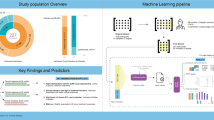
Abstract
Epstein–Barr virus (EBV) exacerbates inflammatory bowel disease (IBD) and is challenging to monitor with invasive or costly tests. We investigated whether explainable machine learning can predict EBV infection from routine clinical data in ulcerative colitis (UC) and Crohn’s disease (CD). In this retrospective study (June 2018–December 2022), EBV status was defined by EBV-DNA > 400 copies/mL. After cleaning, the training cohort (2018–2019) included 174 patients (CD = 122, UC = 52) and the test cohort (2020–2022) included 100 patients. Twenty-one demographic, clinical, and laboratory variables were modeled with ten classifiers; the four best were stacked. Five-fold cross-validation and resampling addressed overfitting and class imbalance. Shapley Additive Explanations (SHAP) provided model interpretability. The ensemble model exhibited high predictive accuracy, achieving area under the ROC curve (AUC) values of 0.93 (overall), 0.97 (CD), and 0.88 (UC) in the training set. In the validation set, AUC values were 0.95 (overall), 0.89 (CD), and 0.97 (UC). SHAP analysis identified age, hemoglobin (HB), total bile acids (TBA), and platelet count (PLT) as significant predictors. Age increased predicted risk in the overall and CD cohorts but decreased risk in UC. TBA emerged as a critical predictor in UC, reflecting its role in bile acid metabolism, while PLT influenced risk across the total patient population, indicating its involvement in coagulation and immune responses. An explainable stacking model using routine biomarkers accurately predicts EBV infection in IBD and reveals subtype-specific determinants. Prospective, multi-center and time-aware validation, and integration into decision-support tools are warranted for clinical deployment.
Similar content being viewed by others
Introduction
Epstein–Barr virus (EBV) is a ubiquitous herpesvirus; lifetime infection exceeds 90%, and reported seroprevalence among adults ≥ 35 years ranges from 20% to 90%1,2. EBV has been linked to gastrointestinal malignancies, particularly in immunocompromised patients and those receiving immunosuppressants (e.g., thiopurines)3. Individuals with inflammatory bowel disease (IBD) face elevated risks of EBV-related complications, including primary intestinal lymphoma, and EBV may aggravate intestinal inflammation, contributing to treatment failure, relapse, and increased surgical need4,5,6,7. Although polymerase chain reaction (PCR) and EBV-encoded small RNA in situ hybridization (EBER-ISH) are effective for detection, their invasiveness and cost limit routine use; peripheral blood EBV-DNA offers a practical, noninvasive biomarker for monitoring EBV-associated complications in IBD8,9,10,11. Chronic active EBV infective enteritis, characterized by severe ulcers and overlapping features with IBD, remains diagnostically challenging, underscoring the need for improved diagnostic strategies12,13,14,15.
Recent advances in epidemic and infection-dynamics modeling provide concepts directly relevant to viral risk prediction in IBD. Age-structured, delay-informed, and nonlocal diffusion frameworks clarify how host age, temporal effects, and spatial heterogeneity shape infection risk and spread16,17. Methods for inferring latent or asymptomatic infection support the use of peripheral biomarkers for EBV monitoring in clinical cohorts18,19. These models also highlight population heterogeneity and nonstationarity, motivating time-aware validation and multi-center generalization testing in predictive studies20,21. Because treatment timing can modify infection dynamics, prospective designs should capture detailed medication exposure and dosing histories22. Contemporary guidance on translational AI emphasizes interpretability, calibration, and decision-analytic evaluation—principles aligned with SHAP-based explanation and decision-curve analysis in this work23,24.
Regulatory frameworks such as the European Regulation on In Vitro Diagnostic Medical Devices (IVDR) underscore stringent validation, transparency, and performance documentation for AI-enabled diagnostics; in parallel, recent medical-imaging studies frequently deploy high-performing architectures (e.g., Vision Transformers, ResNet, MobileNet v2) for classification tasks25,26,27,28,29. Conventional endoscopic approaches have limitations for detecting EBV involvement, suggesting an opportunity for AI-augmented tools to improve diagnostic accuracy and management of EBV-related complications in IBD30,31,32,33,34,35,36,37. Prior gastrointestinal AI research demonstrates gains from heuristic feature selection, optimization-guided networks (e.g., CG-Net with dragonfly optimization), and multi-algorithm segmentation pipelines38,39,40. Additionally, explainable AI integrated with ensemble extreme learning machines, lightweight CNNs, and attention mechanisms has improved both accuracy and interpretability in GI applications41,42. Ensemble strategies (e.g., U-Net/VGG hybrids) and CNNs combined with empirical wavelet transforms further illustrate the promise of model ensembling and signal-aware feature extraction for distinguishing malignant from benign lesions43,44,45,46.
To our knowledge, no prior studies have applied artificial intelligence (AI) to EBV detection in ulcerative colitis or Crohn’s disease. We developed an interpretable machine-learning framework that links peripheral blood EBV-DNA levels with endoscopic findings, using a stacking ensemble with explainable AI to quantify feature contributions. This approach aims to improve detection accuracy and provide clinically actionable risk stratification for EBV-related complications in IBD.
Methods
This retrospective study was approved by the Xiangya Hospital Scientific Research Ethics Committee. All procedures complied with institutional and national guidelines. Informed consent was obtained when applicable; for cases where consent could not reasonably be obtained, the committee granted a waiver due to the retrospective design.
Data preparation
This retrospective study included patients with ulcerative colitis (UC) or Crohn’s disease (CD) diagnosed between June 2018 and December 2022. Clinical and biomarker data were systematically collected. EBV status was determined by real-time quantitative PCR (qPCR), with EBV-positive defined as EBV-DNA > 400 copies/mL. Cohorts were time-split: patients from 2018 to 2019 formed the training set and those from 2020 to 2022 formed the test set. Initially, 293 and 196 patients were enrolled in the training and test datasets, respectively. After removing outliers and excluding records with > 40% missing features, the final training set comprised 174 patients (CD n = 122, UC n = 52) and the test set comprised 100 patients. Given the modest sample size and potential overfitting, we implemented five-fold cross-validation to assess stability and generalization. To mitigate unequal class sizes, we applied resampling procedures. All data collection and analyses adhered to institutional ethical standards, and informed consent was obtained from all participants.
Study diagram
The framework of this study, illustrated in Fig. 1, outlines a four-module framework: data preparation, model training, model integration, and model interpretability. In data preparation, raw data were preprocessed to promote robust feature learning and generalization. To reduce overfitting, Gaussian noise was added to continuous variables using their empirical mean and standard deviation, and discrete noise was applied to categorical variables. Mutual information was then used to rank and select informative features, enabling dimensionality reduction, lowering model complexity, and improving training efficiency and accuracy.
Subsequently, the prepared data was input into ten different models: Decision Tree Classifier (DTC), Gaussian Process Classifier (GPC), K-Nearest Neighbors (KNN), Linear Support Vector Machine (Linear SVM), Logistic Regression, Naive Bayes, Neural Network, Quadratic Discriminant Analysis (QDA), Random Forest Classifier (RFC), and Radial Basis Function Support Vector Machine (RBF-SVM). Hyperparameter tuning for each model was conducted through grid search, with the results visualized in a Sankey diagram. High-performing models are then selected for integration, and the optimal integrated model is identified by plotting its receiver operating characteristic (ROC) curve using five-fold cross-validation.
Shapley Additive Explanations (SHAP)38, attributes to each feature an additive contribution to an individual prediction, based on Shapley values from cooperative game theory47. Averaging absolute SHAP values across observations yields global importance scores that rank features by their influence on model outputs. Larger absolute SHAP values indicate greater predictive impact, enabling transparent instance- and cohort-level interpretation.
For individual predictions, the SHAP value offers a detailed explanation of each feature’s impact, including both the direction (positive or negative influence) and the magnitude of influence on the outcome. This is visually represented through tools such as the force plot, which illustrates how each feature contributes to the prediction for a specific instance. The SHAP value for feature i in a prediction f(x) can be expressed as:
where S represents the subsets of features excluding i, and N is the set of all features. This equation calculates the SHAP value by averaging over all possible contributions of feature i to subsets of the model. By analyzing these values, SHAP provides insights into how each feature influences model predictions and ranks them accordingly, improving interpretability and transparency in model-driven decisions.
Performance evaluation
To evaluate the performance of classification models, we calculate the accuracy using True Positives (TP), False Positives (FP), False Negatives (FN), and True Negatives (TN). The calculation formulas for key metrics, including accuracy and components of the ROC curve, are as follows:
1. Accuracy:
Accuracy represents the proportion of correct predictions (both positive and negative) out of the total number of predictions. A higher accuracy indicates that the model is reliable and can make correct classification decisions in most cases.
2. True Positive Rate (TPR) (also known as Sensitivity or Recall):
TPR measures the model’s ability to correctly identify positive samples.
3. False Positive Rate (FPR):
FPR measures the proportion of negative samples that are incorrectly classified as positive. The ROC curve is generated by plotting the True Positive Rate (TPR) against the False Positive Rate (FPR) at various threshold settings.
Statistical analysis
Group comparisons included the overall cohort versus the UC subset and the CD versus UC subsets. Continuous variables were tested for normality using the Shapiro–Wilk test; normally distributed variables were compared with two-sample t tests, and non-normal variables with Mann–Whitney U tests. Binary variables were analyzed with Fisher’s exact test, and multi-category variables with chi-square tests. For three-group comparisons (overall, UC, CD), one-way ANOVA was applied. All tests were two-sided, with p < 0.05 considered statistically significant. Analyses were performed in R (version 4.4.1).
Results
Patient’s demographics
This study evaluated 21 demographic, clinical, and laboratory variables, including sex, age, disease duration, symptoms (abdominal pain, diarrhea, fever), and blood markers (white blood cell count [WBC], hemoglobin [HB], platelet count [PLT], and total bile acids [TBA]). Values outside reference ranges were labeled abnormal. Records with inconsistencies or > 40% missing data were excluded. CD predominated over UC, mainly in patients aged 0–30 years. Diarrhea and abdominal pain were common. Most laboratory results were within reference limits, though abnormalities in hemoglobin, neutrophil percentage, and C-reactive protein suggested inflammation or nutritional deficits. Most patients received medical rather than surgical treatment; normal levels of blood in stool (OB), WBC, and PLT were typically normal.
Comparative analysis (Table 1) showed age and onset differed significantly across the total cohort and UC/CD subsets (p < 0.05), indicating discriminative value. In UC, sex, age, onset, abdominal pain, diarrhea, fever, surgical history, OB, and ulcers differed from totals and CD (p < 0.05), consistent with mucosal disease of the rectum and colon. By contrast, the segmental, transmural pattern of CD explains distinct presentations. Assessed markers included HB, PLT, red blood cell percentage (RBC%), neutrophil percentage, WBC, erythrocyte sedimentation rate (ESR), CRP, albumin (ALB), total bilirubin (TB), direct bilirubin (DBIL), and TBA.
Ensemble model and performance evaluation
Following data preprocessing and augmentation, ten classical machine learning classifiers were systematically evaluated through grid search optimization. Among these, the RFC, KNN, GPC, and RBF-SVM emerged as the top-performing models, each achieving test accuracies surpassing 0.8 (Fig. 2a). To further optimize predictive performance, an ensemble stacking approach integrating these four classifiers was employed, effectively capitalizing on their complementary strengths. ROC curve analyses demonstrated that ensemble stacking significantly enhanced predictive accuracy compared to single classifiers, evidenced by the Area Under the Curve (AUC) improvements from 0.93 to 0.95 in the training dataset (Fig. 2b) and from 0.95 to 0.96 in the validation dataset (Fig. 2d). These results emphasize the ensemble model’s improved prediction capability and increased stability over individual classifiers.
Subgroup analysis further elucidated performance variability across distinct patient populations when utilizing the four-model ensemble. Specifically, the training dataset exhibited an overall AUC of 0.93, with subgroup AUCs of 0.97 for CD and 0.88 for UC (Fig. 2c). Similar trends were observed in the validation dataset, presenting an overall AUC of 0.95, with values of 0.97 for CD and 0.89 for UC (Fig. 2e). The relatively lower AUC observed within the UC subset may indicate greater heterogeneity in EBV infection patterns or sample size limitations, highlighting inherent disease-specific complexities. Conversely, the consistently high performance in the CD subset suggests more predictable EBV-associated features. The ensemble model’s improved validation performance, particularly within the UC subgroup, signifies robust generalizability and potential clinical utility across both IBD subtypes; underscoring its suitability for accurate and reliable EBV infection risk prediction.
Variable importance in predictions
Machine-learning models can function as “black boxes,” limiting clinical adoption. To enhance transparency, we used SHAP to quantify feature contributions to EBV risk predictions. Positive SHAP values indicate increased predicted risk; negative values indicate decreased risk. Age was a prominent predictor in the total and CD cohorts, with SHAP values increasing with age (Fig. 3a). In contrast, age showed predominantly negative SHAP values in UC (r1 = − 0.5308; Fig. 3b), suggesting lower EBV risk with increasing age—consistent with reports of higher EBV susceptibility in younger UC patients and elevated risk among older CD patients due to diminished immune surveillance48. These subtype-specific patterns (Fig. 3c) support tailored EBV monitoring strategies in IBD.
Model interpretation for CD (a), UC (b), and all patients (c). Left: Feature importance (mean |SHAP|). Middle: Feature effects (feature value vs. SHAP contribution). Right: SHAP vs. standardized age; 0 = EBV−, 1 = EBV+. r0 and r1 are Pearson correlations between age and predicted EBV risk within categories 0 and 1.
SHAP provides global and patient-level interpretability; force plots visualize feature contributions to an individual prediction. In Fig. 4a (CD), the model predicts EBV positivity ($f(x) = 0.93$) driven primarily by hemoglobin (+ 0.724), albumin (+ 0.60), total bilirubin (+ 0.496), and age (+ 0.078), with modest attenuation from CRP (− 0.043) and ESR (− 0.299). In Fig. 4b (UC), EBV positivity is also predicted ($f(x) = 0.90$), dominated by total bile acids (+ 0.878), with added contributions from hemoglobin (+ 0.226), RBC% (+ 0.24), neutrophils (+ 0.203), and direct bilirubin (+ 0.053); WBC (− 0.537) and platelets (− 1.00) reduce the score but are outweighed. In Fig. 4c (overall), a high EBV-positive prediction ($f(x) = 0.90$) is chiefly influenced by platelets (+ 0.877), total bile acids (+ 0.765), neutrophils (+ 0.596), total bilirubin (+ 0.248), and direct bilirubin (+ 0.224), with minimal negative effects. Collectively, TBA is most salient in UC, platelets and neutrophils are influential overall, and hemoglobin/total bilirubin are prominent in CD, illustrating subtype-specific contributors to predicted EBV risk.
Nomogram analyses (Fig. 5) indicated lower odds of EBV positivity in UC versus CD (OR for UC vs. CD 0.478, 95% CI 0.27–0.846) and an age-related increase in risk (OR 2.49, 95% CI 1.63–3.805). Hemoglobin (HB) was an influential predictor of EBV risk (odds ratio [OR] 1.198, 95% CI 0.231–6.202). In the UC cohort, total bile acids (TBA) was the dominant contributor (SHAP 0.878), alongside neutrophil percentage (N%), red blood cell percentage (RBC%), direct bilirubin (DBIL), and HB; TBA’s association was OR 0.914 (95% CI 0.671–1.246). In the overall cohort, total bilirubin (TB), TBA, N%, DBIL, and platelets (PLT) were primary predictors, with PLT contributing most (SHAP 0.877; OR 0.880, 95% CI 0.593–1.307). Findings highlight both shared and subtype-specific mechanisms: HB/TB are prominent in CD, TBA in UC, and PLT and neutrophils are influential overall. These patterns support disease-specific risk stratification while motivating unified protocols targeting common predictors to optimize patient monitoring and management.
Discussion
This study applied machine learning to predict EBV infection in IBD, analyzing UC and CD cohorts. A stacking ensemble of multiple classifiers was used, with SHAP providing model interpretability. Performance was strongest in CD (AUC > 0.9) and remained high in the combined cohort, while UC showed modestly lower accuracy (AUC < 0.9), likely reflecting greater variability and smaller sample size. SHAP identified key predictors of EBV risk across cohorts, supporting transparent, subtype-aware risk stratification.
Significant predictors of EBV infection were age, hemoglobin (HB), total bile acids (TBA), and platelets (PLT), aligning with evidence that immune status and hematologic indices shape EBV pathogenesis. Age correlated positively with EBV risk in the overall and CD cohorts, consistent with age-related immune senescence and impaired T-cell function in older adults49. By contrast, younger UC patients—who often exhibit more active intestinal inflammation—showed greater susceptibility, in line with reports of heightened immune activation in younger IBD populations50. Lower HB was associated with increased EBV risk, particularly in UC, supporting links among chronic inflammatory anemia, iron deficiency, and dysregulated antiviral immunity51. UC patients had higher overall EBV risk than CD patients, with TBA emerging as a key predictor; perturbations in bile acid metabolism—known to influence the microbiome and mucosal immunity—may therefore contribute to EBV susceptibility in IBD52. Elevated PLT in the overall dataset further implicates platelet-mediated inflammatory and coagulation pathways in EBV-related disease processes53.
This study demonstrates the value of interpretable machine learning for clarifying factors linked to EBV infection in IBD, separating shared from disease-specific risks. Clinically, these insights support targeted management for UC and CD while highlighting common markers that could inform unified diagnostic and preventive strategies.
Limitations
Several limitations warrant caution. The retrospective design limits causal inference and may introduce selection bias; prospective, real-time validation is needed. Generalizability is constrained by the lack of external, multi-center testing; future work will include site-level validation with varied case-mix, assessing discrimination (AUC), calibration (intercept/slope, curves), and clinical utility (decision-curve analysis), with harmonization or recalibration as appropriate. The feature set did not include medication exposures, genetic profiles, or microbiome data; forthcoming multi-center studies will add these variables and quantify their incremental value (ΔAUC, NRI) alongside calibration and decision-curve metrics. Temporal differences between cohorts may affect transportability; planned efforts include time-aware validation, statistical monitoring of covariate and calibration drift, evaluation of explanation stability across periods, use of time indicators, and periodic model updating. Finally, model complexity may hinder bedside use. We will develop an EHR-integrated decision-support interface with uncertainty displays and actionable thresholds, and a simpler surrogate model, complemented by usability testing, transparent model cards, and exemplar reports to facilitate clinical adoption.
Conclusion
This study employed machine learning to predict Epstein-Barr Virus (EBV) infection in patients with IBD, specifically focusing on UC and CD. An ensemble model integrating multiple classifiers, and employing Shapley Additive Explanations (SHAP) for interpretability, demonstrated robust predictive accuracy. Within the training dataset, the highest performance was observed in the CD subgroup (AUC = 0.97), followed by the overall dataset (AUC = 0.93). The UC subgroup showed comparatively lower accuracy (AUC = 0.88). Conversely, in the validation dataset, the UC subgroup exhibited the highest performance (AUC = 0.97), followed by the overall dataset (AUC = 0.95). The CD subgroup demonstrating lower performance (AUC = 0.89). Critical predictors of EBV infection identified included age, hemoglobin (HB), total bile acids (TBA), and platelet count (PLT). These findings underscore both common and disease-specific risk factors, facilitating a tailored approach to EBV infection management in IBD and supporting personalized patient care through precise risk assessment.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Levin, L. I. et al. Temporal relationship between elevation of Epstein–Barr virus antibody titers and initial onset of neurological symptoms in multiple sclerosis. JAMA 293, 2496–2500 (2005).
Hjalgrim, H., Friborg, J. & Melbye, M. The epidemiology of EBV and its association with malignant disease. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis (eds Arvin, A. et al.) (Cambridge University Press, 2007).
Macsween, K. F. & Crawford, D. H. Epstein–Barr virus—recent advances. Lancet. Infect. Dis. 3, 131–140 (2003).
Levhar, N. et al. Propagation of EBV-driven lymphomatous transformation of peripheral blood B cells by immunomodulators and biologics used in the treatment of inflammatory bowel disease. Inflamm. Bowel Dis. 26, 1330–1339 (2020).
Kotlyar, D. S. et al. Risk of lymphoma in patients with inflammatory bowel disease treated with azathioprine and 6-mercaptopurine: a meta-analysis. Clin. Gastroenterol. Hepatol. 13, 847–858e4 (2015).
Beaugerie, L. et al. Lymphoproliferative disorders in patients receiving thiopurines for inflammatory bowel disease: a prospective observational cohort study. Lancet 374, 1617–1625 (2009).
Muller, M. et al. Characteristics of lymphoma in patients with inflammatory bowel disease: a systematic review. J. Crohn’s Colitis. 15, 827–839 (2021).
Zhang, H., Zhao, S. & Cao, Z. Impact of Epstein–Barr virus infection in patients with inflammatory bowel disease. Front. Immunol. 13, 1001055 (2022).
Ciccocioppo, R. et al. Human cytomegalovirus and Epstein–Barr virus infection in inflammatory bowel disease: need for mucosal viral load measurement. World J. Gastroenterol. 21, 1915–1926 (2015).
Xu, S. et al. Epstein–Barr virus infection in ulcerative colitis: a clinicopathologic study from a Chinese area. Therapeutic Adv. Gastroenterol. 13, 1756284820930124 (2020).
Wang, W., Chen, X., Pan, J., Zhang, X. & Zhang, L. Epstein–Barr virus and human cytomegalovirus infection in intestinal mucosa of Chinese patients with inflammatory bowel disease. Front. Microbiol. 13, 915453 (2022).
Liu, R. et al. The clinicopathologic features of chronic active Epstein–Barr virus infective enteritis. Mod. Pathol. 32, 387–395 (2019).
Weinberg, I. et al. Epstein–Barr virus–related diarrhea or exacerbation of inflammatory bowel disease: diagnostic dilemma. J. Clin. Microbiol. 47, 1588–1590 (2009).
Pezhouh, M. K. et al. Refractory inflammatory bowel disease: is there a role for Epstein–Barr virus? A case-controlled study using highly sensitive EBER1 in situ hybridization. Hum. Pathol. 82, 187–192 (2018).
Barnes, E. L. & Herfarth, H. H. The usefulness of serologic testing for Epstein–Barr virus before initiation of therapy for inflammatory bowel disease. Gastroenterology 153, 1167 (2017).
Bentout, S. Analysis of global behavior in an age-structured epidemic model with nonlocal dispersal and distributed delay. Math. Methods Appl. Sci. 47, 7219–7242 (2024).
Soufiane, B. & Touaoula, T. M. Global analysis of an infection-age model with a class of nonlinear incidence rates. J. Math. Anal. Appl. 434, 1211–1239 (2016).
Djilali, S., Bentout, S., Kumar, S. & Touaoula, T. M. Approximating the asymptomatic infectious cases of COVID-19 in Algeria and India using a mathematical model. Int. J. Model. Simul. Sci. Comput. 13, 2250028 (2022).
Bentout, S., Chekroun, A. & Kuniya, T. Parameter Estimation and prediction for COVID-19 in Algeria. AIMS Public. Health. 7, 306–318 (2020).
Djilali, S., Chen, Y. & Bentout, S. Dynamics of a delayed nonlocal reaction–diffusion heroin epidemic model in a heterogeneous environment. Math. Methods Appl. Sci. 48, 273–307 (2025).
Mahroug, F. & Bentout, S. Dynamics of a diffusion-dispersal viral epidemic model with infection age in a spatially heterogeneous environment with general nonlinear function. Math. Methods Appl. Sci. 46, 14983–15010 (2023).
Bentout, S. & Djilali, S. Asymptotic profiles of a nonlocal dispersal SIR epidemic model with treat-age in a heterogeneous environment. Math. Comput. Simul. 203, 926–956 (2023).
Zhang, L. & Li, J. Prospects for the application of artificial intelligence in geriatrics. J. Translational Intern. Med. 12, 531–533 (2025).
Chen, K., Li, J. & Li, L. Artificial intelligence for disease X: progress and challenges. J. Translational Intern. Med. 12, 534–536 (2025).
Müller, H. et al. Explainability and causability for AI-supported medical image analysis in the context of the European in vitro diagnostic regulation. New Biotechnol. 70, 67–72 (2022).
Li, C. et al. Development and validation of an endoscopic image-based deep learning model for detection of nasopharyngeal malignancies. Cancer Commun. 38, 1–11 (2018).
Hinata, M. & Ushiku, T. Detecting an immunotherapy-sensitive subtype in gastric cancer using histologic image-based deep learning. Sci. Rep. 11, 22636 (2021).
Le Vuong, T. T., Song, B., Kwak, J. T. & Kwak, J. Prediction of Epstein–Barr virus status in gastric cancer biopsy specimens using a deep learning algorithm. JAMA Netw. Open. 5, e2236408 (2022).
Gao, X., Qian, Y. & Gao, A. COVID-VIT: Classification of COVID-19 from CT chest images based on vision transformer models. arXiv preprint arXiv:2107.01682 (2021).
Habibzadeh, M., Jannesari, M., Rezaei, Z., Baharvand, H. & Totonchi, M. Automatic white blood cell classification using pre-trained deep learning models: ResNet and Inception. In Tenth International Conference on Machine Vision (ICMV 2017), Proc. SPIE 10696, 274–281 (2018).
Wichrowska, O. et al. Learned optimizers that scale and generalize. In Proceedings of the 34th International Conference on Machine Learning (ICML 2017) 3751–3760 (PMLR, 2017).
Keskar, N. S. & Socher, R. Improving generalization performance by switching from Adam to SGD. ArXiv Preprint arXiv :171207628 (2017).
Sae-Lim, W., Wettayaprasit, W. & Aiyarak, P. Convolutional neural networks using MobileNet for skin lesion classification. In 2019 16th International Joint Conference on Computer Science and Software Engineering (JCSSE) 242–247 (IEEE, 2019).
Tan, M. & Le, Q. EfficientNet: Rethinking model scaling for convolutional neural networks. In Proceedings of the 36th International Conference on Machine Learning (ICML 2019) 6105–6114 (PMLR, 2019).
Muti, H. S. et al. Development and validation of deep learning classifiers to detect Epstein–Barr virus and microsatellite instability status in gastric cancer: a retrospective multicentre cohort study. Lancet Digit. Health. 3, e654–e664 (2021).
Magro, F. et al. Epstein–Barr virus in inflammatory bowel disease—correlation with different therapeutic regimens. Inflamm. Bowel Dis. 19, 1710–1716 (2013).
Luo, X. et al. Rapid endoscopic diagnosis of benign ulcerative colorectal diseases with an artificial intelligence contextual framework. Gastroenterology (2024).
Murugesan, R. & Devaki, K. Liver lesion detection using semantic segmentation and chaotic cuckoo search algorithm. Inform. Technol. Control. 52, 761–775 (2023).
Siddiqui, S. et al. CG-Net: A novel CNN framework for Gastrointestinal tract disease classification. Int. J. Imaging Syst. Technol. 34, e23081 (2024).
Sheikdavood, K. & Bala, M. P. Polycystic ovary cyst segmentation using adaptive k-means with reptile search algorithm. Inform. Technol. Control. 52, 85–99 (2023).
Ahamed, M. F. et al. Detection of various Gastrointestinal tract diseases through a deep learning method with ensemble ELM and explainable AI. Expert Syst. Appl. 256, 124908 (2024).
Noor, M. N. et al. GastroNet: a robust attention-based deep learning and cosine similarity feature selection framework for Gastrointestinal disease classification from endoscopic images. CAAI Trans. Intell. Technol. (2023).
Feng, R. et al. A deep learning approach for colonoscopy pathology WSI analysis: accurate segmentation and classification. IEEE J. Biomedical Health Inf. 25, 3700–3708 (2021).
Sivari, E., Bostanci, E., Guzel, M. S., Acici, K. & Asuroglu, T. Ercelebi Ayyildiz, T. A new approach for Gastrointestinal tract findings detection and classification: deep learning-based hybrid stacking ensemble models. Diagnostics 13, 720 (2023).
Mohapatra, S., Pati, G. K., Mishra, M. & Swarnkar, T. Gastrointestinal abnormality detection and classification using empirical wavelet transform and deep convolutional neural networks from endoscopic images. Ain Shams Eng. J. 14, 101942 (2023).
Nissen, L. H. C. et al. Epstein–Barr virus in inflammatory bowel disease: the spectrum of intestinal lymphoproliferative disorders. J. Crohn’s Colitis. 9, 398–403 (2015).
Lundberg, S. M. & Lee, S. I. A unified approach to interpreting model predictions. In Advances in Neural Information Processing Systems 4765–4774 (NeurIPS, 2017).
Zhou, J. Q. et al. Clinical features of Epstein–Barr virus in the intestinal mucosa and blood of patients with inflammatory bowel disease. Saudi J. Gastroenterol. (2020).
Nikolich-Žugich, J. The Twilight of immunity: emerging concepts in aging of the immune system. Nat. Immunol. 19, 10–19 (2018).
Kugathasan, S. et al. Prediction of complicated disease course for children newly diagnosed with crohn’s disease: a multicentre inception cohort study. Lancet 389, 1710–1718 (2017).
Gasche, C., Lomer, M. C. E., Cavill, I. & Weiss, G. Iron, anaemia, and inflammatory bowel diseases. Gut 53, 1190–1197 (2004).
Hu, M. M. et al. Virus-induced accumulation of intracellular bile acids activates the TGR5–β-arrestin–SRC axis to enable innate antiviral immunity. Cell Res. 29, 193–205 (2019).
Danese, S. et al. Inflammation and coagulation in inflammatory bowel disease: the clot thickens. Am. J. Gastroenterol. 102, 174–186 (2007).
Acknowledgements
This work was supported by Scientific Research Fund of the Hunan Provincial Education Department (grant no. 23A0643, 23C0427, 23C0430, and 24A0080), the Key Laboratory of 3D Scene Visualization and Intelligent Education in Hunan Province (grant no. 2023TP1038), and the Research Center for Innovative Development of Teacher Education in the New Era.
Author information
Authors and Affiliations
Contributions
Y.H.Y, X.Y.T. and Y.X. conceived, designed, and/or supervised the project. X.Y.T. and Y.Q.C. performed data analysis, manuscript writing, and figure preparation. Z.W., Y.H.Y, H.Y. and H.H. implemented the development and validation of the system. Y.Q.C, and Y.X. contributed to the collection and process of study data. Z.W, X.Y.T., Y.Q.C and H.Y. assisted with data analysis and/or figure preparation. All authors discussed the results and supervised the manuscript. All authors had full access to all the data in the study and accepted the responsibility to submit it for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, Y., Tang, X., Chen, Y. et al. Explainable ensemble learning for Epstein-Barr virus risk prediction in ulcerative colitis and Crohn’s disease using routine biomarkers. Sci Rep 15, 41919 (2025). https://doi.org/10.1038/s41598-025-25849-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-25849-w