1038/s42255-022-00551-7 (2022).
Mak, T. W. et al. Glutathione primes t cell metabolism for inflammation. Immunity 46, 675–689. https://doi.org/10.1016/j.immuni.2017.03.019 (2017).
Ma, X. et al. Cholesterol induces cd8+ t cell exhaustion in the tumor microenvironment. Cell Metab. 30, 143-156.e5. https://doi.org/10.1016/j.cmet.2019.04.002 (2019).
Park, J. J. et al. Diagnostic accuracy of Raman spectroscopy for prostate cancer: A systematic review and meta-analysis. Transl. Androl. Urol. 10, 574–583. https://doi.org/10.21037/TAU-20-924 (2021).
Correia, N. A. et al. Detection of prostate cancer by Raman spectroscopy: A multivariate study on patients with normal and altered psa values. J. Photochem. Photobiol. B: Biol. 204, 111801. https://doi.org/10.1016/j.jphotobiol.2020.111801 (2020).
Kirpich, A. et al. Variable selection in omics data: A practical evaluation of small sample sizes. PLoS ONE 13, e0197910. https://doi.org/10.1371/journal.pone.0197910 (2018).
Zou, H. & Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B 67, 301–320 (2005).
Geurts, P., Ernst, D. & Wehenkel, L. Extremely randomized trees. Mach. Learn. 63, 3–42. https://doi.org/10.1007/s10994-006-6226-1 (2006).
Ghazwani, M. & Begum, M. Y. Computational intelligence modeling of hyoscine drug solubility and solvent density in supercritical processing: gradient boosting, extra trees, and random forest models. Sci. Rep. 13, 10046. https://doi.org/10.1038/s41598-023-37232-8 (2023).
Lian, G. et al. Glutathione de novo synthesis but not recycling process coordinates with glutamine catabolism to control redox homeostasis and directs murine t cell differentiation. eLife 7, e36158. https://doi.org/10.7554/eLife.36158.001 (2018).
Kurniawan, H. et al. Glutathione restricts serine metabolism to preserve regulatory t cell function. Cell Metab. 31, 920-936.e7. https://doi.org/10.1016/j.cmet.2020.03.004 (2020).
Li, M. et al. Metabolism, metabolites, and macrophages in cancer. J. Hematol. Oncol. 16, 80. https://doi.org/10.1186/s13045-023-01478-6 (2023).
Li, M. et al. Glutathione s-transferase pi is upregulated in the stromal compartment of hormone independent prostate cancer. Prostate 56, 98–105. https://doi.org/10.1002/pros.10249 (2003).
Andersen, M. K. et al. Integrative metabolic and transcriptomic profiling of prostate cancer tissue containing reactive stroma. Sci. Rep. 8, 14269. https://doi.org/10.1038/s41598-018-32549-1 (2018).
Banyard, J. et al. Collagen xxiii expression is associated with prostate cancer recurrence and distant metastases. Clin. Cancer Res. 13, 2634–2642. https://doi.org/10.1158/1078-0432.CCR-06-2163 (2007).
Afik, R. et al. Tumor macrophages are pivotal constructors of tumor collagenous matrix. J. Exp. Med. 213, 2315–2331. https://doi.org/10.1084/jem.20151193 (2016).
O’Mahony, L., Akdis, M. & Akdis, C. A. Regulation of the immune response and inflammation by histamine and histamine receptors. J. Allergy Clin. Immunol. 128, 1153–1162. https://doi.org/10.1016/j.jaci.2011.06.051 (2011).
Igel, P., Dove, S. & Buschauer, A. Histamine h4 receptor agonists. Bioorgan. Med. Chem. Lett. 20, 7191–7199. https://doi.org/10.1016/j.bmcl.2010.10.041 (2010).
Branco, A. C. C. C., Yoshikawa, F. S. Y., Pietrobon, A. J. & Sato, M. N. Role of histamine in modulating the immune response and inflammation. Mediat. Inflamm. 2018, 9524075. https://doi.org/10.1155/2018/9524075 (2018).
Tantawy, A. A. & Naguib, D. M. Arginine, histidine and tryptophan: A new hope for cancer immunotherapy. PharmaNutrition 8, 100149. https://doi.org/10.1016/j.phanu.2019.100149 (2019).
Fritz, V. et al. Abrogation of de novo lipogenesis by stearoyl-coa desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol. Cancer Ther. 9, 1740–1754. https://doi.org/10.1158/1535-7163.MCT-09-1064 (2010).
Dulude, J. P. et al. Intraoperative use of high-speed Raman spectroscopy during soft tissue sarcoma resection. Sci. Rep. 15, 8789. https://doi.org/10.1038/s41598-025-93089-z (2025).
Bali, A. et al. Establishment of a clinical workflow for in vivo Raman spectroscopy during head and neck cancer surgery. Sci. Rep. 15, 24230. https://doi.org/10.1038/s41598-025-08222-9 (2025).
Mannas, M. P. et al. Stimulated Raman histology and artificial intelligence provide near real-time interpretation of radical prostatectomy surgical margins. J. Urol. 213, 609–616. https://doi.org/10.1097/JU.0000000000004393 (2025).
Nargis, H. F. et al. Raman spectroscopy of blood plasma samples from breast cancer patients at different stages. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 222, 117210. https://doi.org/10.1016/j.saa.2019.117210 (2019).
Koster, H. J. et al. Fused Raman spectroscopic analysis of blood and saliva delivers high accuracy for head and neck cancer diagnostics. Sci. Rep. 12, 18464. https://doi.org/10.1038/s41598-022-22197-x (2022).
Schiemer, R. et al. Infrared and Raman spectroscopy of blood plasma for rapid endometrial cancer detection: Clinical studies. Br. J. Cancer https://doi.org/10.1038/s41416-025-03050-0 (2025).
Medipally, D. K. et al. Vibrational spectroscopy of liquid biopsies for prostate cancer diagnosis. Ther. Adv. Med. Oncol. 12, 1758835920918499. https://doi.org/10.1177/1758835920918499 (2020).
Medipally, D. K. et al. Monitoring radiotherapeutic response in prostate cancer patients using high throughput ftir spectroscopy of liquid biopsies. Cancers 11, 925. https://doi.org/10.3390/cancers11070925 (2019).
Cullen, D. et al. Raman spectroscopy of lymphocytes for the identification of prostate cancer patients with late radiation toxicity following radiotherapy. Transl. Biophotonics 2, e201900035. https://doi.org/10.1002/tbio.201900035 (2020).
Andras, D. et al. Sers liquid biopsy in colorectal cancer detection and treatment response: Revealing metabolic memory post-radiochemotherapy. Nanomed.: Nanotechnol. Biol. Med. 68, 102840. https://doi.org/10.1016/j.nano.2025.102840 (2025).
Shrivastava, A. et al. Diagnostic and prognostic application of Raman spectroscopy in carcinoma cervix: A biomolecular approach. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 250, 119356. https://doi.org/10.1016/j.saa.2020.119356 (2021).
Maguire, A. et al. Analyses of ionizing radiation effects in vitro in peripheral blood lymphocytes with Raman spectroscopy. Radiat. Res. 183, 407–416. https://doi.org/10.1667/RR13891.1 (2015).
Gough, M. J. & Crittenden, M. R. The paradox of radiation and t cells in tumors. Neoplasia (United States) 31, 100808. https://doi.org/10.1016/j.neo.2022.100808 (2022).
Gkikoudi, A. et al. Molecular biomarkers for predicting cancer patient radiosensitivity and radiotoxicity in clinical practice. Appl. Sci. (Switzerland) 13, 12564. https://doi.org/10.3390/app132312564 (2023).
McLaren, D. B. & Aitman, T. J. Redefining precision radiotherapy through liquid biopsy. Br. J. Cancer 129, 900–903. https://doi.org/10.1038/s41416-023-02398-5 (2023).
Acknowledgements
Multiplex immunohistochemistry staining was completed by Katy Milne and the Molecular and Cellular Immunology Core at the Deeley Research Centre. This work was supported by grant funding from the Natural Sciences and Engineering Research Council of Canada Discovery (grants no. RGPIN-2020-07232 and RGPIN-2020-04646) and the Canadian Institutes of Health Research (PJT 162279).
Author information
Authors and Affiliations
Contributions
A.J., J.M.C, A.G.B, J.J.L, K.M. and J.L.A. conceived and designed experiments. K.M., J.B., S.Punch., A.R. and S.Popescu acquired, processed and analyzed the data. J.L.A, C.H, A.F., J.B., K.M. and M.W. provided guidance in the analysis and the interpretation of results. S.Popescu wrote the manuscript and prepared the figures. S.Punch. and A.R. provided technical support and expertise in tissue staining. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Popescu, S.N., Milligan, K., Wiebe, M. et al. Modelling of immune infiltration in prostate cancer treated with HDR-brachytherapy using Raman spectroscopy and machine learning. Sci Rep 15, 36248 (2025). https://doi.org/10.1038/s41598-025-20107-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-20107-5