Abstract
Breast ultrasound (BUS) imaging is widely recognized as a non-invasive and cost-effective modality for the timely diagnosis of breast cancer. Despite its clinical importance, automatic tumor segmentation remains a highly challenging task because of speckle noise, varying lesion scale, and inherently indistinct boundaries between malignant and healthy tissue. To address these challenges, we introduce a novel hybrid attention-based segmentation framework, named HA-Net, tailored for BUS images. The proposed HA-Net uses a pre-trained DenseNet-121 backbone in the encoder to extract discriminative features, ensuring robustness against imaging artifacts. At the bottleneck, three complementary modules, Global Spatial Attention (GSA), Position Encoding (PE), and Scaled Dot-Product Attention (SDPA), are incorporated to capture long-range dependencies, preserve structural relationships, and model contextual interactions among features. Moreover, a Spatial Feature Enhancement Block (SFEB) is incorporated within the skip connections to refine spatial detail and emphasize tumor-relevant regions, thereby strengthening the decoder’s reconstruction capability. To further improve segmentation reliability, a composite loss function is employed by combining Binary Cross-Entropy (BCE) with Jaccard Index loss, ensuring balanced optimization across pixel-level classification and region-level overlap. In comparison to current state-of-the-art (SOTA) approaches, extensive experiments on publicly available BUS datasets show that the proposed HA-Net achieves competent performance, highlighting its potential as an efficient decision-support tool for radiologists.
Similar content being viewed by others
Introduction
Breast cancer is a significant health concern for women worldwide, as it is one of the most commonly diagnosed malignancies and the leading cause of deaths1. Timely detection of breast cancer is crucial for improving patient prognosis, and medical imaging serves as a key tool in screening, diagnosis, and treatment planning2,3. Breast ultrasound imaging serves as a commonly adopted supplementary technique to mammography, owing to its non-invasive qualities, low expense, real-time diagnostic capability, and effectiveness in identifying tumors in dense breast tissue4,5. Unlike mammograms, which often struggle with overlapping tissue structures, ultrasound offers superior contrast for distinguishing solid masses from cystic structures and is especially beneficial for younger women with denser breast composition5,6.
Despite its advantages, automated breast lesion segmentation presents substantial challenges. Ultrasound images are inherently characterized by low signal-to-noise ratio, low contrast boundaries, speckle noise, and operator-dependent variability, which collectively hinder the reliable delineation of tumor margins7. Furthermore, intra-class variability and inter-class similarity between malignant and benign masses exacerbate the difficulty of precise segmentation, particularly in small datasets commonly encountered in medical imaging7. These challenges are less pronounced in mammographic images, where tissue structures are generally more consistent and the signal quality is higher8,9,10.
Medical image segmentation has recently seen considerable progress with deep learning approaches, especially convolutional neural networks (CNNs)11,12,13,14. Encoder-decoder frameworks, such as U-Net15 and its variants, have become widely used for biomedical applications due to their efficacy in capturing both fine-grained texture data and high-level semantic information16. Despite their popularity, conventional CNN-based models often struggle to model contextual relationships and long-range spatial dependencies that are necessary for precise segmentation in complex modalities such as breast ultrasound17. Furthermore, when dealing with substantial speckle noise and indistinct tumor boundaries, relying solely on basic skip connections, as adopted in many encoder–decoder architectures, may be insufficient for transmitting detailed spatial information from the encoder to the decoder, which can ultimately hinder segmentation accuracy17.
To overcome these limitations, we introduce a novel hybrid attention-based network for the segmentation of breast ultrasound lesions. It combines a decoder with a pre-trained DenseNet-121 encoder18 for reliable feature extraction with multiple attention mechanisms. At the bottleneck, the model integrates Global Spatial Attention (GSA)19, Position Encoding (PE)20, and Scaled Dot-Product Attention (SDPA)21 to capture global context, spatial dependencies, and relative positional information. Furthermore, the Spatial Feature Enhancement Block (SFEB) is incorporated at the skip connections to refine spatial representations, allowing the model to concentrate more effectively on relevant regions. This architecture improves the precise localization of lesions and sharpens boundary definition, both of which are essential for reliable clinical use18,19. To optimize, a composite loss function is employed that combines jaccard index loss and binary cross-entropy (BCE)18, balancing pixel-level classification with region-level overlap. This strategy improves robustness against class imbalance and accommodates irregular tumor shapes, resulting in more accurate and reliable segmentation outcomes18,19,22.
The key contributions of the proposed HA-Net are outlined below:
-
A hybrid attention network is proposed using an encoder composed of DenseNet-121, pre-trained on ImageNet, specifically designed for precise segmentation of breast ultrasound images (BUSI).
-
A transformer-based attention mechanism is introduced to incorporate spatial, positional, and semantic cues, improving segmentation precision.
-
Spatial Feature Enhancement Block (SFEB) is incorporated in skip connections to refine feature propagation and enhance focus on tumor regions.
-
A combined loss that integrates BCE and Jaccard Index loss is employed to optimize both pixel-level classification and region-level overlap, effectively tackling speckle noise and ambiguous tumor margins.
-
Extensive tests on publicly accessible breast ultrasound datasets show that our proposed HA-Net performs well when evaluated against competent approaches, indicating its potential to help radiologists diagnose breast cancer early and accurately.
The remaining parts of this manuscript are classified as follows. The related work section reviews existing studies with a focus on the limitations of U-Net and conventional CNN-based architectures, the emergence of attention mechanisms, and the recent adoption of Transformer and Mamba-based models in medical imaging. The methodology section presents the proposed framework in detail, including the transformer attention module, transformer self-attention, global self-attention, SFEB, and the hybrid loss functions employed for optimization. The experiments section describes the datasets utilized, preprocessing strategies, implementation details, ablation studies, and evaluation metrics performed to validate the model design. The results section reports both numerical and visual outcomes, supported by ablation results, while the discussion provides statistical analysis and interprets the significance of findings in the context of clinical application. Finally, the conclusion highlights the main contributions, summarizes key insights, acknowledges limitations, and outlines potential ideas for future exploration.
Related work
The segmentation of breast tumors has become a major focus in recent research due to its importance in early detection and treatment planning23. Compared to other modalities like mammography, ultrasound imaging offers a number of benefits, such as real-time imaging, reduced ionizing radiation, cost-effectiveness, and improved visibility in dense breast tissue. However, ultrasound images pose unique challenges for automated analysis because of speckle noise, low contrast, operator variability, and anatomical ambiguities23,24. These difficulties have driven the development of advanced deep learning methods capable of extracting robust features and leveraging contextual information to improve segmentation accuracy23,25.
Limitations of U-Net and CNNs
Initial attempts at breast tumor segmentation relied on classical computer vision techniques such as filtering, active contours, and clustering methods26. For example, threshold-based segmentation and graph-based approaches were used in early studies to delineate lesions in ultrasound images27. However, these methods required extensive domain knowledge and struggled with noise sensitivity and over-segmentation23,24.
The field was revolutionized by CNNs, particularly the U-Net design, which enabled the learning of hierarchical features in an end-to-end manner23,25. Recent studies demonstrate that densely connected U-Net variants with attention mechanisms achieve dice scores exceeding 0.83, outperforming traditional methods23. For instance, ACL-DUNet23 integrates channel attention modules and spatial attention gates to suppress irrelevant regions while enhancing tumor features. Similarly, SK-U-Net24 employs selective kernels with dilated convolutions to adapt receptive fields, gaining a mean dice score of 0.826 in comparison to 0.778 for the standard U-Net.
To address limited contextual awareness, multi-branch architectures have emerged. One approach25 combines classification and segmentation branches, achieving an AUC of 0.991 for normal/abnormal classification and a dice score of 0.898 for segmentation. These models reduce false positives in normal images while maintaining sensitivity advancement for clinical screening25. Hybrid designs like DeepCardinal-5028 further optimize computational efficiency, achieving 97% accuracy in tumor detection with real-time processing capabilities.
However, challenges persist in modeling long-range dependencies for lesions with irregular morphology. While attention mechanisms in ACL-DUNet improve spatial focus23, and scale attention modules enhance multi-level feature integration23, fuzzy boundaries in low-contrast ultrasound images remain difficult24. These constraints are being addressed by ongoing advancements in adaptive kernel selection and boundary-guided networks23,24.
Rise of attention mechanisms
Recent advancements in breast tumor segmentation in ultrasound imaging have been driven by the incorporation of attention mechanisms and hybrid network architectures. Early strategies focused on spatial-channel attention to address challenges such as fuzzy lesion boundaries and variable tumor sizes. For example, SC-FCN-BLSTM29 combined bi-directional LSTM with spatial-channel attention to exploit inter-slice contextual information in 3D automated breast ultrasound. Abraham et al.30 presented hybrid attention mechanisms that adaptively reweigh feature maps based on contextual saliency, improving segmentation performance in noisy ultrasound images. Similarly, adaptive attention modules such as HAAM31 replaced standard convolutions in U-Net variants, allowing dynamic adjustment of the receptive field across spatial and channel dimensions for more robust segmentation.
Further improvements were achieved with CBAM-RIUnet4, which combined convolutional block attention modules with residual inception blocks, yielding intersection-over-union (IoU) and dice scores of 88.71% and 89.38%, respectively, by effectively suppressing irrelevant features. The authors32 presented ESKNet, which integrates particular kernel networks into the U-Net to dynamically modulate receptive fields using attention, enhancing segmentation accuracy across diverse lesion types. Although attention-based models have improved segmentation accuracy, many approaches are still limited in adequately representing long-range spatial relationships, specifically when relying on a single attention strategy. This has led to the exploration of hybrid models that combine multiple attention mechanisms to provide a richer representation of both local and global features.
ARF-Net33 was introduced for breast mass segmentation in both mammographic and ultrasound images, leveraging an encoder–decoder backbone integrated with a Selective Receptive Field Module (SRFM) to adaptively regulate receptive field sizes based on lesion scale, thereby balancing global context and local detail for improved accuracy. In34, the authors presented a lightweight CNN-based model for mammogram segmentation, incorporating feature strengthening modules for enhanced representation, a parallel dilated convolution block for multi-scale context and boundary refinement, and a mutual information loss to maximize consistency with ground truth. These innovations collectively enable accurate and efficient segmentation with low computational cost. ATFE-Net35 employed an Axial-Trans module to efficiently capture long-range dependencies and a Trans-FE module to enhance multi-level feature representations.
Transformer and Mamba-based architectures in medical imaging
Inspired by the breakthroughs of Transformer architectures in natural language processing, Vision Transformers (ViTs) and their variants have gained significant traction in medical image analysis, demonstrating strong capability in modeling global context and capturing long-range dependencies36. Transformers overcome CNNs’ local constraints by enabling global context modeling through self-attention processes. Several studies have successfully incorporated transformers into segmentation pipelines, either as standalone modules or in combination with CNN backbones37,38.
To integrate local convolutional features with long-range contextual information, a hybrid CNN-transformer architecture was presented by He et al.39 and Ma et al.35. While these models demonstrate strong performance, these architectures often face challenges in retaining fine-grained boundary details, which are essential for precise segmentation of medical images. Swin Transformer-based networks address this limitation by employing hierarchical attention and shifted windows to capture features at multiple scales. For instance, DS-TransUNet40 leverages these mechanisms to simultaneously extract coarse and fine features, enhancing segmentation precision. Similarly, Swin-Net41 combines a Swin Transformer encoder with feature refinement and hierarchical multi-scale feature fusion modules to achieve more accurate lesion delineation. SwinHR42 further enhances performance by adopting hierarchical re-parameterization with large kernel convolutions, capturing long-range dependencies efficiently while maintaining high accuracy through shifted window-based self-attention. Cao et al.43 took this further by developing a pixel-wise neighbor representation learning approach (NeighborNet), allowing each pixel to adaptively select its context based on local complexity. This approach is particularly suitable for ultrasound segmentation, where lesion boundaries may be fragmented or ambiguous.
In breast cancer segmentation, a critical research gap lies in the integration of transformer-based models with CNNs, where semantic mismatches between locally extracted CNN features and globally contextualized transformer representations often lead to suboptimal fusion44,45. Inflexible or disjointed fusion strategies, such as rigidly inserting transformer blocks into CNN architectures without addressing feature consistency, result in redundant or insufficient hierarchical representations45. This challenge is exacerbated in noisy or irregular data, such as breast ultrasound images, where speckle artifacts, shadowing, and blurred lesion boundaries create discordance between local texture details and global anatomical structures45,46. Current approaches frequently fail to link the semantic gap between CNNs’ localized feature extraction and transformers’ long-range dependency modeling, particularly in decoder stages where misaligned feature maps reduce segmentation precision for small lesions and complex margins39. Furthermore, the lack of adaptive cross-attention mechanisms to harmonize multi-scale features often diminishes model robustness against ultrasound-specific noise patterns45, highlighting the need for more sophisticated hybrid architectures that enable synergistic local-global feature interaction while maintaining computational efficiency39.
Accurate medical image segmentation is essential for clinical decision-making, but existing CNN-Transformer hybrid models often depend heavily on skip connections, which limit the extraction of contextual features. To address this, MRCTransUNet combines a lightweight MR-ViT with a reciprocal attention module to close the semantic gap and retain fine details. The MR-ViT and RPA modules enhance long-range contextual learning in deeper layers, but skip connections are only utilized in the first layer, in contrast to conventional U-Net variations. Tests on breast, brain, and lung datasets show that MRCTransUNet exceeds the performance of current leading methods on Dice and Hausdorff metrics, demonstrating its potential for reliable clinical use applications47.
The authors proposed HCMNet48, a hybrid CNN–Mamba network that integrates CNN’s strength in local feature extraction with Mamba’s capability for efficient global representation. A wavelet feature extraction module enriches feature learning by combining low- and high-frequency components, reducing spatial information loss during downsampling. Furthermore, an adaptive feature fusion module enhances skip connections by dynamically merging encoder and wavelet features, thereby preserving critical details and suppressing redundancy. The authors introduced AttmNet49, a novel multiscale attention-mamba (MAM) module in a U-shaped model. Using a Mamba unit that combines self-attention and Mamba processes, the MAM block combines multi-level convolutional layers to extract features across various spatial scales. With this design, the model can retain fine structural features.
Methodology
The proposed HA-Net is consists of four key components: an encoder, a decoder, a transformer-based attention module, and a spatial feature enhancement block. For the encoder backbone, DenseNet-12150 is used to effectively capture both complex and fine-grained representations. DenseNet’s used feature–direct connections between all layers within a dense block (DB) encourages feature reuse, improves gradient flow, and supports efficient information propagation. These characteristics are especially valuable in medical image segmentation, where subtle anatomical variations and boundary precision are critical for reliable lesion delineation. In the encoding path, four hierarchical encoding stages are constructed following the standard DenseNet-121 design. Each stage comprises multiple dense blocks interleaved with transition layers (TLs), as illustrated in Fig. 1. This hierarchical organization enables the model to progressively learn low-level texture features alongside high-level semantic information while maintaining spatial continuity. The dense connectivity within DBs strengthens feature propagation, while TLs serve to reduce dimensionality and regulate complexity without discarding critical details. Together, these mechanisms ensure that the encoder produces a rich, multi-scale, and highly discriminative representation suitable for subsequent decoding and attention operations. To further refine extracted features, we append a convolutional block–consisting of a \(3 \times 3\) convolution, a ReLU activation, and batch normalization (BN) after the pre-trained DenseNet-121.
The decoding path follows a simplified U-Net15 inspired design, optimized to maintain strong representational power while reducing the number of parameters for improved computational efficiency. Rather than relying on transposed convolutions, which are prone to introducing checkerboard artifacts and can substantially increase computational complexity, our approach utilizes bilinear upsampling followed by convolutional layers. This combination preserves spatial resolution and fine-grained feature details while minimizing parameter count and inference time. By preserving detailed feature reconstruction and precise boundary delineation, the proposed decoding pathway delivers accurate segmentation while keeping computational demands low.
The proposed HA-Net employs five sequential convolutional blocks in the decoder path as shown in Fig. 1, to progressively extract hierarchical features. Each convolutional block is composed of a \(3 \times 3\) convolution layer, BN, and a ReLU activation function to stabilize training and enhance feature representation. This design ensures stable and efficient training while enabling the network to simultaneously capture high-level semantic representations and fine-grained textural details. BN reduces internal covariate shift, speeds up convergence, and enhances generalization, while ReLU activation adds non-linearity to efficiently represent intricate patterns in breast ultrasound images.
Additionally, skip connections from the encoder are employed to preserve spatial information and facilitate multi-scale feature fusion across different resolution levels. The integration of SFEB within the decoding path further refines the feature representations by selectively emphasizing tumor-relevant regions, thereby improving segmentation accuracy while maintaining a reduced parameter count relative to a conventional U-Net. This optimized architecture not only enables efficient processing of high-resolution medical images but also ensures precise delineation of fine structural details, making the model highly suitable for practical clinical deployment and real-time applications.
Transformer Attention Module (TAM)
To strengthen the method’s capacity to capture and fuse contextual information, we incorporate a self-aware attention module51. There are two main components to this module. Initially, contextual information is captured by the Transformer Self-Attention (TSA) block by taking into account relative positions within the input data. It integrates positional information by concatenating input features with positional embeddings paths to allow the model to understand spatial relationships within the input data. Secondly, the Global Spatial Attention (GSA) block refines local contextual information by aggregating it with global features. By incorporating a broader perspective, this design enhances the model’s ability to retain fine structural details while simultaneously maintaining a holistic understanding of the lesion’s overall morphology. Collectively, these attention mechanisms improve feature representation, helping the model effectively balance local and global information for more precise segmentation.
Figure 2 depicts the Transformer Attention Module (TAM) architecture. The input feature map \(F_{in}\) is first enriched with positional encoding and passed to two parallel branches. In the top branch (TSA), the encoded features are projected into Q, K, and V for calculation of scaled dot-product attention, capturing long-range contextual dependencies. In the bottom branch (GSA), the features are embedded into two complementary representations whose dot product produces a spatial attention map, highlighting global positional relationships. Finally, the outputs of TSA, GSA, and the PE-enriched input are concatenated to generate \(F_{out}\), which jointly preserves local details, global context, and spatial correlations.
Transformer Self-Attention (TSA)
Since multi-head attention effectively captures self-correlation but cannot learn spatial relationships, a common strategy is to introduce positional encoding before applying attention. Specifically, the incoming feature representation \(F\in \mathbb {R}^{h\times w\times c}\) is first enriched with positional information, producing a representation, which is then fed into the multi-head attention block (Fig. 2). F is first reshaped into a two-dimensional representation \(F' \in \mathbb {R}^{c \times (h \times w)}\). Using learnable weight matrices, \(F'\) is then projected into three distinct spaces: queries \(Q \in \mathbb {R}^{c \times (h \times w)}\), keys \(K \in \mathbb {R}^{c \times (h \times w)}\), and values \(V \in \mathbb {R}^{c \times (h \times w)}\), defined as
where \(W_q, W_k, W_v \in \mathbb {R}^{c \times c}\) are learnable projection matrices.
The scaled dot-product attention mechanism computes the similarity between different channels by applying the Softmax-normalized dot-product of Q and the transposed version of K. This matrix represents the contextual attention map \(A \in \mathbb {R}^{c \times c}\). Finally, the contextual attention map A is applied to the value matrix V to produce attention-weighted feature representations. This mechanism allows the multi-head attention module to selectively aggregate relevant features while preserving essential contextual dependencies across spatial positions. Mathematically, the Transformer Self-Attention (TSA) operation can be expressed as:
where \(d_k\) denotes the dimensionality of the key vectors, ensuring proper scaling of the dot-product attention. This formulation allows the TSA block to model long-range dependencies and refine feature aggregation while maintaining spatial coherence in the output representations.
Global Spatial Attention (GSA)
To further enhance contextual learning, the TAM incorporates the Global Spatial Attention (GSA) block, which captures correlations among different spatial positions across the feature map. The initial feature representation \(F\in \mathbb {R}^{h\times w\times c}\) is embedded in \(F^c\in \mathbb {R}^{h\times w\times c}\) and \(F^{cc}\in \mathbb {R}^{h\times w\times c'}\) where c’ = c/2. The latter is reshaped to \(F1^{cc}\in \mathbb {R}^{h\times w\times c'}\) and \(F2^{cc}\in \mathbb {R}^{h\times w\times c'}\). The scaled dot product of these matrices is computed and subsequently passed through a Softmax normalization layer, resulting in an attention map \(GSA \in \mathbb {R}^{(h \times w) \times (h \times w)}\) that encodes the pairwise correlations between different spatial positions. The multi-head attention mechanism is then formulated as:
The outputs from TSA, GSA, and the original input are then concatenated to create the output feature map (\(F_{out}\in \mathbb {R}^{h\times w\times c}\)) of the self-aware attention module. The model’s capacity to extract significant features for precise segmentation is improved by this method, which guarantees that both local spatial relationships and global context are well recorded.
Spatial Feature Enhancement Block (SFEB)
Pooling operations play a critical role in deep learning by reducing the spatial dimensions of feature maps, accelerating computation, and enhancing feature robustness. In lesion segmentation, it is crucial to simultaneously capture fine-grained structural details and global contextual cues, since tumors often exhibit low contrast, small spatial extent, and heterogeneous textural patterns. To address these challenges, we incorporate an SFEB within the skip connections of our network, which strengthens feature fusion, spatial awareness, and residual learning, ultimately improving segmentation accuracy and the preservation of lesion boundaries.
To improve discriminative characteristics and refine spatial features before fusion, the SFEB is integrated into skip connections. Global max-pooling highlights sharp lesion boundaries, while global average-pooling preserves contextual information, and their combination ensures a balance between local detail and global context. The attention pathway further reweights channels to emphasize lesion-relevant features and suppress background noise. Finally, residual fusion preserves fine spatial details, making the SFEB particularly effective for refining skip connection features in noisy ultrasound images with irregular tumor boundaries.
The input tensor is first passed through a \(3 \times 3\) convolutional layer, BN, and a ReLU activation, resulting in an intermediate feature map \(I_1\).
where \({I} = \mathbb {R}^{H\times W\times C}\) represents the input tensor with height H, width W, and channel depth C. To extract global contextual information, the intermediate feature map \(I_1\) is subjected to both global max-pooling (\(G_m\)) and global average-pooling (\(G_a\)), producing the pooled feature maps \(GP_m\) and \(GP_a\), respectively:
These pooled features are concatenated to form a complementary representation Po:
The concatenated pooled features \(P_o\) are then refined by applying a \(3 \times 3\) convolution, BN, and ReLU activation:
In parallel, \(G_a\) is applied to the original input I, followed by a \(1 \times 1\) convolution, BN, and sigmoid activation, producing an attention map Fcc:
where \(\sigma\) represents the sigmoid activation. The attention-enhanced features \(F_{em}\) are computed by element-wise multiplication of the refined feature map \(F_c\) and the attention coefficients \(F_{cc}\):
Finally, to preserve the original spatial information and maintain residual learning, the input tensor I is added element-wise to the attention-enhanced features:
By maintaining fine structural details, this architecture effectively balances local feature intricacy with global contextual information, enabling the model to focus on relevant regions. The SFEB’s architecture is shown in Fig. 3.
Loss functions
An appropriate choice of a loss function is crucial to train the model because it directly influences the convergence behavior, stability, and the balance between pixel-wise accuracy and region-level consistency in segmentation tasks52. BCE loss quantifies the pixel-wise difference between the predicted probability map and the ground truth mask. By computing the negative log-likelihood of the predicted probabilities, it penalizes incorrect classifications and enforces accurate pixel-level segmentation. Formally, for N pixels, BCE loss is defined as:
where \(y_i \in {0,1}\) represents the ground truth label of the i-th pixel, and \(\hat{y}_i \in [0,1]\) denotes the predicted probability. This formulation ensures that confident misclassifications are penalized more heavily, guiding the model toward robust pixel-level discrimination.
Jaccard loss, also known as Intersection-over-Union (IoU) loss, is a region-level metric that evaluates the degree of overlap between the predicted segmentation mask and the ground truth, emphasizing accurate delineation of target regions. Because Jaccard loss prioritizes structural similarity over pixel-wise loss, it works especially well with highly unbalanced datasets in which the lesion or region of interest only takes up a small portion of the image. It has the following mathematical definition:
where \(P_i \in [0,1]\) is the predicted probability for the i-th pixel, and \(G_i \in {0,1}\) is the corresponding ground truth label.
To leverage both pixel-level accuracy (captured by BCE loss) and region-level similarity (captured by Jaccard loss), a hybrid objective is formulated. The final training objective is defined as:
where \(\textrm{Loss}_\textrm{bce}\) ensures fine-grained classification at each pixel, and \(\textrm{Loss}_\textrm{jaccard}\) enforces global shape and boundary consistency. This joint formulation stabilizes convergence and improves segmentation performance across varying lesion sizes and shapes.
Code availability
The source code implementing the proposed Hybrid Attention Network (HA-Net) for breast tumor segmentation in ultrasound images is openly available at GitHub Repository Link: https://github.com/nisarahmedrana/HA-Net. A DOI has been generated via Zenodo to ensure long-term accessibility: https://doi.org/10.5281/zenodo.17190194. The repository includes processed dataset, Jupyter Notebook describing architecture, preprocessing pipelines, training and evaluation scripts and usage instructions required to reproduce the results presented in this study. The code is released for research purposes only under the specified license.
Experiments
Datasets for breast ultrasound image segmentation
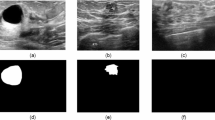
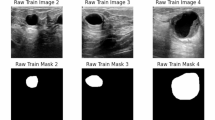
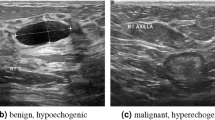
To rigorously evaluate the effectiveness of the HA-Net, we conducted extensive experiments on two publicly available breast ultrasound datasets, BUSI and UDIAT. Both datasets consist of grayscale ultrasound images with corresponding pixel-level annotations provided by clinical experts, serving as reliable benchmarks for tumor segmentation. The BUSI dataset contains ultrasonograms of multiple patients with varying lesion types (benign, malignant, and normal), thereby reflecting the heterogeneity of real clinical scenarios. The UDIAT dataset, on the other hand, offers high-quality ultrasound scans with consistent acquisition settings, enabling controlled evaluation. Together, these datasets provide complementary characteristics, ensuring that the proposed method is validated across diverse imaging conditions and lesion appearances.
BUSI Dataset: The BUSI dataset53 comprises 780 grayscale breast ultrasound images obtained from 600 female patients within the age range of 25 to 75 years. Each image has an approximate spatial resolution of \(500 \times 500\) pixels and is annotated into three diagnostic categories: normal, benign, and malignant. For tumor segmentation, only the benign and malignant categories were retained, as these are accompanied by expert-annotated binary masks delineating tumor regions. Images belonging to the normal class were excluded since they lack lesion annotations. To ensure uniformity in model input, all images and their corresponding masks were resized to \(256 \times 256\). This preprocessing step not only standardizes input dimensions across the dataset but also reduces computational overhead during training and evaluation.
UDIAT Dataset: The UDIAT dataset was introduced by54 and consists of 163 breast ultrasound images. These images are divided into benign and malignant classifications and have a resolution of \(760 \times 570\) pixels. Pixel-wise segmentation masks with expert annotations that identify tumor locations are included with every image. Before training, all images and their corresponding masks were resized to \(256 \times 256\) pixels to ensure consistency. Table 1 provides details of the BUSI and UDIAT datasets’ separation into training and test sets.
Implementation details
To ensure robust training and reliable performance evaluation, 20% of the training set was withheld for validation, enabling effective monitoring of learning progress and guiding hyperparameter adjustments. Model optimization was performed using the Adam optimizer55 with an initial learning rate of 0.001. To promote stable convergence and mitigate the risk of stagnation, the learning rate was reduced by a factor of 0.25 when the validation loss plateaued for four consecutive epochs. In addition, early stopping was employed to prevent overfitting and automatically terminate training once no further improvements were observed.
A hybrid loss function combining Binary Cross-Entropy (BCE) and Jaccard loss was utilized, allowing simultaneous optimization at both the pixel level and the region overlap level. Training was conducted with a batch size of 10, and the model achieved competitive performance without the application of explicit data augmentation strategies. The proposed framework was implemented in Keras with TensorFlow as the backend. All experiments were executed on a workstation equipped with an NVIDIA Tesla K80 GPU, an Intel Xeon 2.20 GHz CPU, 13 GB of system RAM, and 12 GB of dedicated GPU memory
Evaluation metrics
The segmentation performance of the proposed HA-Net was quantitatively assessed using a set of widely adopted evaluation metrics in medical image analysis. These metrics capture both pixel-level accuracy and region-level overlap, providing a comprehensive view of model performance. The definitions and interpretations of all metrics are summarized in Table 2.
Ablation studies
A series of ablation studies were performed on the BUSI dataset to systematically assess the individual contributions of each component within the proposed HA-Net. The pre-trained DenseNet-121 encoder used in the backbone model was chosen for its strong feature extraction and multi-scale representation capabilities. To progressively enhance spatial and contextual understanding, we incrementally integrated three key modules into the baseline: a convolutional block, the SFEB, and TAM.
The results in Table 3 clearly demonstrate the impact of each module, with sequential integration consistently improving performance across all evaluation metrics. In particular, the incorporation of SFEB and TAM yields significant gains in performance, underscoring their effectiveness in refining feature representation and enhancing lesion localization. These findings highlight the critical role of both spatial feature refinement and attention-based contextual modeling in enabling precise delineation of tumor regions, validating the design choices of the proposed architecture.
To further explain the interpretability of the HA-Net, heatmaps of the SEFB are visualized using Grad-CAM56 on the BUSI dataset. The SEFB module is integrated into skip connections across four hierarchical levels of the network, enabling progressive refinement of feature representations. The visualization demonstrates how SEFB adaptively emphasizes salient lesion regions while suppressing background noise throughout the encoding–decoding process. In the presented results in Fig. 4, the first column corresponds to the original ultrasound image, while the second column provides the ground truth segmentation mask. The subsequent columns depict the SEFB attention responses at the four skip-connection stages. These stage-wise heatmaps highlight the evolving focus of the network, where shallow layers capture broad structural context and deeper layers progressively concentrate on more discriminative lesion boundaries. This stage-wise visualization confirms that SEFB effectively guides the network toward lesion-relevant regions, thereby improving the reliability of feature propagation through skip connections and contributing to more accurate segmentation outcomes.
Results and discussion
Comparison with SOTA methods on the BUSI dataset
To comprehensively assess the efficiency of HA-Net, the outcomes of SOTA approaches on the BUSI breast ultrasound dataset are compared with the proposed HA-Net. The selected benchmark models encompass a range of architectures and design strategies, including classical encoder-decoder variants (U-Net, UNet++, Attention U-Net), transformer-based networks (Swin-UNet, Eh-Former, BGRD-TransUNet), and specialized attention-guided frameworks (BGRA-GSA, AAU-Net, MCRNet, DDRA-Net). These models represent the current landscape of approaches for the segmentation task and provide a rigorous reference for evaluating the HA-Net.
The quantitative outcomes are summarized in Table 4. The HA-Net consistently achieves competent performance across all quantitative metrics, including DSC, IoU, sensitivity, precision, specificity, and accuracy. The combined use of SFEB and TAM equips the model with the ability to emphasize detailed boundary information while retaining a broader contextual understanding. This architectural design enables the network to effectively handle common challenges in BUSI.
The improvements are particularly notable in metrics that emphasize overlap and boundary accuracy (DSC and IoU), highlighting the method’s ability to precisely delineate tumor regions. High sensitivity and precision scores further indicate that the model reliably identifies tumor pixels with lower false positives, which is critical in clinical practice for accurate diagnosis and reducing unnecessary interventions. Furthermore, the model maintains high specificity, demonstrating its ability to correctly classify normal tissue and avoid mislabeling background regions as lesions.
By effectively combining dense feature extraction, contextual information based on attention features, and spatial features refinement, the framework consistently outperforms existing SOTA methods, providing reliable and accurate segmentation results that could assist radiologists in early breast cancer detection.
Statistical significance analysis
To validate the observed performance improvements of HA-Net over other segmentation models on the BUSI dataset, we applied the Wilcoxon signed-rank test. Compared to Attention U-Net, HA-Net achieved a p-value of \(1.55 \times 10^{-14}\), and against U-Net, the p-value was \(2.71 \times 10^{-14}\). These highly significant results confirm that the superior performance of HA-Net is statistically robust, highlighting its reliability and effectiveness.
Comparison with SOTA methods on the UDIAT dataset
The generalization capability of HA-Net was further examined through comparative experiments on the UDIAT breast ultrasound dataset. The benchmarked models encompass a wide range of recent SOTA approaches, including BGRA-GSA, AAU-Net, MCRNet, Swin-UNet, Eh-Former, U-Net, BGRD-TransUNet, Attention U-Net, and UNet++. These models collectively represent diverse architectural strategies, from encoder-decoder networks to attention-guided and transformer-based frameworks, providing a robust reference for performance assessment.
As presented in Table 5, HA-Net demonstrates strong and consistent performance across all evaluated metrics. It attains the highest scores in Jaccard Index, Dice coefficient, and specificity, which are critical indicators of precise tumor localization and accurate segmentation boundaries. Although BGRD-TransUNet exhibits slightly higher sensitivity and overall accuracy, our model demonstrates a more balanced performance profile, with notable advantages in overlap-based metrics that are particularly relevant for assessing segmentation quality in medical imaging.
These findings highlight the robustness and adaptability of the model across datasets with diverse imaging conditions and tumor characteristics, thereby demonstrating its strong potential for reliable integration into real-world clinical breast cancer diagnosis and screening workflows. Consistent results on UDIAT further demonstrate the suitability of the proposed HA-Net for clinical deployment, supporting its role in accurate tumor segmentation for early diagnosis and effective treatment planning.
Statistical significance analysis
To evaluate the statistical significance of HA-Net’s performance on the UDIAT dataset, a Wilcoxon signed-rank test was conducted, comparing the proposed model against Attention U-Net and U-Net. The results indicate a p-value of \(1.76 \times 10^{-6}\) when compared to Attention U-Net and \(1.47 \times 10^{-5}\) against U-Net. These highly significant values demonstrate that HA-Net’s superior segmentation performance is statistically robust, confirming its effectiveness and reliability in accurately delineating breast tumors in ultrasound images.
Qualitative visualization results
To complement the quantitative results, we also provide qualitative segmentation examples from both the BUSI and UDIAT datasets. Figure 5 presents side-by-side comparisons between HA-Net and representative SOTA models, including U-Net15, UNet++63, and Attention U-Net62, on the BUSI dataset. These visual results emphasize how different methods perform on challenging conditions, such as poor contrast, heterogeneous lesion boundaries, and varying tumor sizes.
The proposed HA-Net consistently produces more precise segmentation boundaries with higher spatial alignment to the ground truth. It effectively suppresses false positives (highlighted in red) and recovers missed tumor regions (highlighted in blue), resulting in cleaner and more reliable segmentation maps. The SFEB and TAM contribute to these improvements by enhancing both local detail and global contextual understanding.
Similarly, Fig. 6 shows qualitative outcomes on the UDIAT dataset, comparing the proposed HA-Net against U-Net, UNet++, and Attention U-Net. The outcomes indicate that the model maintains high segmentation fidelity even in the presence of noise, low-intensity contrast, and irregular tumor morphology. These visualizations reinforce the quantitative findings reported earlier, particularly improvements in Dice coefficient, Jaccard Index, and specificity, emphasizing the model’s robustness.
Discussion
The HA-Net consistently demonstrates competent performance, outperforming recent SOTA models in critical metrics such as Dice coefficient, Jaccard index, and specificity. These advancements underscore the significance of the proposed HA-Net, which synergistically combines dense feature extraction, a spatial feature enhancement block, and a Transformer-based attention module. The model exhibits a strong ability to accurately delineate lesions even in challenging imaging conditions characterized by low contrast, speckle noise, and irregular tumor morphology, highlighting its robustness and generalizability.
Statistical analyses further validate the significance of these performance gains, particularly when compared against leading segmentation frameworks such as UNet++, Attention U-Net, and BGRD-TransUNet. These findings reinforce the potential clinical utility of HA-Net as a reliable tool for automated breast cancer detection and decision support in real-world scenarios.
Despite these competent results, limitations should be acknowledged. First, the model was trained and evaluated on two datasets of moderate size. While the outcomes are encouraging, further validation on larger, multi-center, or multi-device ultrasound datasets is essential to fully assess generalizability. Second, the proposed method can exhibit reduced performance on images with extremely low contrast or poorly defined lesion boundaries, which may challenge accurate feature extraction and segmentation. Future work could address this limitation by incorporating advanced contrast enhancement techniques, adaptive preprocessing, or specialized attention mechanisms to better handle such challenging cases.
General-purpose backbones such as OverLoCK65, SparX66, TransXNet67, and SegMAN68 have advanced visual recognition through novel architectural strategies, but their validation largely relies on natural image benchmarks. In contrast, HA-Net is explicitly designed for breast ultrasound segmentation, addressing domain-specific challenges including speckle noise, scale variation, and indistinct tumor boundaries. HA-Net combines a DenseNet-121 encoder with hybrid attention modules GSA, PE, and SDPA to model long-range dependencies and contextual feature interactions. The inclusion of SFEB in skip connections strengthens lesion-specific spatial details, while a composite BCE and Jaccard loss ensures balanced optimization across pixel- and region-level accuracy. Compared with OverLoCK’s biologically inspired attention, SparX’s cross-layer aggregation, and TransXNet’s dynamic token mixing, HA-Net adapts these concepts more effectively to the clinical setting by prioritizing feature clarity, spatial refinement, and noise robustness. Unlike SegMAN, which targets large-scale semantic segmentation, HA-Net demonstrates how domain-adaptive design can substantially improve segmentation reliability under the complex conditions of BUS imaging.
Computational complexity
To provide a rigorous evaluation of the proposed HA-Net against SOTA approaches, a computational complexity analysis was conducted on the BUSI dataset. The primary goal of this analysis is to establish a trade-off between segmentation performance and computational complexity, where resource-constrained environments such as portable ultrasound scanners or clinical workstations are common. The comparison considers several complementary metrics. First, the number of trainable parameters is reported, which directly reflects the capacity of the model and its tendency toward overfitting or generalization. Models with fewer parameters generally require less storage and faster inference but may sacrifice representational power if overly simplified. Second, the IoU is adopted as the main performance metric, as it provides a reliable measure of region overlap between the predicted segmentation mask and the ground truth annotation. This metric is especially suitable for medical segmentation tasks, where precise boundary delineation is critical. Alongside segmentation accuracy, we also report the floating-point operations (FLOPs), which represent the theoretical computational cost of a single forward pass through the network. A lower FLOP count reflects reduced arithmetic complexity, thereby improving the model’s suitability for real-time clinical deployment. Finally, the memory footprint is reported, capturing the storage and runtime memory requirements. This measure is crucial in scenarios where computational resources are limited, such as edge devices or cloud-based telemedicine applications. By integrating these four metrics, parameters, IoU score, FLOPs, and memory consumption provide a comprehensive view of model performance that extends beyond accuracy alone. The results, summarized in Table 6, enable a fair comparison between methods and highlight the balance between predictive reliability and computational feasibility, thereby guiding the choice of models for practical medical imaging applications.
Conclusion
This study introduces HA-Net, a hybrid attention-based architecture specifically designed for the automated segmentation of breast tumors in ultrasound images. The architecture leverages a pre-trained DenseNet-121 encoder combined with an attention mechanism incorporating Global Spatial Attention (GSA), Position Encoding (PE), and Scaled Dot-Product Attention (SDPA), thereby allowing the model to effectively capture global contextual relationships while preserving fine-grained spatial details that are critical for precise tumor delineation. The Spatial Feature Enhancement Block was integrated into skip connections to preserve high-resolution information and refine focus on tumor regions. The segmentation process is guided by a combined loss function, thereby effectively mitigating challenges arising from class imbalance and the heterogeneous morphologies of breast lesions. Comprehensive experiments conducted on the BUSI and UDIAT datasets show that HA-Net consistently surpasses existing SOTA segmentation methods across multiple evaluation metrics. Both quantitative and qualitative assessments validate its robustness, high performance, and generalizability, highlighting its potential utility as a clinical tool for facilitating early and precise breast cancer diagnosis.
Future work will aim to improve cross-device and multi-center generalization via domain adaptation, incorporate lesion classification to create a comprehensive diagnostic framework, and enable real-time deployment in clinical workflows to enhance diagnostic efficiency and patient care.
Data availability
The data used in this research is publicly available for research and development purposes at our GitHub repository: https://github.com/nisarahmedrana/HA-Net.
References
Zhang, S., Jin, Z., Bao, L. & Shu, P. The global burden of breast cancer in women from 1990 to 2030: assessment and projection based on the global burden of disease study 2019. Front. Oncol. 14, 1364397 (2024).
Zheng, D., He, X. & Jing, J. Overview of artificial intelligence in breast cancer medical imaging. J. clinical medicine 12, 419 (2023).
Karellas, A. & Vedantham, S. Breast cancer imaging: a perspective for the next decade. Med. physics 35, 4878–4897 (2008).
Benson, S., Blue, J., Judd, K. & Harman, J. Ultrasound is now better than mammography for the detection of invasive breast cancer. The Am. journal surgery 188, 381–385 (2004).
Madjar, H. Role of breast ultrasound for the detection and differentiation of breast lesions. Breast Care 5, 109–114 (2010).
Gonzaga, M. A. How accurate is ultrasound in evaluating palpable breast masses? Pan Afr. Med. J. 7 (2010).
Liu, L. et al. Automated breast tumor detection and segmentation with a novel computational framework of whole ultrasound images. Med. & biological engineering & computing 56, 183–199 (2018).
Ahmed, N., Asif, H. M. S. & Khalid, H. Piqi: perceptual image quality index based on ensemble of gaussian process regression. Multimed. Tools Appl. 80, 15677–15700 (2021).
Khalid, H., Ali, M. & Ahmed, N. Gaussian process-based feature-enriched blind image quality assessment. J. Vis. Commun. Image Represent. 77, (2021).
Ahmed, N. & Asif, H. M. S. Perceptual quality assessment of digital images using deep features. Comput. Informatics 39, 385–409 (2020).
Ahmed, N., Shahzad Asif, H., Bhatti, A. R. & Khan, A. Deep ensembling for perceptual image quality assessment. Soft Comput. 26, 7601–7622 (2022).
Aslam, M. A. et al. Vrl-iqa: Visual representation learning for image quality assessment. IEEE Access 12, 2458–2473 (2023).
Aslam, M. A. et al. Qualitynet: A multi-stream fusion framework with spatial and channel attention for blind image quality assessment. Sci. Reports 14, 26039 (2024).
Aslam, M. A. et al. Tqp: An efficient video quality assessment framework for adaptive bitrate video streaming. IEEE Access (2024).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18, 234–241 (Springer, 2015).
Song, K., Feng, J. & Chen, D. A survey on deep learning in medical ultrasound imaging. Front. Phys. 12, 1398393 (2024).
Huang, Z., Wang, L. & Xu, L. Dra-net: Medical image segmentation based on adaptive feature extraction and region-level information fusion. Sci. Reports 14, 9714 (2024).
Anari, S., Sadeghi, S., Sheikhi, G., Ranjbarzadeh, R. & Bendechache, M. Explainable attention based breast tumor segmentation using a combination of unet, resnet, densenet, and efficientnet models. Scientific Reports 15, 1027 (2025).
Fang, W. & Han, X.-h. Spatial and channel attention modulated network for medical image segmentation. In Proceedings of the Asian conference on computer vision (2020).
Murase, R., Suganuma, M. & Okatani, T. How can cnns use image position for segmentation? arXiv preprint arXiv:2005.03463 (2020).
Shen, X. et al. Dilated transformer: residual axial attention for breast ultrasound image segmentation. Quant. Imaging Medicine Surg. 12, 4512 (2022).
He, J. et al. Sab-net: Self-attention backward network for gastric tumor segmentation in ct images. Comput. Biol. Medicine 169, (2024).
Zhang, H. et al. Acl-dunet: A tumor segmentation method based on multiple attention and densely connected breast ultrasound images. PloS one 19, e0307916 (2024).
Byra, M. et al. Breast mass segmentation in ultrasound with selective kernel u-net convolutional neural network. Biomed. Signal Process. Control. 61, (2020).
Zhang, S. et al. Fully automatic tumor segmentation of breast ultrasound images with deep learning. J. Appl. Clin. Med. Phys. 24, (2023).
Michael, E., Ma, H., Li, H., Kulwa, F. & Li, J. Breast cancer segmentation methods: current status and future potentials. BioMed research international 2021, 9962109 (2021).
Xu, Y., Liu, F., Xu, W. & Quan, R. Overview of graph theoretical approaches in medical image segmentation. In International Conference on Computational & Experimental Engineering and Sciences, 819–835 (Springer, 2024).
Li, L., Niu, Y., Tian, F. & Huang, B. An efficient deep learning strategy for accurate and automated detection of breast tumors in ultrasound image datasets. Front. Oncol. 14, 1461542 (2025).
Pan, P. et al. Tumor segmentation in automated whole breast ultrasound using bidirectional lstm neural network and attention mechanism. Ultrasonics 110, 106271 (2021).
Abraham, N. & Khan, N. M. A novel focal tversky loss function with improved attention u-net for lesion segmentation. In 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), 683–687 (IEEE, 2019).
Chen, G., Li, L., Dai, Y., Zhang, J. & Yap, M. H. Aau-net: an adaptive attention u-net for breast lesions segmentation in ultrasound images. IEEE Transactions on Medical Imaging 42, 1289–1300 (2022).
Chen, G. et al. Esknet: An enhanced adaptive selection kernel convolution for ultrasound breast tumors segmentation. Expert. Syst. with Appl. 246, (2024).
Xu, C. et al. Arf-net: An adaptive receptive field network for breast mass segmentation in whole mammograms and ultrasound images. Biomed. Signal Process. Control. 71, (2022).
Pi, J. et al. Fs-unet: Mass segmentation in mammograms using an encoder-decoder architecture with feature strengthening. Comput. Biol. Medicine 137, (2021).
Ma, Z. et al. Atfe-net: axial transformer and feature enhancement-based cnn for ultrasound breast mass segmentation. Comput. Biol. Medicine 153, (2023).
Xiao, H., Li, L., Liu, Q., Zhu, X. & Zhang, Q. Transformers in medical image segmentation: A review. Biomed. Signal Process. Control. 84, (2023).
Liu, Q. et al. Optimizing vision transformers for medical image segmentation. In ICASSP 2023-2023 IEEE international conference on acoustics, speech and signal processing (ICASSP), 1–5 (IEEE, 2023).
Zhang, J., Li, F., Zhang, X., Wang, H. & Hei, X. Automatic medical image segmentation with vision transformer. Appl. Sci. 14, 2741 (2024).
He, Q., Yang, Q. & Xie, M. Hctnet: A hybrid cnn-transformer network for breast ultrasound image segmentation. Comput. Biol. Medicine 155, (2023).
Lin, A. et al. Ds-transunet: Dual swin transformer u-net for medical image segmentation. IEEE Transactions on Instrumentation Meas. 71, 1–15 (2022).
Zhu, C. et al. Swin-net: a swin-transformer-based network combing with multi-scale features for segmentation of breast tumor ultrasound images. Diagnostics 14, 269 (2024).
Zhao, Z. et al. Swinhr: Hemodynamic-powered hierarchical vision transformer for breast tumor segmentation. Comput. biology medicine 169, (2024).
Cao, W. et al. Neighbornet: Learning intra-and inter-image pixel neighbor representation for breast lesion segmentation. IEEE J. Biomed. Heal. Informatics (2024).
Zhang, H. et al. Hau-net: Hybrid cnn-transformer for breast ultrasound image segmentation. Biomed. Signal Process. Control. 87, (2024).
Wu, R., Lu, X., Yao, Z. & Ma, Y. Mfmsnet: A multi-frequency and multi-scale interactive cnn-transformer hybrid network for breast ultrasound image segmentation. Comput. Biol. Medicine 177, (2024).
Tagnamas, J., Ramadan, H., Yahyaouy, A. & Tairi, H. Multi-task approach based on combined cnn-transformer for efficient segmentation and classification of breast tumors in ultrasound images. Vis. Comput. for Ind. Biomed. Art 7, 2 (2024).
Zhang, Z. et al. A novel deep learning model for medical image segmentation with convolutional neural network and transformer. Interdiscip. Sci. Comput. Life Sci. 15, 663–677 (2023).
Xiong, Y., Shu, X., Liu, Q. & Yuan, D. Hcmnet: A hybrid cnn-mamba network for breast ultrasound segmentation for consumer assisted diagnosis. IEEE Transactions on Consumer Electron. 1–1, https://doi.org/10.1109/TCE.2025.3593784 (2025).
Zhu, H. et al. Attmnet: a hybrid transformer integrating self-attention, mamba, and multi-layer convolution for enhanced lesion segmentation. Quant. Imaging Medicine Surg. 15, 4296–4310, https://doi.org/10.21037/qims-2024-2561 (2025).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, 4700–4708 (2017).
Chen, B., Liu, Y., Zhang, Z., Lu, G. & Kong, A. W. K. Transattunet: Multi-level attention-guided u-net with transformer for medical image segmentation. IEEE Transactions on Emerg. Top. Comput. Intell. (2023).
Jadon, S. A survey of loss functions for semantic segmentation. In 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), 1–7 (IEEE, 2020).
Al-Dhabyani, W., Gomaa, M., Khaled, H. & Fahmy, A. Dataset of breast ultrasound images. Data in brief 28, 104863 (2020).
Yap, M. H. et al. Breast ultrasound region of interest detection and lesion localisation. Artif. intelligence medicine 107, (2020).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014).
Selvaraju, R. R. et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international conference on computer vision, 618–626 (2017).
Hu, K. et al. Boundary-guided and region-aware network with global scale-adaptive for accurate segmentation of breast tumors in ultrasound images. IEEE J. Biomed. Heal. Informatics 27, 4421–4432 (2023).
Lou, M., Meng, J., Qi, Y., Li, X. & Ma, Y. Mcrnet: Multi-level context refinement network for semantic segmentation in breast ultrasound imaging. Neurocomputing 470, 154–169. https://doi.org/10.1016/j.neucom.2021.10.102 (2022).
Cao, H. et al. Swin-unet: Unet-like pure transformer for medical image segmentation. In Computer Vision–ECCV 2022 Workshops: Tel Aviv, Israel, October 23–27, 2022, Proceedings, Part III, 205–218 (Springer, 2023).
Qu, X. et al. Eh-former: Regional easy-hard-aware transformer for breast lesion segmentation in ultrasound images. Inf. Fusion 109, (2024).
Ji, Z. et al. Bgrd-transunet: A novel transunet-based model for ultrasound breast lesion segmentation. IEEE Access (2024).
Oktay, O. et al. Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 (2018).
Zhou, Z., Rahman Siddiquee, M. M., Tajbakhsh, N. & Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4, 3–11 (Springer, 2018).
Sun, J. et al. Ddra-net: Dual-channel deep residual attention upernet for breast lesions segmentation in ultrasound images. IEEE Access (2024).
Lou, M. & Yu, Y. Overlock: An overview-first-look-closely-next convnet with context-mixing dynamic kernels. In Proceedings of the Computer Vision and Pattern Recognition Conference, 128–138 (2025).
Lou, M., Fu, Y. & Yu, Y. Sparx: A sparse cross-layer connection mechanism for hierarchical vision mamba and transformer networks. In Proceedings of the AAAI Conference on Artificial Intelligence 39, 19104–19114 (2025).
Lou, M. et al. Transxnet: learning both global and local dynamics with a dual dynamic token mixer for visual recognition. IEEE Transactions on Neural Networks Learn. Syst. (2025).
Fu, Y., Lou, M. & Yu, Y. Segman: Omni-scale context modeling with state space models and local attention for semantic segmentation. In Proceedings of the Computer Vision and Pattern Recognition Conference, 19077–19087 (2025).
Acknowledgements
All authors thank the School of Information Engineering, Xi’an Eurasia University, Xi’an, Shaanxi, China, for their Financial Support and Funding.
Funding
The study is supported by the School of Information Engineering, Xi’an Eurasia University, Xián, Shanxi, China. The authors declare no conflict of interest.
Author information
Authors and Affiliations
Contributions
M.A.A. contributed to the interpretation of results and participated in manuscript review and editing. A.N. conceived the research idea, designed the methodology, conducted the experiments, performed data analysis, and led the manuscript drafting. N.A. contributed to methodology development, assisted in data analysis, and supported manuscript preparation. Z.K. assisted with experiments, contributed to data analysis, and supported manuscript review and editing. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Aslam, M.A., Naveed, A., Ahmed, N. et al. A hybrid attention network for accurate breast tumor segmentation in ultrasound images. Sci Rep 15, 39633 (2025). https://doi.org/10.1038/s41598-025-23213-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-23213-6