Abstract
Although berberine exhibits anticancer activity, its mechanistic role in esophageal cancer (ESCA) remains unclear. Long non-coding RNAs (lncRNAs) expression profiling was conducted on berberine-treated and control ESCA TE-1 cells, identifying differentially expressed lncRNAs. Prognostic lncRNAs were selected through integration with the TCGA database, followed by the construction of a prognostic nomogram. In vitro assays and bioinformatics analyses were performed to validate berberine’s impact on cell proliferation, metastasis, and key lncRNAs. Berberine treatment significantly altered lncRNA expression in TE-1 cells, with 344 upregulated and 235 downregulated lncRNAs. The expression of CCDC18-AS1, TBILA, and LINC02084 was associated with poorer overall survival (OS) and served as independent prognostic factors in ESCA. The nomogram accurately predicted 1-, 3-, and 5-year OS rates. Berberine inhibited TE-1 cell proliferation, migration, and invasion, primarily through the upregulation of CCDC18-AS1 expression. Genes co-expressed with CCDC18-AS1 were enriched in mRNA processing, RNA splicing, spliceosome, and mRNA surveillance pathways. Among these, overexpression of PCMTD2 and AL391840.2 represented risk factors, while RBM43 and LRIG2 were identified as protective factors in ESCA. Berberine may inhibit ESCA progression by modulating CCDC18-AS1 expression, thereby improving patient prognosis.
Similar content being viewed by others
Introduction
Berberine, a naturally occurring isoquinoline alkaloid, is well known for its broad-spectrum antimicrobial activity and widespread use in treating gastrointestinal infections. Increasing evidence supports its potent anticancer properties across various tumor types1,2,3,4. For instance, Liu et al. demonstrated that berberine inhibits proliferation and migration while promoting apoptosis in oral squamous cell carcinoma (OSCC) by modulating the RAGE/PI3K/AKT/mTOR signaling pathway2. Zhao et al. further showed that berberine suppresses non-small cell lung cancer (NSCLC) progression through downregulation of CDCA5 and CCNA2, thereby inhibiting cell proliferation, migration, invasion, and xenograft tumor growth. Notably, overexpression of CDCA5 or CCNA2 counteracts the antitumor effects of berberine4.
Emerging studies highlight the pivotal role of non-coding RNAs in digestive system cancers5,6,7. For example, Wang et al. found that METTL3 silencing inhibited ESCA cell proliferation and induced apoptosis through YTHDF1-mediated m6A modification of the long non-coding RNA (lncRNA) FOXD2-AS1, which regulates ESCA cell behavior7. Despite these advances, the relationship between berberine and lncRNAs in cancer remains largely uncharted. Specifically, no research has explored whether berberine exerts antitumor effects in ESCA through lncRNA regulation. This study aims to fill this gap by employing transcriptome sequencing to identify key differentially expressed lncRNAs (DelncRNAs) after berberine treatment in ESCA and elucidate their functional roles and underlying mechanisms. The findings aim to provide novel therapeutic targets and a mechanistic basis for berberine-based treatment strategies in ESCA.
Materials and methods
Cell culture and model construction of ESCA
ESCA TE-1 cells were sourced from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI-1640 medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Gibco) at 37 °C in a humidified 5% CO2 atmosphere. For experimental purposes, TE-1 cells were divided into two groups: the experimental group treated with 40µM berberine (Sigma-Aldrich) and the control group, which received vehicle treatment only.
Differentially expressed LncRNAs (DelncRNAs) in ESCA cells
Total RNA was extracted from the TE-1 cells of both the berberine-treated and control groups when the cells were in optimal growth conditions. Four RNA samples were collected from each group and sent to Wuhan RuiSheng Biotechnology Co., Ltd. for lncRNA expression analysis. Raw sequencing reads were processed with DESeq2 to identify DelncRNAs between the two groups. Differential expression was defined by a fold change (FC) ≥ 2 or ≤ 0.5 and a false discovery rate (FDR) < 0.05.
DelncRNAs related to prognosis in patients with ESCA
Transcriptome data from tissues of patients with ESCA were obtained from the TCGA database. DelncRNA expression data were extracted using the Xiantao academic platform. Based on the median expression levels, DelncRNAs were classified into high- and low-expression groups, and their association with overall survival (OS) was analyzed via Kaplan-Meier (K-M) survival analysis, with prognostic DelncRNAs identified at a significance level of P < 0.05.
COX regression analysis and nomogram construction
Cox regression analysis was performed on the DelncRNAs (such as RMDN2-AS1, EPB41L4A-AS1, CCDC18-AS1, MIR9-1HG, LINC00365, L3MBTL2-AS1, and ADAMTSL4-AS2) identified as significant in the K-M survival analysis in ESCA. Univariate and multivariate Cox regression analyses were conducted to assess the relationship between these lncRNAs and OS, with a significance threshold set at P < 0.05 for identifying relevant lncRNAs. A nomogram was constructed based on the results of the multivariate Cox regression analysis.
DelncRNAs were verified by RT-PCR technology
For transcriptome sequencing, total RNA from both cell groups underwent cDNA synthesis followed by PCR amplification to quantify the expression levels of CCDC18-AS1, TBILA, and LINC02084. The primer sequences used were as follows: CCDC18-AS1: 5′-AAACTGTCGTCCTGGTGGG-3′; TBILA: 5′-CTGAGAGCCACTTCCAC CAT-3′; GAPDH: 5′-AACGGATTTGGTCGTATTG-3′; and LINC02084: 5′-TACATTCCTCCCCCTACGCA-3′.
CCK-8
Berberine was added to ESCA cells at an appropriate concentration and incubated at 37 °C in 5% CO2. After 24 h, 3 × 103 cells from each group were harvested, centrifuged, and counted. The cells were then plated in 96-well plates, and absorbance values were measured at 24 h post-adhesion, with three replicate wells for each condition.
Transwell
Diluted Matrigel was added to the membrane surface of the upper chamber of a Transwell. TE-1 cells were prepared and incubated with berberine to generate a cell suspension, followed by cell counting. A total of 1 × 105 cells were then inoculated into the Transwell chamber. To the lower chamber, 600 µL of complete culture medium (containing serum or specific chemokines) was added as an attractant. After 24 h of incubation at 37 °C in a 5% CO2 atmosphere, cells were fixed, stained, and subsequently counted and photographed.
Wound healing
Two sets of pre-incubated cell suspensions were seeded into 6-well plates. The plates were gently swirled to ensure even distribution of the cells and incubated overnight at 37 °C under 5% CO2. Once the cells reached 100% confluence, the original medium was aspirated and discarded. Parallel lines were drawn on the underside of the plates using a marker to guide wounding and serve as reference points for imaging. Using a sterile 200 µL pipette tip, straight lines perpendicular to the plate bottom were scratched along the guides, applying consistent pressure. After aspirating the medium, this time was marked as 0 h, and images were acquired. The plates were returned to the CO2 incubator for continued culture. Imaging was repeated at the predetermined 24-hour time point, and cell migration was quantified based on the recorded images.
Co-expressed gene of CCDC18-AS1
Co-expressed genes of CCDC18-AS1 in the cancer tissue data of patients with ESCA from the TCGA database were analyzed using Pearson correlation analysis via the Xiantao academic platform. A higher absolute value of the correlation coefficient, closer to 1, indicates a stronger correlation. Co-expressed genes were defined as having an absolute correlation coefficient greater than 0.5 and a P-value < 0.001, with results visualized using scatter plots.
Functional mechanisms and protein network of CCDC18-AS1 co-expressed genes
To explore the functional mechanisms and protein network of CCDC18-AS1 co-expressed genes, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed, widely used to investigate gene functions and potential signaling pathways8,9. The analysis was conducted in R, with a FDR < 0.05 set as the criterion for statistical significance. Additionally, a protein network of the co-expressed genes was constructed using the STRING database.
Prognostic analysis and nomogram of the co-expressed gene of CCDC18-AS1
For prognostic analysis and nomogram construction of the co-expressed genes of CCDC18-AS1, transcriptome data from tissues of patients with ESCA were retrieved from the TCGA database. The expression data for co-expressed genes of CCDC18-AS1 were extracted using the Xiantao academic platform, and the median expression value was used to classify genes into high- and low-expression groups. K-M survival analysis was employed to assess the relationship between the expression of these co-expressed genes and OS, with significance determined at P < 0.05. Both univariate and multivariate Cox regression analyses were applied to evaluate the prognostic value of co-expressed genes with respect to OS. A significance threshold of P < 0.05 was set, and a gene interaction network was constructed.
Statistical analysis
The t-test was applied to evaluate significant differences between the berberine and control groups. K-M survival analysis, Cox regression analysis, and nomogram construction were used to identify prognostic lncRNAs and genes, with FDR and P < 0.05 as significance criteria.
Results
DelncRNAs associated with Berberine in ESCA cells
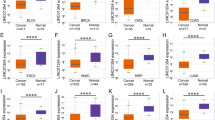
The FCs in lncRNA expression following berberine treatment of TE-1 cells were visualized using heatmaps and volcano plots (Fig. 1 A, B). Specifically, 344 lncRNAs were upregulated, and 235 were downregulated (Fig. 1 A, B). After alignment and deduplication, a total of 294 DelncRNAs were identified. A scatter plot highlighted the top 6 lncRNAs, including LINC01558, LINC01512, HIF1A-AS3, MIR210HG, LUCAT1, and LINC01449, ranked by FC (Fig. 1C–H).
Prognostic LncRNAs and nomogram associated with berberine in ESCA
K-M survival analysis revealed that the expression levels of RMDN2-AS1, EPB41L4A-AS1, CCDC18-AS1, MIR9-1HG, LINC00365, L3MBTL2-AS1, ADAMTSL4-AS2, NRSN2-AS1, TBILA, ZFAS1, and LINC02084 were associated with poorer OS in patients with ESCA (Fig. 2). Both univariate and multivariate Cox regression analyses identified CCDC18-AS1, TBILA, and LINC02084 as independent risk factors for OS in these patients (Table 1). Based on these results, a nomogram incorporating CCDC18-AS1, TBILA, and LINC02084 was constructed (Fig. 3).
Berberine inhibits the growth of ESCA by promoting CCDC18-AS1
CCK-8 assay results demonstrated a significant reduction in TE-1 cell proliferation following berberine treatment (Fig. 4A). Transwell and wound healing assays confirmed that 40 μM berberine inhibited TE-1 cell migration and invasion, with statistically significant results (Fig. 4B–E). Additionally, RT-PCR analysis revealed an increased expression of CCDC18-AS1 and TBILA in both sequenced and newly incubated berberine-treated ESCA cells, while the expression of LINC02084 remained unchanged. Notably, the FC of CCDC18-AS1 was the most significant (Fig. 4F,G).
Berberine inhibits the growth and metastasis of ESCA by promoting CCDC18-AS1 expression. (A) Cell growth; (B,C) Invasion capabilities; (D,E) Migration capabilities; (F,G) Expression of lncRNAs in TE-1 cells associated with berberine in cell sequencing and subsequent experimental samples. ESCA, esophageal cancer.
Functional mechanisms and protein networks of co-expressed genes of CCDC18-AS1
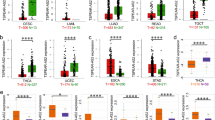
Based on screening criteria, 376 co-expressed genes associated with CCDC18-AS1 were identified. The top 20 co-expressed genes are presented in a heatmap (Fig. 5A), while a scatter plot illustrates the relationships between CCDC18-AS1 and SRSF11, ZNRD1ASP, AC138393.1, ZRANB2, AL138831.3, and BBS1 (Fig. 5B–G). GO and KEGG analyses revealed that the co-expressed genes of CCDC18-AS1 are involved in processes such as mRNA processing, RNA splicing, ribonucleoprotein complex assembly, miRNA binding, methylated histone binding, spliceosome activity, and the mRNA surveillance pathway. The protein network among these co-expressed genes is depicted in (Fig. 6).
The prognostic values of co-expressed genes
K-M survival analysis examined the relationship between the expression of CCDC18-AS1 co-expressed genes and the prognosis of patients with ESCA. The results showed that abnormal expression of PCMTD2, ZCWPW1, AL391840.2, PJVK, FAM227B, HMGN2P28, RBM43, and LRIG2 was correlated with poor prognosis in these patients (Table 2). Univariate and multivariate Cox regression analyses identified PCMTD2 and AL391840.2 overexpression as risk factors, while RBM43 and LRIG2 overexpression were recognized as protective factors. Nomograms in Figs. 7 and 8 illustrate the relationship between the expression levels of PCMTD2, AL391840.2, RBM43, CCDC18-AS1, and LRIG2 and the OS of patients with ESCA.
Discussion
Traditional Chinese medicine, including natural compounds, exerts anti-cancer effects across various cancers, influencing cancer growth, metastasis, and poor prognosis1,2,4,10,11,12,13,14,15,16,17. For instance, Wang et al. reported that berberine inhibits the expression of KIF20A and CCNE2, thereby suppressing the PI3K/AKT pathway. This results in reduced growth, invasion, tumor formation in nude mice, and glycolysis in LUAD H1299 and A549 cells, while promoting apoptosis16. Chen et al. demonstrated that berberine induces apoptosis in NSCLC cells in a dose- and time-dependent manner via ROS-mediated ASK1/JNK activation and the mitochondrial pathway12. These findings collectively highlight the anti-cancer efficacy of berberine. However, to date, no published literature has explored the role of berberine in ESCA. Thus, this study presents the first report on the effects of berberine in ESCA, with preliminary results indicating that berberine inhibits the proliferation of ESCA cells, further confirming its anti-cancer properties.
LncRNAs have long been recognized as closely associated with ESCA5,6,7,18,19,20. For instance, Xue et al. found significant upregulation of the lncRNA LUESCC in esophageal squamous carcinoma (ESCC) tissues. Overexpression of LUESCC correlates with factors such as gender, infiltration, lymph node metastasis, and poor prognosis in patients with ESCC. Inhibition of LUESCC expression suppresses proliferation, colony formation, migration, invasion, and tumor growth in ESCC cells. LUESCC mediates its oncogenic effects in ESCC through the miR-6785-5p/NRSN2 axis18. Recent studies also confirm that berberine modulates cancer progression through lncRNAs21,22,23,24,25,26. However, the relationship between berberine and lncRNAs in ESCA remains unexplored. In this study, transcriptome sequencing and in vitro experiments were conducted to validate that berberine promotes the expression of CCDC18-AS1. Additionally, elevated expression of CCDC18-AS1 in patients with ESCA is associated with better prognosis. These results further suggest that berberine enhances CCDC18-AS1 expression, inhibiting ESCA progression and improving patient prognosis, thus providing new insights and theoretical foundations for ESCA treatment.
In recent years, nomograms have been widely used to illustrate the relationship between prognostic factors and patient outcomes in cancer27,28,29. For example, Yang et al. reported that patients with upper ESCA exhibit significantly worse prognoses. Cox regression analysis identified age, gender, marital status, histological type, grade, T stage, lymph node metastasis, and distant metastasis as key risk factors for poor prognosis in patients with ESCA. The nomogram constructed based on these factors showed enhanced predictive performance for the prognosis of patients with upper ESCA27. This study also utilized data from the TCGA database to develop a nomogram illustrating the relationships between the expression levels of CCDC18-AS1, TBILA, and LINC02084, as well as PCMTD2, AL391840.2, RBM43, CCDC18-AS1, and LRIG2, with OS in patients with ESCA. These nomograms clearly depict the associations between these prognostic factors and the 1-, 3-, and 5-year OS rates, facilitating the assessment and prediction of OS to guide precision treatment strategies.
The anti-cancer efficacy of berberine in ESCA was confirmed through bioinformatics analysis, transcriptome sequencing, and in vitro experiments, yielding valuable insights. However, this study has some limitations. The relationships between berberine and CCDC18-AS1, as well as the mechanisms underlying CCDC18-AS1’s role in ESCA, require further verification through additional in vitro experiments. This remains an area for future investigation. Overall, berberine appears to enhance the expression of CCDC18-AS1, inhibiting cancer progression and potentially improving the prognosis of patients with ESCA.
Data availability
The sequencing data are currently confidential and cannot be uploaded. However, the data are available upon reasonable request from the corresponding author.
References
Li, J. et al. Berberine triggers apoptosis through the PI3K/Akt pathways and Nrf2 by inducing ROS in papillary thyroid cancer. Arch. Biochem. Biophys. 771, 110481. https://doi.org/10.1016/j.abb.2025.110481 (2025).
Liu, D. et al. Berberine promotes apoptosis and inhibits the migration of oral squamous carcinoma cells through Inhibition of the RAGE/PI3K/AKT/mTOR pathway. Biomed. Pharmacother. 187, 118147. https://doi.org/10.1016/j.biopha.2025.118147 (2025).
Zhang, X. et al. Berberine inhibits metastasis of ovarian cancer by blocking lipid metabolism, alleviating aging of adipose tissue and increasing tumor infiltrating immune cells. Transl Oncol. 56, 102380. https://doi.org/10.1016/j.tranon.2025.102380 (2025).
Zhao, X., Ha, M., Zhou, L., Wang, Y. & Li, P. Berberine diminishes the malignant progression of non-small cell lung cancer cells by targeting CDCA5 and CCNA2. J. Nat. Med. 79 (3), 530–542. https://doi.org/10.1007/s11418-025-01885-8 (2025).
Ge, H. et al. Circular RNA hsa_circ_0005939 regulates UHRF1BP1L expression by targeting miR-4693-3p to promote colorectal cancer progression. Protein Pept. Lett. 31 (6), 437–446. https://doi.org/10.2174/0109298665297110240611115010 (2024).
Xue, Y., Yang, R., Gong, P. & Zhu, H. LncRNA FENDRR predicts adverse prognosis and regulates the development of esophageal squamous cell carcinoma through negatively modulating miR-495-3p. Turk. J. Gastroenterol. 1 (1). https://doi.org/10.5152/tjg.2025.24350 (2025).
Wang, Z., Liu, X. C., Gao, Z. G., Shi, W. D. & Wang, W. C. FOXD2-AS1 is modulated by METTL3 with the assistance of YTHDF1 to affect proliferation and apoptosis in esophageal cancer. Acta Pharm. 75 (1), 69–86. https://doi.org/10.2478/acph-2025-0009 (2025).
Xiang, Q. M. et al. Overexpression of SH2D1A promotes cancer progression and is associated with immune cell infiltration in hepatocellular carcinoma via bioinformatics and in vitro study. BMC Cancer. 23 (1), 1005. https://doi.org/10.1186/s12885-023-11315-1 (2023).
Li, R. C. et al. ELMOD2 overexpression predicts adverse outcomes and regulates tumor progression in gliomas. Curr. Med. Sci. https://doi.org/10.1007/s11596-025-00057-9 (2025).
Liu, L. et al. Berberine in combination with cisplatin induces necroptosis and apoptosis in ovarian cancer cells. Biol. Res. 52 (1), 37. https://doi.org/10.1186/s40659-019-0243-6 (2019).
Zhang, P. et al. Berberine inhibits growth of liver cancer cells by suppressing glutamine uptake. Onco Targets Ther. 12, 11751–11763. https://doi.org/10.2147/OTT.S235667 (2019).
Chen, Q. et al. Berberine induces non-small cell lung cancer apoptosis via the activation of the ROS/ASK1/JNK pathway. Ann. Transl Med. 10 (8), 485. https://doi.org/10.21037/atm-22-1298 (2022).
Han, X. et al. Based on network Pharmacology and experimental validation, Berberine can inhibit the progression of gastric cancer by modulating oxidative stress. Transl Cancer Res. 14 (1), 554–568. https://doi.org/10.21037/tcr-24-732 (2025).
Zeng, X. et al. Effect of low dose of Berberine on the radioresistance of cervical cancer cells via a PI3K/HIF-1 pathway under nutrient-deprived conditions. Int. J. Radiat. Biol. 96 (8), 1060–1067. https://doi.org/10.1080/09553002.2020.1770358 (2020).
Yang, W. et al. Berberine improved the microbiota in lung tissue of colon cancer and reversed the bronchial epithelial cell changes caused by cancer cells. Heliyon 10 (2), e24405. https://doi.org/10.1016/j.heliyon.2024.e24405 (2024).
Wang, Q., Wu, H., Wu, Q. & Zhong, S. Berberine targets KIF20A and CCNE2 to inhibit the progression of nonsmall cell lung cancer via the PI3K/AKT pathway. Drug Dev. Res. 84 (5), 907–921. https://doi.org/10.1002/ddr.22061 (2023).
Bailon-Moscoso, N., Cevallos-Solorzano, G., Romero-Benavides, J. C. & Orellana, M. I. Natural compounds as modulators of cell cycle arrest: application for anticancer chemotherapies. Curr. Genomics. 18 (2), 106–131. https://doi.org/10.2174/1389202917666160808125645 (2017).
Xue, S. T. et al. LncRNA LUESCC promotes esophageal squamous cell carcinoma by targeting the miR-6785-5p/NRSN2 axis. Cell. Mol. Life Sci. 81 (1), 121. https://doi.org/10.1007/s00018-024-05172-9 (2024).
Fu, C. et al. AP001885.4 promotes the proliferation of esophageal squamous cell carcinoma cells by histone lactylation- and NF-κB (p65)-dependent transcription activation and METTL3-mediated mRNA stability of c-myc. Anim. Cells Syst. (Seoul). 28 (1), 536–550. https://doi.org/10.1080/19768354.2024.2417458 (2024).
Shen, G. Y. et al. FOXP4-AS1 promotes CD8 + T cell exhaustion and esophageal cancer immune escape through USP10-stabilized PD-L1. Immunol. Res. 72 (4), 766–775. https://doi.org/10.1007/s12026-024-09482-9 (2024).
Yan, X. et al. Berberine modulates ovarian cancer autophagy and Glycolysis through the LINC01123/P65/MAPK10 signaling axis. Phytomedicine 135, 156121. https://doi.org/10.1016/j.phymed.2024.156121 (2024).
Zhao, Z. et al. The long non-coding RNA keratin-7 antisense acts as a new tumor suppressor to inhibit tumorigenesis and enhance apoptosis in lung and breast cancers. Cell. Death Dis. 14 (4), 293. https://doi.org/10.1038/s41419-023-05802-3 (2023).
Li, X., Su, Y., Li, N., Zhang, F. R. & Zhang, N. Berberine attenuates MPP+-Induced neuronal injury by regulating LINC00943/miR-142-5p/KPNA4/NF-κB pathway in SK-N-SH cells. Neurochem Res. 46 (12), 3286–3300. https://doi.org/10.1007/s11064-021-03431-w (2021).
Zheng, F. et al. Novel regulation of miR-34a-5p and HOTAIR by the combination of Berberine and gefitinib leading to Inhibition of EMT in human lung cancer. J. Cell. Mol. Med. 24 (10), 5578–5592. https://doi.org/10.1111/jcmm.15214 (2020).
Dai, W. et al. Long non–coding RNA CASC2 enhances berberine–induced cytotoxicity in colorectal cancer cells by Silencing BCL2. Mol. Med. Rep. 20 (2), 995–1006. https://doi.org/10.3892/mmr.2019.10326 (2019).
Dai, W. et al. Berberine promotes apoptosis of colorectal cancer via regulation of the long Non-Coding RNA (lncRNA) cancer susceptibility candidate 2 (CASC2)/AU-Binding factor 1 (AUF1)/B-Cell CLL/Lymphoma 2 (Bcl-2) axis. Med. Sci. Monit. 25, 730–738. https://doi.org/10.12659/MSM.912082 (2019).
Yang, D., Liu, Z. & Xie, J. Clinical characteristics, prognosis, and nomogram for upper esophageal cancer: a SEER database analysis. Sci. Rep. 15 (1), 15155. https://doi.org/10.1038/s41598-025-00289-8 (2025).
Zhang, L. et al. Construction of LncRNA prognostic model related to Cuproptosis in esophageal carcinoma. Front. Genet. 14, 1120827. https://doi.org/10.3389/fgene.2023.1120827 (2023).
Guo, Q., Peng, Y., Yang, H. & Guo, J. Prognostic nomogram for postoperative patients with gastroesophageal junction cancer of no distant metastasis. Front. Oncol. 11, 643261. https://doi.org/10.3389/fonc.2021.643261 (2021).
Funding
This study was provided by Natural Science Foundation of Hubei (No. 2024AFB694), and Science and Technology Bureau Project of Shiyan City (No. 24Y084).
Author information
Authors and Affiliations
Contributions
Chuang-Yan Wu and Liang Liu wrote the manuscript together and visualized the data together with Yan-Jiao Huang. Jiu-Ling Chen and Xiang-Yu Luo supervised the implementation of the project and edited the manuscript together with Yan-Jiao Huang and Qiang Guo. Qiang Guo verified the data and carried out data quality control. All authors agree to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, CY., Liu, L., Huang, YJ. et al. Berberine alleviates the proliferation and metastasis of ESCA by promoting CCDC18-AS1 expression based on bioinformatics and in vitro experimental verification. Sci Rep 15, 37647 (2025). https://doi.org/10.1038/s41598-025-21581-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-21581-7