Abstract
Serine/threonine phosphatase (Stp1) modulates the expression of Staphylococcus aureus (S. aureus) by regulating cysteine phosphorylation. Therefore, Stp1 is a promising target for inhibiting S. aureus infection. In this study, the natural compound momordin Ic was found to have significant inhibitory activity against Stp1 through virtual screening and phosphatase assays. Molecular dynamics simulations and enzyme kinetics experiment demonstrated that momordin Ic binds to the active center of Stp1, thereby reducing its affinity for the substrate. Radius of gyration, solvent-accessible surface area, and root mean square fluctuation analyses showed that drug-bound Stp1 was more stable than free protein. Moreover, Gly41, His42, Lys43, Thr102, Asn162, and Ile164 played crucial roles in the binding of Stp1 to momordin Ic. The binding sites were analyzed using phosphatase and fluorescence quenching experiments. This study demonstrates that momordin Ic is an effective Stp1 inhibitor and provides a foundation for the development of highly effective antitoxic drugs.
Similar content being viewed by others
Introduction
S. aureus is an important foodborne pathogen that causes food poisoning and can be transmitted to humans through animals and animal-derived foods1. This bacterium is widely distributed in nature and can cause a variety of infections in humans and animals2, including general skin infections3, food poisoning4, and life-threatening diseases5. Antibiotics are important drugs for the clinical treatment of S. aureus infections6. Under normal circumstances, antibiotics have a high degree of specificity for bacterial targets7, mainly by inhibiting bacterial growth and reproduction. S. aureus faces great selection pressure owing to growth inhibition, and it is easy to produce drug-resistant strains. Penicillin-resistant S. aureus appeared by the mid-1940s8. Subsequently, methicillin was administered to inhibit S. aureus infection9. A methicillin-resistant strain of S. aureus was discovered in 1961. Methicillin-resistant Staphylococcus aureus (MRSA) is a recognized superbug resistant to all beta-lactam drugs10. Since its discovery, the rate of isolation of MRSA has increased annually11. The increasing incidence of MRSA infections has led to an increase in vancomycin use12. With the emergence of vancomycin-resistant S. aureus (VRSA) strains, there is an urgent need to discover new strategies to combat S. aureus infections.
S. aureus has a conserved pair of eukaryotic-like Ser/Thr kinase/phosphatases (Stk1/Stp1)13. Stk1/Stp1 function as global regulators of bacteria through the phosphorylation/dephosphorylation of substrate proteins. Gene expression analyses have demonstrated that Stp1 is a critical factor in toxin production and virulence of S. aureus14. Pathogenicity of the Δstp1 strain was markedly diminished in a murine model of abscess infection. Specifically, the number of S. aureus isolated from the livers and kidneys of infected mice was reduced 100-fold and 10,000-fold, respectively, compared to the wild-type strain15. Additionally, Stp1 has been identified as a crucial regulator of cell wall biosynthesis, and its deficiency leads to cell wall thickening16. These findings underscore the critical role of Stp1 in S. aureus pathogenicity, and suggest its potential as a therapeutic target.
Structural analysis revealed that Stp1 is a 244-amino acid protein with an overall fold resembling that of the PP2C phosphatase17. The core structure of Stp1 consists of a central beta sandwich formed by two antiparallel five-stranded beta sheets positioned beneath the active site and flanked on either side by pairs of antiparallel alpha helices18. Stp1 harbors four metal ions, three of which are commonly found in bacterial PP2C phosphatases19. The four metal ions in Stp1 were modeled as Mn2+. Notably, the binuclear metal centers (M1 and M2) of Stp1 are strictly conserved, whereas the third metal ion (M3) is in close proximity to this center. The most notable structural distinction between Stp1 and other bacterial PP2C phosphatases is the presence of a fourth metal ion (M4) near the active site of Stp1, which plays a pivotal role in its catalytic activity.
However, there are limited reports on Stp1 inhibitors. This study used AutoDock Vina software20 to identify novel compounds that inhibit Stp1 from the natural compound library of the Zinc database21. Through virtual screening, momordin Ic was identified as a potential Stp1 inhibitor, with subsequent experiments confirming its effectiveness. Molecular simulations and binding free energy analyses revealed that momordin Ic could stably bind to Stp1 through hydrogen bonding and hydrophobic interactions. These findings underscore the significance of integrating theoretical and experimental approaches in the development of antimicrobials for the treatment of S. aureus infections.
Materials and methods
Virtual screening
AutoDock Vina has many advantages such as fast calculation speed, high precision, and easy operation22; therefore, it is widely used in molecular docking and virtual screening research23. The Zinc database (https://zinc15.docking.org/) is a publicly accessible resource commonly used for ligand screening24. The natural molecules were downloaded from the Zinc database. AutoDock Vina was employed on the Windows operating system platform for virtual screening of a total of 5,172 purchasable natural compounds derived from plant sources. These small molecules exhibit significant pharmacological value and biological activity, thereby enhancing the likelihood of targeting Stp1. The main natural products include flavonoids, glycosides, alkaloids, saponins, coumarins, and other natural compounds. The structural file (.sdf) of the small molecule was converted into a 3D structure file (.pdbqt), which is suitable for use in AutoDock Vina. The 3D structure of Stp1 was obtained from the Protein Data Bank (PDB ID: 5F1M )18 and served as the target structure for the virtual screening. The receptor protein Stp1 was used before screening. The AutoDock tool25 was used to add polar hydrogen atoms to Stp1 and to assign partial charges using Kollman joint atoms26. Subsequently, the structural file for Stp1 was converted to a 3D format (.pdbqt) using the AutoDock Tools. The target protein Stp1 remained rigid, whereas all torsion bonds of the inhibitor were allowed to rotate freely27. A grid box was constructed around the hydrolyzed active region centered on Mn2+ as the ligand-docking site28. To ensure the docking accuracy and calculation efficiency, the volume of the constructed box was reduced as much as possible while covering the active region.
Molecular dynamics simulation
The molecular dynamics simulations and trajectory analyses were performed using Gromacs 4.5.129. Modified Amber99SB field parameters30 were used for both proteins and ligands in the system. At an initial temperature of 300 K, the initial velocities of all atoms were determined using the Maxwell distribution. Bond length of the hydrogen atom was bound using the LINCS algorithm to improve the efficiency of the molecular dynamics simulations31. Tip3p water molecular configuration was maintained using the SETTLE algorithm32. Simultaneously, the remote electrostatic interactions were calculated using the PME method33.
In the molecular simulation process, the complex system was initially optimized using the steepest descent method for 2000 steps, followed by the conjugate gradient method until convergence of the system energy was achieved. The protein complex was then simulated with molecular dynamics of 500 ps using a position-binding algorithm, and the simulation system was gradually relaxed. Finally, molecular dynamics simulations of the complex system were performed at 200 ns34.
Calculation of the binding free energy
In this study, the MM/PBSA method35 was employed to calculate the binding free energy between the inhibitor and Stp1 using the Amber 10 software36. According to previous research reports37, this calculation method has good accuracy and precision, and has been successfully applied to complex systems. The calculation formula is as follows:
ΔGbind = ΔEMM + ΔGsol - TΔS
Where ΔGbind represents the total combined free energy, ΔEMM is the enthalpy changes in the gas phase upon complex formation, ΔGsol refers to the solvated free energy contribution, and -TΔS indicates the entropy contribution to the binding.
Plasmid construction
In this study, stp1 was amplified by PCR using the S. aureus USA300 genome as a template. The stp1 gene and pET-28a plasmid were digested with BamHI and XhoI before ligation. The ligated product (pET-28a-Stp1) was introduced into E. coli BL21 (DE3) cells38. Primers used in the experiments are listed in Table S1.
Expression and purification of Stp1 protein
E.coli BL21 (pET28a-Stp1) was inoculated into the LB medium and cultured to OD600 nm = 0.6. Isopropyl β-D-thiogalactoside was added to LB liquid medium and cultured overnight at 16 °C and 180 r/min to induce Stp1 expression. Bacteria were collected by centrifugation. After resuspension, bacteria were lysed by sonication. The supernatants were collected for protein purification. The proteins were purified and concentrated using a centrifugal filter column39.
Phosphatase assay
Phosphatase hydrolysis experiments were performed to verify the inhibitory activity of the selected candidate compounds against Stp1. Purified Stp1 protein and candidate compounds at different concentrations were successively added to pNPP buffer in 96-well plates. A negative control group with only drugs and no proteins was established, and a positive control group with no drugs was established. After the solution was thoroughly mixed and incubated for 10 min, pNPP was added, and the mixture was incubated at 25 °C for 20 min. The absorbance was measured at 405 nm using an enzyme-labeled instrument40.
Fluorescence quenching assay
In this assay, proteins acted as fluorophores whereas drugs served as quenchers. Binding constant (Kd) values were calculated using the following equation:
r / Df = n Kd - r Kd
Results and discussion
Structural analysis
Stp1 belongs to the eSTP/protein phosphatase 2 C (PP2C) family. The core structure of Stp1 consists of a central beta sandwich formed by two antiparallel five-stranded beta sheets positioned beneath the active site and flanked on either side by pairs of antiparallel alpha helices (Fig. S1A). Stp1 has four metal ions located at the active center, three of which are common in PP2C phosphatases of other bacteria (Fig. S1B). However, human PP2Cα only contains the two-metal center (M1 and M2). The canonical dinuclear metal center (M1 and M2) of Stp1, including the direct ligands of all coordinating metal ions, are strictly conserved in the PP2C members (Fig. S1C). As expected, the Stp1 structure also revealed a third metal ion (M3) close to the dinuclear metal center as in the bacterial PP2C phosphatases. All metal ions were hexa-coordinated. The coordination sphere of M3 was composed of the side-chain oxygen of Asp120 and Asp194 and four water molecules18 (Fig. S1D). Compared with the PP2C phosphatases of other bacteria, the most significant structural difference of Stp1 was the fourth metal ion (M4) near the active site pocket, located between the β1-β2 loop and the β3-α1 turn. M4 is coordinated by Glu18 and five water molecules18 (Fig. S1E).
By searching the RCSB database, we found that an Arg161 variant also exists in addition to the wild-type Stp1 protein structure. The phosphatase activity experiment revealed that the variants of R161A and R161E almost completely lost their phosphatase activity41. In addition, Arg161 forms hydrogen bonds by contacting the main chain of Ala155 and the side chain of Asp198 (Fig. S2). These two residues are respectively located on the helix of the flap domain and the helix of the catalytic domain. Similarly, Arg161 also closely combines the flap domain and the catalytic domain by interacting with the backbone of Phe156 and Asp198 in the stp1 structure41. These results indicate the role of Arg161 in the structure and catalysis of Stp1.
Molecular docking studies
In recent years, S. aureus has developed resistance to multiple antibiotics, and the emergence of methicillin-resistant S. aureus (MRSA) poses challenges to human health and food safety. Owing to the overuse of antibiotics and increased international transportation, the infection rate of MRSA around the world is constantly rising. Therefore, researchers are searching for new targets to combat S. aureus infections. At present, many targets related to the growth, virulence, and drug resistance of S. aureus have been discovered, and their effective inhibitors have been explored. Dalal et al. identified four potential inhibitors of S. aureus FemC through molecular simulation42. In 2021, five potential inhibitors of Lipophilic membrane (LLM) were discovered43. Kumari et al. discovered five small molecules that were tightly bound to YsxC through virtual screening10. Furthermore, potential inhibitors of S. aureus FmtA were screened through theoretical calculation methods44. This study conducted virtual screening of Stp1 inhibitors through molecular docking.
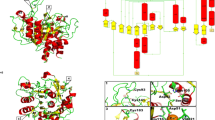
To identify potential inhibitors of Stp1, natural compounds from the Zinc database were used for molecular docking studies. The binding affinity (kcal/mol) of the complex formed by each ligand and receptor protein was determined. Binding affinity is a key indicator of the binding ability of ligands to receptor proteins. After virtual screening based on molecular docking, the binding affinities of all ligands for Stp1 were ranked from lowest to highest, and the top 30 compounds were selected for further analysis. The binding affinity of the first 30 compounds were all less than − 7.0 kcal/mol, thereby indicating that the complex system formed by these compounds and proteins was relatively stable. The complex structures formed by the 30 compounds and the receptor protein Stp1 were analyzed using PyMOL and LigPlot software, respectively. Small molecules tightly bound to the active center of the protein were selected. Finally, six molecules were identified as candidate compounds, which underwent subsequent experimental validation to assess their inhibitory activities. The complexes formed by the six candidate compounds and Stp1 are shown in Fig. 1. All selected candidate compounds demonstrated tight binding within the active region of Stp1, with binding energies recorded at less than − 7.2 kcal/mol (Table S2).
In addition, the discovered inhibitors aurintricarboxylic acid (ATA) and Stp1 have also undergone molecular docking experiments10,43. Our results show that ATA lies in a clover-like pocket located above the dinuclear metal center and near the flap domain (Fig. S3). The binding position is similar to that of other candidate compounds. The binding energy of Stp1 and ATA is -7.1 kcal/mol. The results show that momordin Ic binds more closely to Stp1.
Inhibitory effect of momordin Ic on Stp1 activity
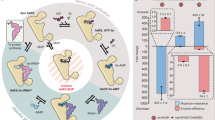
Through virtual screening, six compounds were found to potentially inhibit the Stp1 activity. Notably, momordin Ic (Fig. 2A) inhibited Stp1 expression in phosphatase assays. The results indicated that momordin Ic significantly inhibited Stp1 (Fig. 2B). When the concentration was 32 µg/mL or higher, it was found that the inhibition effect on Stp1 was greater than 60%. As shown in Fig. 2C, KM of the reaction have been changed. This means that momordin Ic is a competitive inhibitor of stp1. These results indicate that virtual screening is an effective method for high-throughput screening.
Molecular dynamics simulation
A 200 ns molecular dynamics simulation was conducted using Gromacs 4.5.1 software to reveal the binding mechanism of momordin Ic and Stp145. The results demonstrated that momordin Ic can bind to the active site of Stp1 (Fig. 3A), which suggested that momordin Ic competes with the substrate for binding to this region, thereby reducing substrate binding to Stp1.
After molecular dynamics simulation, the stability of the complex was measured by the root mean square deviation (RMSD). As shown in Fig. 3B, the RMSD value of Stp1-Momordin Ic complex fluctuates between 0.35 and 0.40 nm after 100 ns, which means that the system reached equilibrium after 100 ns. Additionally, the RMSD fluctuations of the Stp1-momordin Ic complex were notably lower during the 100–200 ns period than those of the free protein, which indicated enhanced stability upon ligand binding.
Radius of gyration (Rg) and solvent-accessible surface area (SASA) analysis
The stability of the Stp1-Momordin Ic complex was investigated by analyzing the Rg and SASA. In Fig. 4A, the Rg value of the Stp1-Momordin Ic complex was lower than that of the free protein. The mean Rg values of free protein and Stp1-Momordin Ic complex were 1.85 nm and 1.84 nm, respectively (Table S3). As shown in Fig. 4B, the SASA of the protein bound to momordin Ic was lower than that of the unbound protein. Rg and SASA analyses confirmed the increased stability of Stp1 after combining with the drug.
Binding free energy and its components
To analyze the contribution of each binding energy, the MM/PBSA method was used to calculate the binding free and decomposition energies. Energy decomposition revealed that the contribution of van der Waals (ΔEvdw), electrostatic (ΔEele), and non-polar solvation (ΔGNP) energies were favorable for complex binding. Table S4 shows that the total binding free energies of the Stp1-Momordin Ic and Stp1-ATA complexes are − 19.34 and − 15.43 kcal/mol respectively, thereby indicating that Stp1 can stably bind with momordin Ic and ATA. It affirms that inhibitors bind efficiently at the active site of Stp1 and form stable Stp1-inhibitor complexes. Further, the energetic contribution of important amino acid residues of Stp1 in Stp1-Momordin Ic and Stp1-ATA complexes were estimated by using MM/PBSA43. Binding energy contributed by important residues to momordin Ic is higher as compared to ATA as shown in Table S5. Overall, MM/PBSA results conclude that Gly41, His42, Lys43, Thr102, Asn162, and Ile164 residues play an important role in the binding of inhibitor(s) to Stp1 and results in the formation of stable complexes.
Identification of binding sites in complexes
To further analyze the binding site of Stp1 with momordin Ic, the binding free energy of each residue is decomposed into the electrostatic (ΔEele), van der Waals (ΔEvdw), and solvation interaction (ΔEsol). According to the binding free energy decomposition (Fig. 5), the amino acid residue with the highest total binding energy was Asn162 (ΔEtotal = -1.40 kcal/mol) primarily due to the van der Waals (ΔEvdw = -1.61 kcal/mol) and electrostatic forces (ΔEele = -1.04 kcal/mol). This indicated that the Asn162 residue of Stp1 strongly interacted with the oxygen atom of momordin Ic. Owing to strong electrostatic interactions, there may be a hydrogen bond between Asn162 and momordin Ic. The total binding free energy of Gly41, His42 and Lys43 are − 1.00, -0.95, and − 1.01 kcal/mol, respectively. Combined with Fig. 3A, it can be found that the benzene ring of momordin Ic is firmly anchored to Stp1 by these three amino acids through hydrophobic interactions. Simultaneously, Ile164 and Thr102 of Stp1 strongly interact with the six-membered ring of momordin Ic, and the total binding free energy is -0.98 and − 0.63 kcal/mol, respectively. This was mainly because of the better van der Waals contributions of Ile164 and Thr102. The electrostatic interaction of Thr102 also implied a hydrogen-bonding force between Thr102 and the ligand. Gly41, His42, Lys43, Thr102, Asn162, and Ile164 anchor momordin Ic to the active center of Stp1.
The root mean square fluctuation (RMSF) describes the fluctuation of each residue during molecular dynamics simulation, thereby providing insights into the flexibility of the system. The flexibility of residues at the binding sites of the complex system was lower than that of free proteins, which indicated that these residues exhibited reduced flexibility and increased rigidity upon binding (Fig. 6A,B).
Concurrently, by calculating the distance between Stp1 residues and momordin Ic, it was found that Gly41, His42, Lys43, Thr102, Asn162, and Ile164 of Stp1 were within 0.2 nm of momordin Ic (Fig. 7A,B). These results indicate that Gly41, His42, Lys43, Thr102, Asn162, and Ile164 are key amino acids involved in the binding of momordin Ic to Stp1.
Determination of hydrogen bonds and coordinate bonds
By observing the structural characteristics of protein side chain groups and ligands, the amino acid residues of proteins may interact with ligands to form hydrogen bonds. As shown in Fig. 8A, the number of hydrogen bonds formed between momordin Ic and Stp1 was between one and two. As illustrated in Fig. 8B, momordin Ic formed two strong hydrogen bonds with the specific amino acid residues of Stp1. The O4 atom of the ether bond in momordin Ic formed a hydrogen bond with the ND2 atom of Asn162 in Stp1 at a distance of 2.89 Å, while the O2 atom formed a hydrogen bond with OG1 of Thr102 at 2.93 Å. This conclusion was consistent with the analysis of the number of hydrogen bonds.
Confirmation of the binding site of momordin Ic to Stp1
To validate the molecular dynamics simulation results, amino acids Gly41 and Ile164 were mutated to alanine, and molecular dynamics simulations of the G41A -Stp1-Momordin Ic and I164A -Stp1-Momordin Ic complexes were performed. Table S6. shows that the ΔGtotal values of wild type (WT) -stp1-, G41A -Stp1-, and I164A -Stp1-Momordin Ic complexes were − 19.34, -17.91, and − 14.74 kcal/mol, respectively. This indicated that the wild-type protein bound more strongly to momordin Ic than the mutant protein. Additionally, fluorescence quenching experiments were performed using wild-type Stp1 and two different Stp1 mutants. The results showed that the Kd values for momordin Ic and Stp1 decreased in the following order: WT (3.32 × 106) ˃ G41A (2.07 × 106) ˃ I164A (4.63 × 105). Phosphatase activity experiments showed that G41A -Stp1 and I164A -Stp1 mutant proteins retained catalytic activity for pNPP, but the inhibitory effect of momordin Ic on these mutants was reduced (Fig. S4). These results indicate that the affinity between WT Stp1 and momordin Ic was higher than that between the mutant protein and momordin Ic. In Fig. S5, the distance between M3 and M4 of Stp1-Momordin Ic is longer than that of free Stp1. Upon binding with the inhibitor, M4 expands outward, resulting in a change in the active site and subsequently affecting the catalytic activity of the active region.
Conclusion
Momordin Ic is an active ingredient extracted from plants and has anticancer46 and anti-inflammatory47 properties. Natural medicines have the advantages of low toxicity, harmlessness, and low cost. Virtual screening and phosphatase experiments confirmed that momordin Ic effectively inhibited Stp1. Molecular dynamics simulations revealed that momordin Ic binds to the active center of Stp1, thereby preventing substrate binding and reducing its catalytic activity. Further studies revealed that Stp1 binds momordin Ic mainly through hydrogen bonding and hydrophobic forces. Gly41, His42, Lys43, Thr102, Asn162, and Ile164 play crucial roles in complex formation. This work offers a novel approach to studying natural drug inhibitors and provides a foundation for future inhibitor research.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding authors upon request.
References
Qian, C., Castañeda-Gulla, K., Sattlegger, E. & Mutukumira, A. N. Enterotoxigenicity and genetic relatedness of Staphylococcus aureus in a commercial poultry plant and poultry farm. Int. J. Food Microbiol. 363, 109454 (2022).
Ahmad-Mansour, N. et al. Staphylococcus aureus toxins: An update on their pathogenic properties and potential treatments. Toxins 13 677 (2021).
Hindy, J. R., Haddad, S. F. & Kanj, S. S. New drugs for methicillin-resistant Staphylococcus aureus skin and soft tissue infections. Curr. Opin. Infect. Dis. 35, 112–119 (2022).
Yu, G., Huang, T. Y. & Li, Y. Kanamycin promotes biofilm viability of MRSA strains showing extremely high resistance to Kanamycin. Microb. Pathog. 196, 106986 (2024).
Tiwari, H., Saha, S. & Ghosh, M. In silico hybridization and molecular dynamics simulations for the identification of candidate human microRNAs for inhibition of virulent proteins’ expression in Staphylococcus aureus. J. Cell Biochem. e30684 (2024).
Wu, K. Y. et al. Multi-target anti-MRSA mechanism and antibiotic synergistic effect of marine alkaloid ascomylactam A in vitro and in vivo against clinical MRSA strains. Biochem. Pharmacol. 232, 116697 (2025).
Rayner, C. & Munckhof, W. J. Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Internal Med. J. 35 S3-S16 (2005).
Marin, M. Methicillin resistant Staphylococcus. Med. (B Aires). 62 (Suppl 2), 30–35 (2002).
Jevons, M. P. & Parker, M. T. The evolution of new hospital strains of Staphylococcus aureus. J. Clin. Pathol 17 243 – 50. (1964).
Kumari, R., Rathi, R., Pathak, S. R. & Dalal, V. Structural-based virtual screening and identification of novel potent antimicrobial compounds against YsxC of Staphylococcus aureus. J. Mol. Struct. 1255, 132476 (2022).
Chambers, H. F. & Deleo, F. R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641 (2009).
Peng, H. et al. Monitoring Vancomycin blood concentrations reduces mortality risk in critically ill patients: a retrospective cohort study using the MIMIC-IV database. Front. Pharmacol. 15, 1458600 (2024).
Sun, F. et al. Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc. Natl. Acad. Sci. 109 15461–15466 (2012).
Chen, F. et al. The enzyme activity of sortase A is regulated by phosphorylation in Staphylococcus aureus. Virulence 14 2171641 (2023).
Cameron, D. R. et al. Serine/threonine phosphatase Stp1 contributes to reduced susceptibility to Vancomycin and virulence in Staphylococcus aureus. J. Infect. Dis. 205, 1677–1687 (2012).
Cheung, A. & Duclos, B. Stp1 and Stk1: the Yin and Yang of Vancomycin sensitivity and virulence in Vancomycin-Intermediate Staphylococcus aureus strains. J. Infect. Dis. 205, 1625–1627 (2012).
Burnside, K. et al. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS One. 5, e11071 (2010).
Zheng, W. et al. Structure-Based identification of a potent inhibitor targeting Stp1-Mediated virulence regulation in Staphylococcus aureus. Cell. Chem. Biology. 23, 1002–1013 (2016).
Liu, T. T. et al. The inhibitory mechanism of aurintricarboxylic acid targeting serine/threonine phosphatase Stp1 in Staphylococcus aureus: insights from molecular dynamics simulations. Acta Pharmacol. Sin. 40, 850–858 (2019).
Trott, O. & Olson, A. J. AutoDock vina: improving the speed and accuracy of Docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Sterling, T. & Irwin, J. J. ZINC 15 – Ligand discovery for everyone. J. Chem. Inf. Model. 55, 2324–2337 (2015).
Ramos, R. S. et al. Potential inhibitors of the enzyme acetylcholinesterase and juvenile hormone with insecticidal activity: study of the binding mode via Docking and molecular dynamics simulations. J. Biomol. Struct. Dyn. 38, 4687–4709 (2020).
Bucinsky, L. et al. Machine learning prediction of 3CLpro SARS-CoV-2 Docking scores. Comput. Biol. Chem. 98, 107656 (2022).
Irwin, J. J., Sterling, T., Mysinger, M. M., Bolstad, E. S. & Coleman, R. G. ZINC: A free tool to discover chemistry for biology. J. Chem. Inf. Model. 52, 1757–1768 (2012).
Forli, S. et al. Computational protein–ligand Docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 11, 905–919 (2016).
Hosseini, M., Chen, W., Xiao, D. & Wang, C. Computational molecular Docking and virtual screening revealed promising SARS-CoV-2 drugs. Precis Clin. Med. 4, 1–16 (2021).
Pantaleão, S. Q. et al. Virtual screening and in vitro assays of novel hits as promising DPP-4 inhibitors. Biochimie 194, 43–50 (2022).
He, J. et al. Identification of selective MtbDHFR inhibitors by virtual screening and experimental approaches. Chem. Biol. Drug Des. 100, 1005–1016 (2022).
Van Der Spoel, D. et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005).
Showalter, S. A. & Brüschweiler, R. Validation of molecular dynamics simulations of biomolecules using NMR spin relaxation as benchmarks: application to the AMBER99SB force field. J. Chem. Theory Comput. 3, 961–975 (2007).
Thallmair, S., Javanainen, M., Fábián, B., Martinez-Seara, H. & Marrink, S. J. Nonconverged constraints cause artificial temperature gradients in lipid bilayer simulations. J. Phys. Chem. B. 125, 9537–9546 (2021).
Stojceski, F. et al. Influence of dexamethasone on the interaction between glucocorticoid receptor and SOX9: A molecular dynamics study. J. Mol. Graph. Model. 125, 108587 (2023).
Sheng, Q. et al. A new function of thymol nanoemulsion for reversing colistin resistance in Salmonella enterica serovar typhimurium infection. J. Antimicrob. Chemother. 78, 2983–2994 (2023).
Zhao, H. et al. Virtual screening and molecular dynamics simulation for identification of natural antiviral agents targeting SARS-CoV-2 NSP10. Biochem. Biophys. Res. Commun. 626, 114–120 (2022).
Bhati, S. K. et al. In Silico screening and molecular dynamics analysis of natural DHPS enzyme inhibitors targeting acinetobacter baumannii. Sci. Rep. 15, 7723 (2025).
Genheden, S. & Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 10, 449–461 (2015).
Yau, M. Q., Liew, C. W. Y., Toh, J. H. & Loo, J. S. E. A head-to-head comparison of MM/PBSA and MM/GBSA in predicting binding affinities for the CB1 cannabinoid ligands. J. Mol. Model. 30, 390 (2024).
Zhao, X., Liu, B., Liu, S., Wang, L. & Wang, J. Anticytotoxin effects of Amentoflavone to Pneumolysin. Biol. Pharm. Bull. 40, 61–67 (2017).
Li, H. et al. β-sitosterol interacts with Pneumolysin to prevent Streptococcus pneumoniae infection. Sci. Rep. 5, 17668 (2015).
Zheng, W., Liang, Y., Zhao, H., Zhang, J. & Li, Z. 5,5’-Methylenedisalicylic acid (MDSA) modulates SarA/MgrA phosphorylation by targeting Ser/Thr phosphatase Stp1. Chembiochem 16, 1035–1040 (2015).
Yang, T. et al. Structural insight into the mechanism of Staphylococcus aureus Stp1 phosphatase. ACS Infect. Dis. 5, 841–850 (2019).
Dalal, V. & Kumari, R. Screening and identification of natural product-like compounds as potential antibacterial agents targeting FemC of Staphylococcus aureus: An in-Silico approach. ChemistrySelect 7 e202201728 (2022).
Kumari, R. & Dalal, V. Identification of potential inhibitors for LLM of Staphylococcus aureus: structure-based pharmacophore modeling, molecular dynamics, and binding free energy studies. J. Biomol. Struct. Dynamics. 40, 9833–9847 (2022).
Dalal, V. et al. Structure-Based identification of potential drugs against FmtA of Staphylococcus aureus: virtual Screening, molecular Dynamics, MM-GBSA, and QM/MM. Protein. J. 40, 148–165 (2021).
Faraji, N., Daly, N. L., Arab, S. S. & Khosroushahi, A. Y. In Silico design of potential Mcl-1 peptide-based inhibitors. J. Mol. Model. 30, 108 (2024).
Wang, J., Liu, Q., Xiao, H., Luo, X. & Liu, X. Suppressive effects of Momordin Ic on HepG2 cell migration and invasion by regulating MMP-9 and adhesion molecules: involvement of p38 and JNK pathways. Toxicol. In Vitro. 56, 75–83 (2019).
Yoo, S. R., Jeong, S. J., Lee, N. R., Shin, H. K. & Seo, C. S. Quantification analysis and In vitro Anti-Inflammatory effects of 20-Hydroxyecdysone, Momordin Ic, and oleanolic acid from the fructus of Kochia scoparia. Pharmacognosy Magazine. 13, 339–344 (2017).
Acknowledgements
This work was supported by the Natural Science Research of Jiangsu Higher Education Institutions of China [Grant no. 25KJD230001], Special Funding for Suzhou Polytechnic Institute of Agriculture Innovative Research Team [Grant no. CXTD202408], the Doctoral Promotion Program Research Initiation Fund of Suzhou Polytechnic Institute of Agriculture [Grant no. BS[2022]21] and Jiangsu double Innovation PhD project [Grant no. JSSCBS20221014].
Author information
Authors and Affiliations
Contributions
Yanan Yang: Methodology, Investigation, Data curation, Writing—original draft. Xuenan Li: Data curation, Methodology. Pingping Hou: Validation, Software, Formal analysis. Na Wei: Data curation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, Y., Li, X., Hou, P. et al. Exploring the inhibition mechanisms of momordin Ic on S. aureus serine/threonine phosphatase (Stp1) using theoretical and experimental approaches. Sci Rep 15, 39054 (2025). https://doi.org/10.1038/s41598-025-24255-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-24255-6