Abstract
Global foodborne Salmonella outbreaks pose a serious concern for food safety and public health underscoring the need for isolation of lytic phages with potential as biocontrol agents. A lytic bacteriophage vB_Sal_S6 was isolated which targets Salmonella serovars - Typhimurium, Bovismorbificans, Infantis, Typhi, Paratyphi, Weltevreden, Poona and as well as Escherichia coli. It exhibited strong bacteriolytic activity across a range of MOIs (0.01–10), with stability across pH 4–10 and temperature conditions (25–55 °C). In artificially contaminated milk, vB_Sal_S6 reduced Salmonella counts by 3.9log at RT and 2.4log at 4 °C within 9 and 6 h respectively (MOI 10). Genomic characterization revealed a dsDNA genome of 36,127 bp length and the absence of virulence, integrase, excisionase, and antibiotic resistance genes, supporting its suitability for biocontrol. Phylogenetic analysis indicated that vB_Sal_S6 clustered with other Salmonella/E.coli phages in Tequintavirus genus within Demerecviridae family. Docking studies identified that ORF123-encoded receptor-binding tail protein binds with ferrichrome receptor FhuA in Salmonella and E.coli as putative receptor. These findings establish vB_Sal_S6 as a promising candidate for biocontrol applications against Salmonella and E. coli.
Similar content being viewed by others
Introduction
Salmonella is a gram-negative, facultative anaerobic, zoonotic bacterium responsible for one-fourth of the diarrheal diseases worldwide1. An estimated 1.35 million infections resulting in 26,500 hospitalizations, and 420 deaths are attributed to Salmonella annually in the United States2 alone. Non-typhoidal Salmonella infections cause salmonellosis that manifests as symptoms including vomiting, nausea, diarrhea, fever, and abdominal cramps occurring within 12–72 h of infection and last for 2–7 days1. Environmental sources such as contaminated water, soil, animal manure, abattoir equipment and direct contact with animal or human carriers introduce Salmonella into food products3 ultimately leading to human infection upon consumption. S. Typhimurium and S. Enteritidis are the predominant serovars globally, commonly linked to meat, poultry, eggs, dairy, fruits, leafy greens, and fresh vegetables4.
The widespread use of broad-spectrum antibiotics against Salmonella has driven the rapid emergence of resistant strains5 and raised concerns about antibiotic residues in the food chain. Consequently, bacteriophages, which are viruses that specifically infect and lyse bacteria, are being explored as alternative therapeutic and control strategy for Salmonella. They offer several advantages, including host specificity, lack of toxicity, rapid bacteriolytic activity, and self-replication to sustain effective populations6.
Phages, the most abundant biological entities on earth, with approximate 1031 particles in existence6, are present in natural microflora in various foods7. Despite their abundance, it is estimated that less than 0.0002% of the global phage metagenome has been identified8 of which, only about 37,882 complete phage genomes are sequenced and available in NCBI as of June 20259. This underscores the fact that a substantial portion of their population remains unexplored and the paramount significance of comprehensive genomic characterization for any practical phage application, encompassing phage therapy and biocontrol. Genomic characterization facilitates the identification of putative receptors and undesirable genes, such as those encoding lysogeny, virulence factors, or antibiotic resistance, which could potentially be transferred to bacterial hosts10. This, coupled with phenotypic characterization such as a one-step growth curve, high lytic ability, and host range determination, enables the selection of safe, strictly lytic phages, thereby minimizing the burden of extensive downstream safety evaluations.
The present study was aimed to isolate and characterize bacteriophages with lytic activity against Salmonella. A novel phage, designated vB_Sal_S6, was isolated and subjected to phenotypic and genomic characterization. The host range was determined to assess its lytic spectrum, while its bacteriolytic activity was evaluated across varying multiplicities of infection (MOIs). Additional studies included one-step growth curve analysis, pH and thermal stability studies, and evaluation of its lytic potential in milk. Furthermore, whole-genome sequencing was performed, followed by genome annotation, generation of a genomic map, and phylogenetic analysis to determine its taxonomic placement and evolutionary relationships.
Materials and methods
Bacterial strains and their culture conditions
Salmonella Typhimurium ATCC 14028 was used as the host strain for phage isolation. In total, 43 bacterial strains obtained from the culture collection of Defence Institute of Bio-Defence Technologies (DIBT), Mysore (Table 1) were used for the determination of host specificity. These strains were preserved in 25% glycerol stocks (v/v) at -80 ˚C and revived overnight in Brain Heart Infusion Broth (Himedia, India) at 37 ˚C. Each bacterial strain was confirmed biochemically using an automated microbial analyzer BD Phoenix™ M50 (BD, USA).
Isolation of phage
The phage was isolated from drainage samples obtained from locations in Mysuru, Karnataka, India. Liquid drainage samples (25 ml) were centrifuged at 15,285 g for 15 min at 4 ˚C and the supernatant containing phages was filtered using a 0.45 μm PTFE membrane syringe filter (Membrane Solutions, US) to remove solid particles. For phage enrichment, overnight culture (100 µl) of S. Typhimurium ATCC 14028 was inoculated to 5 ml of double-strength BHI broth along with 5 ml of the above filtrate and incubated overnight at 37 ˚C. After overnight phage enrichment, the samples were centrifuged at 15,285 g for 10 min at 4 ˚C, followed by filtration with a 0.22 μm PTFE membrane syringe filter (Membrane Solutions) to remove bacterial cells. The presence of lytic phages in the enriched solution was evaluated by spot test. First, a top agar (0.75% BHI agar) kept molten at 55 °C, was combined with 40 µl of Salmonella culture (108 CFU/ml) and overlaid on BHI bottom agar plates (1.5% agar), so that it can grow into a bacterial lawn. After the top agar set, 10 µl of the enriched filtered phage supernatant was spotted on the top agar, allowed to dry and then incubated at 37 °C overnight. If a clear lysis zone was observed the next day, isolation of phages was carried out from the enriched phage solution as described below.
Phages were purified and isolated by double agar overlay method. Briefly, logarithmic serial dilution (100 µl) of overnight enriched phage supernatant in 1X sterile Phosphate-Buffered Saline (PBS, pH 7.4) were mixed with 5 ml of warm BHI top agar (0.75% agar) and 40 µl of Salmonella culture (108 CFU/ml) and overlaid on BHI bottom agar (1.5% agar) plates. Individual phage plaques observed after overnight incubation were picked using a cut pipette tip and enriched again as described previously. Three rounds of purification were performed and one lytic phage was isolated.
For phage solution preparation, 10 ml of sterile SM buffer (5.8 g NaCl, 2 g MgSO4.7 H2O, 50 ml 1 M pH 7.5 Tris-Cl, per liter) was poured on top of a nearly confluent lysed plate with only a web-like pattern of visible bacterial lawn and kept for overnight gentle shaking at room temperature. After 16 h of incubation, the phage-containing buffer was carefully removed, centrifuged and purified with 0.22 μm syringe filter as described above. The phage solution thus obtained was used for all subsequent experiments.
Host range assay
To determine the host range of the phage, the phage solution was applied to bacterial lawns of a total of 43 strains – 25 Salmonella strains and 18 non-Salmonella strains, as listed in Table 1. A 10 µl aliquot of phage lysate solution (109 PFU/ml) was spotted onto BHI double agar plates inoculated with appropriate bacterial strain. The plates were incubated at 37 °C for 16–18 h. Following incubation, the plaques formed on the bacterial lawns were observed and scored based on their characteristics. Plaques were categorized as follows: +++ for complete lysis, ++ for complete lysis with individual colonies at the center, + for turbid lysis, and - for no lysis.
Transmission electron microscopy
The physical morphology of the phage was determined using Transmission electron microscopy (TEM). 10 µl of phage solution (~ 109 PFU/ml) in SM buffer were fixed onto copper grids, negatively stained with 2% uranyl acetate (pH 4.2) for 10 min, and then air-dried for 1 h at room temperature. HT7700 Transmission Electron Microscope (Hitachi, Japan) with accelerating voltage of 50 kV and 12,000 X magnification was used for acquiring the phage images. Images were analyzed using Fiji11.
Bacteriolytic ability of phage on Salmonella
In order to assess the bacteriolytic activity of the phage, Salmonella Typhimurium ATCC 14028 was infected with the phage at varying Multiplicity of Infections (MOIs) ranging from 0.01 to 10. Briefly, in a 96-well plate, 100 µl of phage solution at concentrations ranging from 106 to 1010 PFU/ml was mixed with 100 µl of overnight Salmonella culture (108 CFU/ml) in BHI broth (OD600 0.2) to achieve MOIs ranging from 10 to 0.01. The mixture was incubated at 37 °C for 7 h with orbital shaking at 244.5 rpm (settle time: 5 ms) in a microplate reader (Tecan Infinite 200Pro, Switzerland). Using kinetic cycle setting in Magellan software version 7.2 SP1 INK, bacterial growth curve was monitored by OD600 measurements (No of reads − 25) and results were analyzed using GraphPad Prism v 5. The control line indicates bacterial culture with 100 µl of 1X PBS.
One-step growth curve of phage
For the determination of one-step growth curve of the phage, 10 µl of phage solution (108 PFU/ml) was added to 1 ml S. Typhimurium ATCC 14028 (108 CFU/ml) and incubated at 37 °C. After 15 min of incubation, the pellet obtained by centrifugation at 7000 g for 2 min was washed with BHI broth, resuspended in 50 ml BHI broth and incubated at 37 °C for 100 min. 200 µl aliquots were removed at 10 min intervals in triplicates and centrifuged at 13,000 g for 30 s. From the supernatant, enumeration of phages was done by double agar overlay method as described above. For the determination of phage growth kinetics, the graph plotted of phage count in log10 (PFU/ml) vs. time (in minutes) was used to determine the latent period as well as the burst size using the following formula: Burst size = (PFU/ml at plateau) / (PFU/ml at latent period). This experiment was repeated independently three times.
pH and temperature stability of phage
The stability of phage when exposed to different temperature and pH conditions was examined as follows. To estimate the pH stability of phage, 100 µl of 107 PFU/ml phage solution was added to 900 µl 1X PBS pH ranging from 2 to 10 (adjusted using 5 M HCl or 10 M NaOH) in triplicates and incubated for 2 h at 37 °C. After the incubation, phage count was enumerated by double agar overlay method as described above. The experiment was repeated independently three times.
For temperature stability, 100 µl of phage solution (107 PFU/ml) was mixed with 900 µl sterile SM buffer in triplicates and incubated in a water bath at temperatures ranging from 25 °C to 75 °C for 60 min. After the incubation, phage count was enumerated by double agar overlay method as described above. The experiment was repeated independently three times. One way ANOVA with Dunnett’s test was performed using GraphPrism v 5.0 in order to analyse the significance of the phage log difference in both experiments. α = 0.05. p < 0.05 (*), p < 0.01(**).
Lytic ability of phage in milk
Lytic ability of the phage to reduce Salmonella contamination in milk was analyzed by measuring the log reduction of the bacterial count in artificially spiked milk. Pasteurized milk (8.9 ml) (Nandini, Karnataka Milk Federation, India) was inoculated with 1 ml of 106 CFU/ml S. Typhimurium ATCC 14028 followed by addition of 100 µl 108 PFU/ml phage solution at an MOI of 10. The milk samples were incubated at Room Temperature (RT) (25 °C) as well as 4 °C and samples drawn for microbiological analysis at 3 h intervals from 0 to 9 or 12 h. 100 µl of samples were drawn in triplicates and were serially diluted with 1X PBS and plated on Salmonella-Shigella (SS) agar (Himedia, India) for determining Salmonella count. After overnight incubation in 37 °C, colorless colonies with black centers were enumerated from the SS agar plates to determine the bacterial count. The experiment was repeated independently two times.
DNA extraction and library Preparation
Phage genomic DNA was extracted from phage solution (108 PFU/ml) using Qiagen Blood and Tissue Kit (69506, Qiagen, Germany) following the protocol detailed by Jakociune and Moodley12. Briefly, 450 µL of the filter-sterilized lysate was incubated with 50 µL DNase I 10X buffer (Thermo Fisher Scientific, US), 1 µL DNase I (1 U/µL) (Thermo Fisher Scientific), and 1 µL RNase A (10 mg/mL) (Qiagen) for 1.5 h at 37 °C. Thereafter, 20 µL of 0.5 M EDTA was added to inactivate DNase I and RNase A. Further DNA was extracted as per the manufacturer’s protocol. DNA for library preparation as well as the prepared library was quantified by Qubit 3 Fluorometer (Thermo Fisher Scientific) using ds-DNA High Sensitivity (HS) Kit (Thermo Fisher Scientific). The DNA library preparation for whole genome sequencing was carried out from the above phage genomic DNA using QiaSeq FX DNA Library Kit (Qiagen) according to the manufacturers’ instructions. The amplified products were cleaned up by using AMPure XP beads (A63880, Beckman Coulter, USA) and the final DNA library was eluted in 0.1 X TE buffer. The fragment analysis of the prepared library was performed on Agilent 2100 Bioanalyzer (Agilent Techologies, Inc., USA). After library preparation, Next Generation Sequencing was performed in Illumina Hiseq™ sequencing system (Illumina, USA). Library preparation, sequencing and assembly was performed by Biokart India Pvt. Ltd., India.
Genome assembly and annotation
FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess sequencing quality. Adapter removal and trimming was done by TrimGalore (https://github.com/FelixKrueger/TrimGalore). The primary assembly was performed through a de novo approach (SPAdes13 tool) and the assembled files were saved in FASTA format. After primary assembly, the reads were filtered using PHASTER14. CONTIGuator15 was used for the secondary assembly. RASTtk pipeline16 and ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder.com) were used for the annotation of the phage genome. The putative ORFs were manually curated in UGENE v.40.017 using NCBI BLASTp, InterPro, UniProtKB, and HHPred. SMART v.6 and PDBmmCIF databases were selected for protein prediction using HHpred with a probability cutoff of > 20%. DeepTMHMM v1.018 analysis was used to predict the transmembrane topology in putative holin protein. The genome was also screened for the presence of tRNAs using tRNAscan-SE19. CARD analysis was performed (https://card.mcmaster.ca/) on the phage sequence to identify the presence of antibiotic-resistant genes. UGENE software v.40.0 was used to generate a Genbank Flatfile (.gbk) for the construction of circular genome map through CGView (https://proksee.ca/). The average nucleotide identity (ANI) of the phage against the top BLASTn hits was calculated using JSpeciesWS20,21.
Docking studies of receptor binding tail protein (ORF 123) of phage
Homology modeling of the receptor binding tail protein (ORF 123) from vB_Sal_S6 phage was performed using MODELLER v 10.5 with PDB ID: 8B14 as template. Briefly, the target sequence of ORF123 was searched against the Protein Data Bank (PDB) database using the PSI-BLAST tool. Two hits identified from the search were subsequently screened based on the resolution of their structures. PDB ID: 8B14:B (resolution: 2.6Å – electron microscopy), was selected as the template for homology modeling of ORF123.
The structural 3D model of the receptor binding tail protein (ORF 123) of vB_Sal_S6 phage was constructed using MODELLER v.10.5. This model was then employed for subsequent docking studies. The amino acid sequence for the Salmonella Typhimurium homolog of the Escherichia coli receptor, FhuA, was retrieved from existing literature and UniProtKB (UniProt ID: Q8ZRQ2; 729 amino acids). PDB ID: 8B14:A was used as a template to model Salmonella Typhimurium FhuA using MODELLER v.10.5.
Docking scores were calculated using the HDOCK server (https://hdock.phys.hust.edu.cn/), with Salmonella Typhimurium FhuA serving as the receptor and ORF 123 as the ligand (RMSD Threshold of 4Å). The resulting docking interactions were visualized using Discovery Studio Visualizer (Biovia) and PyMol 3.0.
Codon usage bias of Salmonella Typhimurium and phage
The Relative Synonymous Codon Usage analysis of primary host and vB_Sal_S6 phage genome was carried out using RSCU calculator (https://jamiemcgowan.ie/bioinf/rscu.html) and spider chart generated using Microsoft Excel. Further, the Codon Adaptation Index (CAI) of phage and host were calculated using the codon usage table which was generated using Codon Usage Database (www.kazusa.or.jp/codon/). This was then used for estimating the CAI values against Salmonella Typhimurium and Escherichia coli K12 codon usage bias using Biopython (SeqUtils module) (https://github.com/biopython/biopython) in Jupyter Notebook (https://github.com/jupyter/notebook).
Phylogenetic analysis of phage
To infer the evolutionary history of the isolated phage vB_Sal_S6, phylogenetic analysis based on the terminase large subunit (terL) (ORF 124) was performed. The ORF for terminase large subunit protein of the phage, which is one of the major conserved regions in phages22, was analyzed using BLASTp and the closest neighbors was used for construction of phylogenetic tree using MEGA-1123. The evolutionary history was inferred by using the Neighbor Joining method24 and evolutionary distances were computed using Poisson correction method25. The bootstrap consensus tree was inferred from 1000 replicates.
Statistical analysis
Results are represented as Mean + standard deviations (s.d). Two-way ANOVA with Sidak’s multiple Comparison Test was performed using GraphPad Prism v 8.0.2 to analyze the statistical significance between the control and phage treatment groups for studying the lytic effect of phage against Salmonella in milk (Fig. 2). The same test was also used to assess bacteriolytic ability of phage against Salmonella (Fig. 1C). Significance levels were represented as follows: p < 0.0332 (*), p < 0.0021 (**), p < 0.0002 (***), p < 0.0001 (****). α = 0.05.
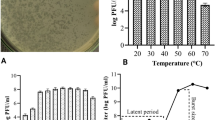
Characterization of vB_Sal_S6 phage (A) Plaque morphology image of vB_Sal_S6 on a double agar layer plate (B) Transmission Electron Microscopy (TEM) image of vB_Sal_S6 obtained by negative staining indicating an icosahedral head and a tail. Images were analyzed and lengths of head and tail were measured using Fiji. Scale bar – 50 nm. (C) Bacteriolytic activity of vB_Sal_S6 against Salmonella Typhimurium ATCC 14028 at different Multiplicity of Infection (MOI) (0.01-10). Control (Cntrl) indicates no phage treatment. (D) One-step Growth curve of vB_Sal_S6 phage indicating the latent period and burst size. (E) pH stability of vB_Sal_S6 at a pH range of 2–10 after 2 h. p ≤ 0.01(**). (F) Temperature stability of vB_Sal_S6 at temperature range of 25–75 °C after 60 min. p ≤ 0.01(**). Each point in all the graphs above are the mean of three replicates. All error bars indicate standard deviation (s.d).
Lytic effect of vB_Sal_S6 phage in milk (MOI 10) at (A) 4 °C and (B) Room temperature (RT)- Bacterial count in milk that was artificially contaminated with S. Typhimurium ATCC 14028 are represented in red whereas, bacterial count in phage treated sample are represented in blue at various time points. Each point is the average of three replicates and error bars indicate standard deviation (s.d). Two-way ANOVA with Sidak’s multiple Comparison Test was performed to analyze the statistical significance between the control and phage treatment groups. p < 0.0332 (*), p < 0.0021 (**), p < 0.0002 (***), p < 0.0001 (****). α = 0.05.
Results
Host range assay
The Salmonella phage vB_Sal_S6 was isolated from drainage source using the method followed in Sect. Isolation of phage. The host range of the phage vB_Sal_S6 was determined by spot testing against 43 different host strains (Table 1). The phage exhibited lytic activity against 18 of the 25 Salmonella serovars that were analyzed. The phage showed prominently high lytic activity against the serovars S. Typhimurium (ATCC 14028 (+++), MTCC 98 (+++), BCH 103 (++), MTCC 1251 (+++), IMTECH 1 (+++), NICED 1025 (+), KMC 2 (++), Gwalior 1253 (++), Manipal 2501 (+++)), S. Bovismorbificans MTCC 1162 (+++), S. Infantis MTCC 1167 (+++), S. Typhi ATCC 6539 (++) and E.coli (ATCC 10536 (++), MTCC 98 (++)). All 11 tested strains of S. Typhimurium were susceptible to the phage. The phage exhibited turbid lysis of S. Typhimurium (Kerala S1 (+), ATCC 13311 (+)), Paratyphi (ATCC 9150 (+), MTCC 735 (+)), Weltevreden MTCC 1169 (+), and Poona NCTC 4840 (+). The host range assay did not indicate lysis of other non-Salmonella or non-E. coli strains (Table 1).
Transmission electron microscopy and bacteriolytic ability of phage on Salmonella
The plaque morphology of vB_Sal_S6 indicated clear rounded plaques (Fig. 1A) with an average plaque diameter of 1.19 mm ± 0.15. Morphology of the phage vB_Sal_S6 was observed by TEM. Negatively stained TEM images revealed a phage virion with an icosahedral head of average diameter 81.88 ± 5.03 nm and a tail of 100.81 `± 0.23 nm length and tail diameter of 21.53 ± 0.54 nm (Fig. 1B).
The bacteriolytic activity of the phage was assessed by addition of phage solution to Salmonella Typhimurium ATCC 14028 at various MOIs (0.01-10) (Fig. 1C). In the control without phage treatment, bacterial growth progressed logarithmically for up to 2 h, reaching an optical density at 600 nm (OD600) of 0.6. At MOI 0.01, log phase lasted for approximately 1 h, reaching an OD600 of 0.45, followed by a plateau phase persisting from 1 to 7 h. For MOI 0.1, the log phase also lasted for about 1 h, attaining an OD600 of 0.41 before gradually declining to 0.24. At MOI 1, the initial OD600 remained constant for the first hour, followed by a decline, indicating effective bacterial lysis. At MOI 10, the log phase was insignificant with OD600 values remaining low throughout the observation period, though a slight increase was noted after 3 h. In all cases (MOIs (0.01-10)), the bacterial growth was found to be significantly less than control without phage treatment (p < 0.0001 (****) for all time points 2–7 h).
One-Step growth curve of phage
Phage growth kinetics and burst size were determined, as described previously (Sect. One-step growth curve of phage). The one-step growth curve of the vB_Sal_S6 assayed for 100 min at 37 °C indicated that the phage had a latent period of 40 min followed by an ascent from 5.56 log10 (PFU/ml) to 7.60 log10 (PFU/ml) (Fig. 1D). By 80 min, the phage growth curve had achieved stationary phase at an average of 7.5 log10 (PFU/ml). Burst size was estimated to be 123 PFU/cell (Fig. 1D) based on the phage PFU values calculated.
pH and temperature stability of phage
Stability of phages at dynamic ranges of pH and temperature are crucial for efficient phage administration for food safety applications. The phage vB_Sal_S6 (in solution) was exposed to various temperature and pH conditions, and the phage count was enumerated using double agar overlay method to determine the pH and temperature stability. The phage titer decreased slightly from 5.9 log (PFU/ml) to 5.5 log (PFU/ml) for the pH range of 10 to 4 (Fig. 1E). When exposed to pH 2, phage titer was significantly reduced to 0 (p < 0.01(**)) indicating sensitivity of the phage to high acidic environments.
In the temperature stability experiment, vB_Sal_S6 showed maximum titer stability (around 7.5 log10 (PFU/ml)) at temperatures 25˚C, 35 °C, and 55 °C beyond which the phage titer progressively reduced (Fig. 1F). At 65˚C and 75 °C, the log10 (PFU/ml) was observed to be 4.77 and 3 log10 (PFU/ml) respectively (Fig. 1F) (p < 0.01(**)), indicating that the plaque formation of the phage is affected at temperatures greater than 55 °C. Therefore, the phage vB_Sal_S6 was observed to be stable at a temperature range of 25–55 °C.
Lytic ability of phage in milk
To determine the lytic ability of phage vB_Sal_S6 in milk, pasteurized milk samples artificially contaminated with Salmonella were treated with phage at an MOI of 10 (Fig. 2). Phage-treated milk samples maintained at 4 °C showed 1 log bacterial count reduction at 3 h (p < 0.0021 (**)), 2.4 log at 6 h (p- <0.0001 (****)), and 1.6 log at 12 h (p < 0.0021 (**)) (Fig. 2A) whereas milk samples that were incubated at RT, it was observed that bacterial growth reduced 0.4 log in 3 h, 3.3 log in 6 h and 3.9 log in 9 h (p < 0.0021(**)) (Fig. 2B).
General features of the phage genome
Genomic analysis
The sequencing data passed quality control by FastQC: mean per-base Phred scores ranged from ~ 34 to 36 across read positions. The primary assembly of vB_Sal_S6 phage nucleotide sequences resulted in 1611 contigs. Primary assembly was screened with PHASTER, which identified one ~ 5 kb contig of apparent bacterial origin; this contig was excluded from subsequent analyses. Based on PHASTER results, Escherichia phage_fp01 was used as reference sequence for the secondary assembly using CONTIGuator. The secondary assembly resulted in a dsDNA 36,127 bp genome with a GC content of 38.6% and a coding density of 84.77% (Table 2).
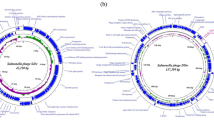
Annotation of the phage genome revealed 161 putative ORFs through ORFFinder (Table 2). The nested ORFs were ignored and alternate start codons (ATG, TTG, CTG, GTG, ATT, ATC) were selected by which 48 ORFs were assigned putative functions with any of NCBI BLASTp, UniProtKB, InterPro and HHPred (Supplementary Table S1). The phage was observed to be devoid of genes encoding recombinases, integrases, excisionases, or repressors associated with integration and excision of phage genome (Supplementary Table S1 and Table 2). The circular genomic map generated from these ORFs using Proksee CGView revealed 14 and 34 ORFs in the negative and positive strands respectively (Fig. 3). Among 48 ORFs with known functions, 17 were associated with DNA, RNA and nucleotide metabolism; 2 with transcriptional regulation, 4 with host lytic activity; 4 with head structure and packaging; 6 with tail structure; 2 were predicted as membrane proteins, and 13 were identified to have other functions (Table 2). The remaining 113 ORFs were inferred as hypothetical proteins among which 50 were found to be conserved amongst the phages (Table 2).
Circular genome map of vB_Sal_S6 indicating color-coded genome regions constructed using CGView. The outer ring denotes the ORFs on the positive strand of vB_Sal_S6 genome. The next ring illustrates ORFs on the complementary strand. The inner rings show G + C content and G + C skew, where peaks represent the positive (outward) and negative (inward) deviation from the mean G + C content and G + C skew, respectively. All the annotations indicated in Supplementary Table S1 are illustrated in the map.
CARD analysis of the phage genome proved the absence of antibiotic resistance genes (Table 2). tRNAscan-SE demonstrated the presence of 21 tRNAs among which 12 were predicted to code for standard amino acids, while 1 was undetermined and 8 were predicted as tRNAs with mismatch isotypes (Supplementary Table S2 and S3). BLASTn analysis revealed highest similarity with Salmonella phage Citrobacter phage vB_CfrD-Cit2 on the basis of query coverage (Accession number: OP745948.1, Query coverage: 92%, Percentage identity: 97.75%). The top hits obtained from BLASTn (e-value = 0, query coverage = 87–92%, identity = 95–99%) are shown in Supplementary Table S4. The average nucleotide identity of vB_Sal_S6 was determined against top 29 hits from BLASTn using JSpeciesWS based on whole genome alignment. The highest ANIb (BLASTn-based Average Nucleotide Identity) values was observed to be 94.34% with Salmonella phage HMD-P4 (PQ462666.1) with 67.3% aligned genome (Supplementary Table S5).
Structural and packaging proteins
The ORF annotation in Supplementary Table S1 was analyzed in detail. The phage icosahedral head encapsulating DNA is composed of major capsid protein (ORF 81) enclosing a nucleocapsid comprising nucleocapsid protein (ORF 38) (Supplementary Table S1).
Six ORFs were identified as encoding tail-associated structural proteins critical for host receptor recognition and specificity. These included 6 ORFs corresponding to tail fiber protein (ORF 46), baseplate wedge protein (ORF 89), tail completion or Neck1 protein (ORF 91), tail terminator (ORF 92), receptor binding tail protein (ORF 123), tail tube protein/major tail protein pb6 (ORF 148), which are structural proteins found at the distal end of tail assembly and mediate phage attachment to receptors in host thereby determining the host range26. All the 6 tail protein ORFs showed 85.38% to 100% similarity with orthologous genes in Salmonella and Escherichia phages belonging to Tequintavirus genus (Supplementary Table S1).
The DNA packaging system of tailed phages comprises a heterodimeric terminase constituted of large and small subunits where the small subunit facilitates DNA binding and the large subunit (terminase) facilitates prohead binding and cleavage of the phage concatameric DNA into individual genome units27. ORF 124 and ORF 147 were predicted as terminase large and small subunits respectively with 100% and 98.12% similarity with respective proteins in Escherichia phage T5 (Supplementary Table S1).
Further, ORF 87 and ORF 113 were putatively identified as membrane proteins with 93.83% and 85.29% similarity with those of Escherichia phage SP15 belonging to Tequintavirus genus and Salmonella phage Rutana belonging to Epseptimavirus genus respectively (Supplementary Table S1).
Cell wall Lysis proteins
For Caudovirales of Gram-negative hosts, the holin-endolysin system plays a crucial role in disrupting the host cell wall leading to lysis28. The host cell lysis mechanism is putatively comprised of 4 ORFS comprising one holin and three endolysins. ORF 79 encodes holin which form pores in the host’s inner and outer membrane, allowing endolysins to reach and degrade peptidoglycan, a component of bacterial cell walls29. ORF 79 showed 100% identity with that of Escherichia phage vB_EcoS_FFH_1 belonging to Tequintavirus genus (Supplementary Table S1). DeepTMHMM v1.0 analysis reveals that ORF 79 is a Class 3 holin with a single highly hydrophilic transmembrane domain (Supplementary Figure S1), which is generally found in T4-like and T5-like phages30.
ORF 112 (N-acetylmuramoyl-L-alanine amidase), ORF 137 (endolysin) and ORF 154 (cell wall hydrolase) encode potential endolysins that are involved in hydrolysis of peptidoglycan of the bacterial host from within, resulting in cell lysis and release of progeny virions31. The presence of 3 endolysins alongside a Class 3 holin suggests a robust lysis module with possible redundancy, potentially increasing host cell wall disruption efficiency. Spanin that is engaged in host cell lysis by forming bridges spanning both membranes of the host cell was absent28 indicating that vB_Sal_S6 has an alternate way of disrupting the outer membrane .
Replication and regulation proteins
Genome annotation revealed several ORFs associated with facilitating viral replication and the suppression of host defenses. The ORF 88 and ORF 121 (A1 protein) and ORF 86 (A2 protein) proteins work in tandem at the initial stages of infection. The A1 protein (ORFs 88 and 121) is associated with host DNA degradation and suppression of host gene expression, as well as the shutoff of phage pre-early genes32. The A2 protein, encoded by ORF 86, facilitates the second-step transfer of phage DNA into the host cell, ensuring complete genome delivery33.
ORF 10 was found to encode the D5 protein, a transcription factor implicated in the replication of the phage genome and in suppression of early phage gene expression34. Additionally, ORF 78 encodes an ATP-dependent protease, likely involved in the selective degradation of host regulatory proteins to fine-tune phage gene expression35.
The genome also harbored genes encoding endonucleases. ORF 65 and ORF 103 code for HNH and homing endonucleases, respectively, which are implicated in horizontal gene transfer and DNA packaging36. Genes involved in DNA replication were also identified, including ORF 42 and ORF 63 encoding NAD-dependent DNA ligases, with ORF 152 encoding an ATP-dependent DNA helicase (Hel308) that facilitates the unwinding of DNA, enabling both transcription and replication37. The Cor superinfection exclusion protein, encoded by ORF 45, was identified and is likely involved in preventing lytic development of superinfecting phages, thereby protecting the primary phage infection38.
The transcriptional regulator encoded by ORF 62, which shares 100% identity with the transcriptional regulator of Salmonella phage Spc35 (Supplementary Table S1), suggests a conserved mechanism of gene regulation among phages within similar host environments.
Docking studies of receptor binding tail protein (ORF 123) of phage
The vB_Sal_S6 ORF 123 annotated as Receptor binding tail protein (RBTP) (Supplementary Table S1) was further investigated to identify the putative receptor in Salmonella and E.coli. Protein Data Bank search with ORF 123 as probe using PSI-BLAST algorithm led to the complex structure of E.coli T5 phage- receptor binding protein pb5 bound to E.coli ferrichrome transporter (FhuA) (e value = 8.097 × 10–56) (PDB: 8B14) which implied that FhuA could be the putative receptor for vB_Sal_S6 in bacterial host. Using MODELLERv.10.5, 5 models of ORF 123 were generated and the best model chosen based on the DOPE score (-53,144.58) was subjected to Loop Refining.
Considering that the vB_Sal_S6 host range included Salmonella Typhimurium as well as E.coli (Table 1), the presence of FhuA homologs in Salmonella was investigated and the amino acid sequence for Salmonella Typhimurium FhuA was obtained (Uniprot ID: Q8ZRQ2, 729 aa). Using the sequence, PDB: 8B14 was used as a template and Salmonella Typhimurium FhuA was modelled using MODELLER v.10.5 and the best model was chosen on the basis of DOPE score (-66,919.31).
The docking studies of vB_Sal_S6 ORF 123 (RBTP) and Salmonella Typhimurium FhuA receptor using HDOCK revealed a docking score of -371.1 with 0.38 Å RMSD and 98.81% confidence (Fig. 4A). The interactions spanned from hydrogen bonding to van der Waals forces, with distances ranging from approximately 2.085 Å to 4.969 Å. The interaction between the FhuA receptor and ORF 123 (RBTP) is mediated by specific residues at their respective surfaces. The interacting residues of ORF 123 (RBTP) with Salmonella FhuA receptor were found to be 112ALA, 113ASP, 118PHE, 164TYR, 169ARG, 171GLY, 172ALA, 175PRO, 187LEU, 188GLY, 189PHE, 234TYR, 238ARG, 267ASN, 539ASN. At the docking interface; 188GLY, 171GLY, 169ARG and 112ALA (RBTP) interacting with 410ALA, 409SER, 711TYR, 518PHE (Salmonella FhuA) respectively are shown in Fig. 4A.
Molecular docking of Receptor Binding Tail Protein (RBTP) bound to the pocket region of FhuA receptor. (A) Overall structure of vB_Sal_S6 phage (ORF 123) (red) with FhuA (Ferrichrome porin) (yellow) receptor of Salmonella Typhimurium and their interactions (shown in yellow and blue text respectively). (B) Overall structure of vB_Sal_S6 phage (ORF 123) (red) with FhuA receptor of E.coli (cyan) (C) Overall structure of T5 phage (green) with FhuA receptor of Salmonella (yellow). The hydrophobic interaction sites in FhuA are represented as surface view (blue to brown gradient). All images were visualized using Discovery Studio Visualizer. (S6 phage – vB_Sal_S6).
Alternatively, a docking score of -511.68 with 0.73 Å RMSD and 99.93% confidence was observed in the docking between ORF 123 and Escherichia coli FhuA receptor (Fig. 4B). The interacting residues of ORF 123 (RBTP) with Escherichia coli FhuA receptor were found to be 110THR, 115PHE, 116ARG, 165HIS, 166GLY, 168VAL, 169ARG, 183ALA, 185ALA, 188GLY, 190PRO, and 238ARG (interactions not shown).
Docking studies were also carried out on the components of the complex PDB ID: 8B14 (i.e. E.coli FhuA and T5 phage pb5) as a reference and a docking score of -852.11 with 100% confidence and 0.33Å RMSD was obtained (Fig. 4C).
Codon usage bias of Salmonella Typhimurium and phage
The codon usage profiles of bacteriophage vB_Sal_S6 and its bacterial host Salmonella enterica serovar Typhimurium were compared using relative synonymous codon usage (RSCU) values (Fig. 5). Overall, vB_Sal_S6 exhibited codon usage patterns broadly similar to its host, consistent with potential phage adaptation to the translational machinery of S. Typhimurium. Several codons, such as ATG (Met, RSCU = 1.0), TGG (Trp, RSCU = 1.0), and GAA (Glu, RSCU = 1.291), showed identical usage between phage and host, suggesting evolutionary optimization for essential amino acids (Supplementary Table S6).
Codon Adaptation index (CAI) was used to quantify the extent of similarities between synonymous codon usage patterns of phage genes and host reference set39. CAI analysis revealed CAI values of 0.5225 for E. coli and 0.6386 for S. Typhimurium. These results indicate that the phage genome exhibits a higher level of codon usage optimization toward S. Typhimurium than E. coli, suggesting a stronger translational compatibility with Salmonella.
Phylogenetic analysis of phage
Phylogenetic tree constructed based on terminase large subunit (ORF 124) using MEGA-11 indicated 2 major clades – Clade 1 and Clade 2 (Fig. 6). vB_Sal_S6 was found to cluster very closely with phages belonging to Tequintavirus genus (Clade 1a) of family Demerecviridae. The other neighboring clades belong to Epseptimavirus (Clade 1b) and Sugarlandvirus (Clade 2) both belonging to Demerecviridae. It can be observed within the Tequintavirus clade that TerL protein of vB_Sal_S6 clustered predominantly with Salmonella phages (42%) and Escherichia phages (50%). Alternatively, the entire tree comprised of 42% Salmonella phages, 27% (7/26) E.coli phages, 27% (7/26) Klebsiella phages and 4% (1/26) Citrobacter phages (Fig. 6).
Discussion
Non-typhoidal Salmonella serovars remain a leading cause of gastroenteritis globally, with contaminated natural waterways serving as critical transmission routes and frequent sources of agricultural produce contamination40. The rapid increase in Salmonella food safety burden globally along with increasing concerns in view of resistance to frontline antibiotics has placed renewed emphasis on phages that are selectively bacteriolytic with robust proliferative capacity to control the intended bacterial population.
In this study, a novel phage vB_Sal_S6 was isolated from sewage using Salmonella Typhimurium ATCC 14028 as host bacterium. Host range analysis revealed bacteriolytic activity against Salmonella Typhimurium, Bovismorbificans, Infantis, Weltevreden, Poona, Paratyphi, and Typhi (Table 1) all of which are associated with food-borne transmission to humans worldwide41,42,43,44,45,46. In addition to these Salmonella serovars, bacteriolytic effect could be seen with E.coli ATCC 10536 and MTCC 98 (Table 1), demonstrating vB_Sal_S6 as a putative polyvalent phage entering host bacteria (E.coli and Salmonella) through a common receptor protein. E.coli is an indicator of fecal contamination and coexists with Salmonella in contaminated waters47 which leads to the evolution of phages with bacteriolytic effect on both the bacteria. Isolation of polyvalent phages lysing closely related bacterial species have been reported48,49 and in this case, it provides the possibility of using E.coli that has a lesser scale of safety requirements than Salmonella for amplification of phages for possible large scale requirements in food safety applications.
The burst size of 123 PFU/cell (Fig. 1D) indicates a high number of virions being released for the propagation of phage infections and was comparable with the Salmonella phages reported earlier50. The latent period of 40 min, which indicates the time between adsorption and host lysis, is higher than the doubling time of 20 min of Salmonella51 and E. coli and this facilitates continuous availability of host cells and prevent extinction of the phage population in the natural environment. Studies on the phage bacteriolytic activity revealed that Salmonella growth was significantly suppressed from the start, maintaining a low OD600 throughout the experiment (Fig. 1C). Maximum lytic effect at MOI of 10 and 1 shows that phage application to bacterial cultures has a dose-dependent lytic effect of the phage on bacterial cultures, where higher MOIs lead to more effective bacterial killing. The findings support the rationale of using higher MOI phage treatments for rapid and efficient bacterial clearance in biocontrol/food applications.
Environmental factors like pH and temperature influence phage stability and their ability to infect bacterial hosts, which is crucial for their effective use in food safety and biocontrol applications52. vB_Sal_S6 maintained its effective bacteriolytic property as indicated by uniform phage titer through the dynamic pH range 4–10, typically observed in the majority of food matrices (Fig. 1E). A comparable pH stability range (pH 4–10) has been reported for other well-characterized Salmonella phages, including P22 and LPSTE, which also retain high bacteriolytic activity across diverse food matrices such as milk (pH 6.7–6.9), apple juice (pH ~ 4), chicken breast and mince (pH 5.3–6.5), liquid eggs (pH 6.5–6.7), and energy drinks (pH < 4)53,54,55,56. This indicates the possible application of our phage across a wide range of food matrices with varying pH conditions. The reduction in vB_Sal_S6 phage titer observed at pH levels below 2 can be attributed to irreversible coagulation and precipitation of phages at highly acidic pH, leading to a loss of lytic activity7.
The key aspects of phage infectivity that include attachment to bacterial hosts, entry and the subsequent replication of phage genome inside the host are affected by temperature52. Hence phages for possible food safety applications should be able to sustain their infectivity in the “temperature danger zone” (5–60 °C or 40–140 °F) that supports rapid growth of food borne bacteria57. In the temperature stability experiment, we observed a stable average phage titer of 7.5 log10 (PFU/ml) for the 25–45 °C temperature range followed by a slight decline at 55 °C (Fig. 1F). However, a significant impact on phage titer was observed at 65 °C and 75 °C with progressive 2.75 and 4.53 log reduction in phage titer respectively. The ability to retain infectivity across the temperature range of 25–55 °C varies among phages. Here, vB_Sal_S6 demonstrates stability within this temperature danger zone indicating its potential for biocontrol of Salmonella in food matrices. However, subsequent reheating of foods may reduce both Salmonella and phage populations, thereby supporting the safety of phage application in food systems.
In our study, milk was evaluated as a matrix to assess the bacteriolytic activity of vB_Sal_S6 application owing to its frequent contamination with Salmonella, either directly from the udder or indirectly through contact with manure during the milking process58. Furthermore, economic constraints and energy costs often compel farmers to store milk at suboptimal temperatures (around 6 °C), which exceed the recommended 2–4 °C storage conditions within the critical 2–3 h window post-milking59 which risks Salmonella proliferation. Our study revealed a marked reduction in Salmonella viable count in both refrigeration temperature (4 °C) as well as room temperature (RT). A maximum log reduction of about 3.9 log and 2.4 log reduction of viable Salmonella count was observed after 9 h incubation at RT and 6 h at 4 °C (Fig. 2) which is relevant in the light of the International Microbiological Criteria for Dairy Products criteria which stipulates that the acceptable levels of Salmonella in raw milk to be pasteurized is 105 CFU/ml60.
Here, we followed an MOI of 10 which was followed as per previous reports on phage application on milk61. Whey proteins present in milk can impede bacteriophage–host interactions by obstructing access to specific bacterial surface receptors, thereby diminishing phage infection efficiency61. To overcome this interference, the higher MOI allows bacteriophages to attach better to the host bacteria in food system by increasing probability of phage infecting the bacteria62. For future studies, the applications of varied phage MOIs can be explored to further improve bacterial log reduction. Our experiments demonstrating significant reduction in Salmonella counts with application of phage in milk indicates potential application in ensuring milk safety and quality, and support the development of effective phage-based interventions for Salmonella control in the milk industry.
The host range of a phage is determined by the specific binding of tail fiber proteins with the host receptors26. The putative receptor for vB_Sal_S6 has been identified as ferrichrome transporter (FhuA) by Protein Data Bank search with ORF123 (Receptor Binding Tail Protein (RBTP)). FhuA (Ferrichrome porin) is a trans-membrane receptor found in Escherichia coli and Salmonella sp. consisting beta-barrels, a distinguished characteristic of porins used by several other well-characterized phages such as T1, T5 and phi80 for host recognition63,64,65. The characteristic of phage vB_Sal_S6 (isolated with Salmonella Typhimurium ATCC 14028 as host) to infect the Salmonella serovars Paratyphi, Typhi, and E.coli correlates with the presence of FhuA receptor in these bacteria66 suggesting putative interaction with vB_Sal_S6 ORF123. This provides a molecular basis for the putative polyvalent nature of vB_Sal_S6, a feature often reported among Salmonella phages67. The interactions spanned from hydrogen bonding to van der Waals forces, with distances ranging from approximately 2.085 Å to 4.969 Å (Sect. Docking studies of receptor binding tail protein (ORF 123) of phage), indicating a mix of strong and weak molecular contacts. Docking (HDOCK) of vB_Sal_S6 phage RBTP ORF123 with S. Typhimurium FhuA and E.coli FhuA yielded a docking score of -371.1 and − 511.68 respectively (Sect. Docking studies of receptor binding tail protein (ORF 123) of phage). Relatively higher docking score for the former interaction makes us speculate the possibility for preferential binding of vB_Sal_S6 with E. coli when present along with Salmonella.
The terL phylogenetic tree constructed using sequences obtained by ORF 124 BLAST revealed 2 distinct clusters of Demerecviridae family comprising the phage genera Tequintavirus (cluster 1a), Epseptimavirus (cluster 1b), and Sugarlandvirus (cluster 2). vB_Sal_S6 phage clustered with phages of Tequintavirus genus that have Salmonella and E.coli as their principal hosts (Fig. 6). The neighboring cluster 2, Sugarlandvirus genus is comprised of Klebsiella phages. This phylogenetic placement of the phages appears to corroborate with gyrA phylogeny of Salmonella, Shigella, E.coli and Klebsiella wherein the former three are closely related in comparison with Klebsiella68 suggesting coevolution of phages with their hosts.
Clustering of vB_Sal_S6 phage within the Tequintavirus genus based on TerL phylogeny is supported by JSpeciesWS ANIb analysis, which identified Salmonella phage HMD-P4 (PQ462666.1) of Tequintavirus genus, as the closest match with 94.34% ANIb (Supplementary Table S5). As 94.34% ANIb falls below the species threshold of 95%, but above the genus threshold of 70% accepted by International Committee on Taxonomy of Viruses (ICTV)69, vB_Sal_S6 could possibly be assigned to a unique species within Tequintavirus genus. It may be seen that the genome size of vB_Sal_S6 (36,127 bp) is smaller than the typical genome size of Tequintavirus phages, which generally ranges from 89 to 121 kb70. However, smaller phage genomes are not uncommon, as the majority of complete phage genomes in NCBI fall within the 40–45 kbp range, followed closely by those in the 35–40 kbp range71 placing vB_Sal_S6 well within the common genome size distribution.
In conclusion, this study highlights the isolation and characterization of a novel Salmonella phage, vB_Sal_S6, with the ability to infect both Salmonella and E. coli through interaction with the putative FhuA receptor. Importantly, vB_Sal_S6 demonstrated lytic activity against clinically significant Salmonella serovars transmitted through contaminated food (Typhimurium, Weltevreden, Bovismorbificans, Infantis, Poona, Paratyphi, Typhi), which are increasingly implicated in outbreaks worldwide. Its host range, burst size, and stability across diverse pH and temperature conditions underscore its potential as a practical tool for food safety applications. The demonstrated reduction of Salmonella counts in milk further emphasizes its applicability in real food systems where contamination risk is high. Genomic and phylogenetic analyses not only confirm the lytic nature and taxonomic placement of vB_Sal_S6 within the Tequintavirus genus but also point to its uniqueness at the species level, expanding our understanding of phage diversity within Demerecviridae. Taken together, these findings position vB_Sal_S6 as a promising candidate for development into phage-based interventions targeting Salmonella in food matrices, while also providing valuable insights into phage–host coevolution.
For future work, testing the host range of vB_Sal_S6 against additional strains of E. coli and other related bacterial species will be important to assess and confirm its polyvalent nature. Its bacteriolytic efficacy can also be evaluated in other food matrices associated with Salmonella infections such as poultry and salad vegetables. In addition, in vivo studies and food safety trials are required for further food-related or potential therapeutic applications. Finally, while docking studies provided insights into phage-receptor interactions, these findings warrant experimental validation to confirm the molecular basis of host recognition.
Data availability
The vB_Sal_S6 phage genome has been submitted to NCBI GenBank and assigned an accession number PV934232 (submission ID: BankIt2971270). The genome will be made publicly available upon completion of the NCBI verification process. All other data generated or analyzed during this study are included in this published article and its supplementary files. Further inquiries or requests for data should be directed to the corresponding author Dr. Joseph Kingston J. at joseph.dfrl@gov.in .
References
Salmonella (non-typhoidal)|WHO. https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal) (2025).
Salmonella Infection | Salmonella Infection | CDC. https://www.cdc.gov/salmonella/index.html (2025).
Mkangara, M. Prevention and Control of Human Salmonella enterica Infections: An Implication in Food Safety. Int. J. Food Sci. 2023, 8899596 (2023).
Salmonella (ed) (Salmonellosis) | FDA. https://www.fda.gov/food/foodborne-pathogens/salmonella-salmonellosis (2025).
Wang, Y. et al. A global atlas and drivers of antimicrobial resistance in Salmonella during 1900–2023. Nat. Commun. 16, 4611 (2025).
Mushegian, A. R. Are There 10(31) Virus Particles on Earth, or More, or Fewer? J. Bacteriol. 202, e00052-20 (2020).
Ranveer, S. A. et al. Positive and negative aspects of bacteriophages and their immense role in the food chain. Npj Sci. Food. 8, 1 (2024).
Rohwer, F. Global Phage Divers. Cell 113, 141 (2003).
NCBI -List. of complete sequenced phage genomes. https://www.ncbi.nlm.nih.gov/nuccore/?term=(bacterial+virus%5BOrganism%5D)+AND+(complete+genome%5Bti%5D)+NOT+shotgun%5Bti%5D+NOT+plasmid%5Bti%5D+NOT+bacteria%5BOrganism%5D (2025).
Casey, E., van Sinderen, D. & Mahony, J. In Vitro Characteristics of Phages to Guide ‘Real Life’ Phage Therapy Suitability. Viruses 10, 163 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9, 676–682 (2012).
Jakočiūnė, D., Moodley, A. A. & Rapid Bacteriophage DNA extraction method. Methods Protoc 1, 27 (2018).
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A. & Korobeynikov, A. Using spades de Novo assembler. Curr. Protoc. Bioinforma. 70, e102 (2020).
Arndt, D. et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–21 (2016).
Galardini, M., Biondi, E. G., Bazzicalupo, M. & Mengoni, A. CONTIGuator: a bacterial genomes finishing tool for structural insights on draft genomes. Source Code Biol. Med. 6, 11 (2011).
Brettin, T. et al. RASTtk: a modular and extensible implementation of the RAST algorithm for Building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365 (2015).
Okonechnikov, K., Golosova, O. & Fursov, M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167 (2012).
Hallgren, J. et al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv https://doi.org/10.1101/2022.04.08.487609 (2022).
Lowe, T. M. & Chan, P. P. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57 (2016).
Richter, M., Rosselló-Móra, R., Glöckner, O., Peplies, J. & F. & JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931 (2016).
Zaki, B. M., Fahmy, N. A., Aziz, R. K., Samir, R. & El-Shibiny, A. Characterization and comprehensive genome analysis of novel bacteriophage, vB_Kpn_ZCKp20p, with lytic and anti-biofilm potential against clinical multidrug-resistant Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 13, 1077995 (2023).
Leavitt, J. C., Gilcrease, E. B., Wilson, K. & Casjens, S. R. Function and horizontal transfer of the small terminase subunit of the tailed bacteriophage Sf6 DNA packaging nanomotor. Virology 440, 117–133 (2013).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Zuckerkandl, E. & Pauling, L. Molecules as documents of evolutionary history. J. Theor. Biol. 8, 357–366 (1965).
Taslem Mourosi, J. et al. Understanding bacteriophage tail fiber interaction with host surface receptor: the key ‘Blueprint’ for reprogramming phage host range. Int J. Mol. Sci 23, 12146 (2022).
Turner, D. et al. Characterisation and genome sequence of the lytic acinetobacter baumannii bacteriophage vB_AbaS_Loki. PLoS One. 12, e0172303 (2017).
Cahill, J. & Young, R. Phage lysis: multiple genes for multiple barriers. Adv. Virus Res. 103, 33–70 (2019).
Abeysekera, G. S., Love, M. J., Manners, S. H., Billington, C. & Dobson, R. C. J. Bacteriophage-encoded lethal membrane disruptors: advances in Understanding and potential applications. Front. Microbiol. 13, 1044143 (2022).
Shi, Y. et al. Characterization and determination of Holin protein of Streptococcus suis bacteriophage SMP in heterologous host. Virol. J. 9, 70 (2012).
Abdelrahman, F. et al. Phage-Encoded endolysins. Antibiot (Basel Switzerland) 10, 124 (2021).
Bujak, K., Decewicz, P., Rosinska, J. M. & Radlinska, M. Genome study of a novel virulent phage vB_SspS_KASIA and Mu-like prophages of Shewanella sp. M16 provides insights into the genetic diversity of the Shewanella Virome. Int J. Mol. Sci 22, 11070 (2021).
Davison, J. Pre-early functions of bacteriophage T5 and its relatives. Bacteriophage 5, e1086500 (2015).
McCorquodale, D. J. et al. Gene D5 product of bacteriophage T5: DNA-binding protein affecting DNA replication and late gene expression. J. Virol. 29, 322–327 (1979).
Kobiler, O., Oppenheim, A. B. & Herman, C. Recruitment of host ATP-dependent proteases by bacteriophage lambda. J. Struct. Biol. 146, 72–78 (2004).
Kala, S. et al. HNH proteins are a widespread component of phage DNA packaging machines. Proc. Natl. Acad. Sci. 111, 6022–6027 (2014).
Tafel, A. A., Wu, L. & McHugh, P. J. Human HEL308 localizes to damaged replication forks and unwinds lagging strand structures. J. Biol. Chem. 286, 15832–15840 (2011).
Bucher, M. J. & Czyż, D. M. Phage against the machine: the SIE-ence of superinfection exclusion. Viruses 16, 1348 (2024).
Chithambaram, S., Prabhakaran, R. & Xia, X. Differential codon adaptation between DsDNA and SsDNA phages in Escherichia coli. Mol. Biol. Evol. 31, 1606–1617 (2014).
Liu, H., Whitehouse, C. A. & Li, B. Presence and persistence of Salmonella in water: the impact on microbial quality of water and food safety. Front. public. Heal. 6, 159 (2018).
Gopinath, G. R. et al. Phylogenomic Analysis of Salmonella enterica subsp. enterica serovar bovismorbificans from clinical and food samples using whole genome wide core genes and kmer binning methods to identify two distinct polyphyletic genome pathotypes. Microorganisms 10, 1199 (2022).
Georganas, A., Graziosi, G., Catelli, E. & Lupini, C. Salmonella enterica Serovar Infantis in Broiler Chickens: A Systematic Review and Meta-Analysis. Animals 14, 3453 (2024).
Zhang, J. et al. Genomic characterization of Salmonella enterica serovar Weltevreden associated with human diarrhea. Microbiol. Spectr. 11, e0354222 (2023).
Jain, P., Nandy, S., Bharadwaj, R., Niyogi, S. K. & Dutta, S. Salmonella enterica serovar Weltevreden ST1500 associated foodborne outbreak in Pune, India. Indian J. Med. Res. 141, 239–241 (2015).
Laughlin, M. et al. Multistate outbreak of Salmonella Poona infections associated with imported cucumbers, 2015–2016. Epidemiol. Infect. 147, e270 (2019).
Popa, G. L. & Papa, M. I. Salmonella spp. infection - a continuous threat worldwide. Germs 11, 88–96 (2021).
Cho, S., Jackson, C. R. & Frye, J. G. The prevalence and antimicrobial resistance phenotypes of Salmonella, Escherichia coli and Enterococcus sp. in surface water. Lett. Appl. Microbiol. 71, 3–25 (2020).
Hou, P. F., Tang, R. J., Huang, J. & Luo, D. Isolation, characterization and application of a novel polyvalent bacteriophage SF02 for the control of Salmonella and Escherichia coli O157:H7 in foods. LWT 203, 116383 (2024).
Hamdi, S. et al. Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci. Rep. 7, 40349 (2017).
Stante, M. et al. Four novel caudoviricetes bacteriophages isolated from Baltic sea water infect colonizers of Aurelia aurita. Viruses 15, 1525 (2023).
Perez-Sepulveda, B. M. & Hinton, J. C. D. Microbe profile: Salmonella typhimurium: the master of the Art of adaptation. Microbiology 171, 001521 (2025).
Jończyk, E., Kłak, M., Międzybrodzki, R. & Górski, A. The influence of external factors on bacteriophages–review. Folia Microbiol. (Praha). 56, 191–200 (2011).
Islam, M. S. et al. Application of a broad range lytic phage LPST94 for biological control of salmonella in foods. Microorganisms 8, 247 (2020).
Jeon, G. & Ahn, J. Evaluation of phage adsorption to Salmonella typhimurium exposed to different levels of pH and antibiotic. Microb. Pathog. 150, 104726 (2021).
Battistelli, N. et al. In vitro characterization and genome sequencing of two novel lytic phages against Salmonella infantis isolated from poultry feces. Front Microbiol 15, 1479700 (2024).
Zinno, P., Devirgiliis, C., Ercolini, D., Ongeng, D. & Mauriello, G. Bacteriophage P22 to challenge Salmonella in foods. Int. J. Food Microbiol. 191, 69–74 (2014).
How Temperatures Affect Food | Food Safety and Inspection Service. USDA https://www.fsis.usda.gov/food-safety/safe-food-handling-and-preparation/food-safety-basics/how-temperatures-affect-food (2020).
Prevalence of Salmonella and Listeria in Bulk Tank Milk and In-line Filters on U.S. Dairies, 2007. http://nahms.aphis.usda.gov (2009).
O’Connell, A., Ruegg, P. L., Jordan, K., O’Brien, B. & Gleeson, D. The effect of storage temperature and duration on the microbial quality of bulk tank milk. J. Dairy. Sci. 99, 3367–3374 (2016).
Institute of Medicine (US) and National Research Council (US). Committee on the Review of the Use of Scientific Criteria and Performance Standards For Safe Food. Scientific Criteria To Ensure Safe Food. Appendix F, International Microbiological Criteria Fo (in (National Academies Press (US), 2003).
Phongtang, W., Choi, G. P., Chukeatirote, E. & Ahn, J. Bacteriophage control of Salmonella typhimurium in milk. Food Sci. Biotechnol. 28, 297–301 (2019).
García, P., Martínez, B., Obeso, J. M. & Rodríguez, A. Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 47, 479–485 (2008).
Braun, V., Schaller, K. & Wolff, H. A common receptor protein for phage T5 and colicin M in the outer membrane of Escherichia coli B. Biochim. Biophys. Acta. 323, 87–97 (1973).
Wayne, R. & Neilands, J. B. Evidence for common binding sites for ferrichrome compounds and bacteriophage phi 80 in the cell envelope of Escherichia coli. J. Bacteriol. 121, 497–503 (1975).
Killmann, H., Videnov, G., Jung, G., Schwarz, H. & Braun, V. Identification of receptor binding sites by competitive peptide mapping: phages T1, T5, and phi 80 and colicin M bind to the gating loop of FhuA. J. Bacteriol. 177, 694–698 (1995).
Wang, Y. et al. Evolution and sequence diversity of FhuA in Salmonella and Escherichia. Infect Immun 86, e00573–18 (2018).
Li, S. H. et al. Biological and genomic characterization of a polyvalent phage PSH-1 against multidrug-resistant Salmonella enteritidis. BMC Microbiol. 24, 349 (2024).
Fukushima, M., Kakinuma, K. & Kawaguchi, R. Phylogenetic analysis of Salmonella, Shigella, and Escherichia coli strains on the basis of the GyrB gene sequence. J. Clin. Microbiol. 40, 2779–2785 (2002).
Turner, D., Kropinski, A. M. & Adriaenssens, E. M. A roadmap for genome-based phage taxonomy. Viruses 13, 506 (2021).
List of Tequintavirus complete genome seqeunces - Nucleotide. - NCBI. https://www.ncbi.nlm.nih.gov/nuccore/?term=complete+genome%2C+tequintavirus (2025).
Zrelovs, N., Dislers, A. & Kazaks, A. Motley crew: overview of the currently available phage diversity. Front. Microbiol. 11, 579452 (2020).
Kumar, G., Kumar, S., Jangid, H., Dutta, J. & Shidiki, A. The rise of non-typhoidal Salmonella: an emerging global public health concern. Front Microbiol 16, (2025).
Acknowledgements
The authors thank Centre Head, DIBT for providing all the facilities for conducting the research. The authors also acknowledge Sophisticated Analytical Instrument Facility (SAIF) at Indian Institute of Horticultural Research – Indian Council of Agricultural Research (IIHR-ICAR), Bengaluru, India for providing their support in carrying out the TEM work. The authors also acknowledge Biokart India Pvt. Ltd, Bengaluru, India for providing their support in the NGS sequencing activities and primary data analysis. A.M.I and C.N.M acknowledge the University of Mysore for facilitating their Ph. D work. A.M.I is funded by Senior Research fellowship from Defence Institute of Bio-Defence Technologies (DRDO), Govt. of India. C.N.M is funded by University Grants Commission, Govt. of India.
Author information
Authors and Affiliations
Contributions
J.K.J – supervision, manuscript revision; A.M.I - planning and execution of all the experiments, data analysis and curation, manuscript writing (original draft), manuscript revision; P.K.P- genome data analysis, phylogenetic analysis, docking; C.N.M – aided in phage isolation and purification. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Isaac, A.M., K.P, P., Mhatre, C.N. et al. Genomic characterization of novel lytic phage vB_Sal_S6 with putative host FhuA interaction and its application for Salmonella biocontrol in milk. Sci Rep 15, 40777 (2025). https://doi.org/10.1038/s41598-025-24573-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-24573-9