Abstract
Antiviral peptides (AVPs), produced by all living organisms, play a vital role as the first line of immune defense against viral infections. AVPs present a promising path for developing novel antiviral therapies that target diverse viruses, including those resistant to existing drugs. However, identifying AVPs using wet lab methods is often costly and requires significant effort, and existing computational methods still have certain limitations. In this study, a novel attention-based Gated Recurrent Unit framework, named GRUATT-AVP, is proposed for accurate and fast AVPs identification. In GRUATT-AVP, several Natural Language Processing (NLP) based encoding mechanisms, including One-Hot Encoding, Word2Vec, GloVe, FastText, and ProtBert, are adopted to encode the peptide sequences. Sequentially, different embedding dimensions based on the k-mer with fixed lengths (1–6) and pooling were explored, aiming to capture the local context within the sequences. After that, we conducted another experiment to determine the best feature selection technique and integrated the SHAP technique to eliminate noise and less important encoded features, thereby improving the model’s generalization performance. Finally, the most informative subset was fed into our developed GRUATT-AVP model to construct the GRUATT-AVP for classification. To understand the contribution of each component in the GRUATT-AVP model, an ablation study was performed, and the outcomes showed that our proposed model outperforms its other variants, establishing the model’s stability and efficacy. In terms of AVP prediction results, GRUATT-AVP demonstrated better performance compared to several state-of-the-art classifiers, with an accuracy of 94.8% and an AUC of 0.986, suggesting promising therapeutic potential against viral infections. To ensure wide accessibility and practical usage, the GRUATT-AVP web server is available at https://gruatt-avp.vercel.app/.
Similar content being viewed by others
Introduction
Viruses, which are super tiny agents that can only reproduce within living cells, cause many severe diseases in humans, such as Hepatitis, AIDS, Pneumonia, Cancer, Dengue, etc.1,2. The impact of viral infections is enormous, affecting the economy and leading to high rates of illness and death. Recently, antiviral peptides (AVPs) have shown significance in antiviral-related drug construction3. AVPs are antimicrobial peptides that combat viruses and exhibit a wide range of antimicrobial effects, making them essential for the development of new treatments. AVPs target viruses or host cells to stop the virus from replicating, prevent it from fusing with cells, or disrupt different stages of its life cycle. Identifying AVPs accurately is crucial for developing new antiviral drugs, but traditional experimental methods are slow, costly, and labor-intensive. With the rapid increase in newly discovered peptide sequences, conventional methods are no longer efficient enough. To overcome these challenges, machine learning (ML) and deep learning (DL) techniques are now utilized to quickly and reliably predict AVPs4.
A variety of ML models have been constructed for classifying AVPs. Researchers have leveraged quantifiable properties of peptides, alongside ML algorithms to develop AVP classifiers. Thakur et al.5 introduced AVP-pred, a computational tool designed to predict AVPs by curating a dataset of experimentally validated peptides targeting key human viruses such as influenza, HIV, HCV, and SARS. The methodology involved extracting sequence-based features, including motif analysis, sequence alignment, amino acid composition, and physicochemical properties, which were then used to train an SVM classifier under five-fold validation. Pang et al.6 proposed the AVPiden, a two-stage classification framework that identifies AVPs and characterizes their antiviral activity. In the first stage, the model distinguishes AVPs from various peptides, including both non-antimicrobial and antimicrobial peptides without antiviral activity. In the second stage, it predicts the virus families or species targeted by the AVPs. The model uses multiple peptide descriptors to capture sequence characteristics and employs explainable ML with Shapley value analysis to interpret each descriptor’s influence on antiviral activity. Qureshi et al.7 developed AVP-ic50pred, a regression-based ML approach designed to predict the antiviral activity of peptides in terms of their IC50 values (\(\mu\)M). The model was trained using 759 non-redundant peptides. Several sequence-derived features were employed, and their hybrid combinations with four ML algorithms: SVM, RF, Instance-based classifier, and K-Star were utilized, with the model’s performance assessed using 10-fold validation. In addition, Kurata et al.8 introduced iACVP, a computational model developed to predict anti-coronavirus peptides (ACVPs) using a combination of conventional features, a binary profile, and word-embedding techniques. The model systematically explores five different machine learning methods: Transformer, CNN, BiLSTM, RF, and SVM. The RF classifier with W2V consistently outperformed other methods by utilizing a dataset-specific W2V dictionary generated from the training and independent test datasets and optimizing the k-mer value in W2V. This approach enhances the discrimination between positive and negative samples, improving prediction performance. Schaduangrat et al.9 introduced Meta-iAVP, a sequence-based meta-predictor that provides an effective feature representation strategy by extracting features from a combination of prediction scores generated by various ML algorithms and multiple feature types. This ensemble-style method leverages the strengths of individual classifiers to enhance the reliability of predictions. Yan et al.10 developed the PreTP-Stack model, a stacking-based framework for predicting multiple types of therapeutic peptides. The model integrates ten distinct feature types and employs an ensemble of four base classifiers: RF, LDA, XGBoost, and SVM. In addition to these, an auto-weighted multi-view learning model serves as a meta-classifier, combining the predictions from the base models to enhance overall performance. This architecture is designed to utilize diverse feature representations and capture complementary predictive signals effectively. Furthermore, Chowdhury et al.11 introduced the FIRM-AVP. It is a ML approach designed to classify AVPs based on the physicochemical and structural properties of amino acid sequences. The model utilizes feature selection techniques to identify and retain the most informative features, with a particular emphasis on secondary structure, which is highly indicative of antiviral activity. By focusing on the most relevant sequence attributes, FIRM-AVP aims to enhance model efficiency and interoperability. However, a significant drawback of ML models is the need to craft, collect, and refine hand-engineered features as input, adding complexity. Additionally, these models tend to underperform compared to DL approaches, especially with large datasets.
Beyond ML algorithms, there are many advanced models developed using DL methods. Sharma et al.12 introduced the Deep-AVPpred model, which utilizes transfer learning with DL algorithms to identify novel AVPs. This model was used successfully to identify antiviral peptides in human interferon-\(\alpha\) family proteins and can be used to predict novel AVPs for the development of antiviral compounds in human and veterinary medicine. In contrast, Li et al.13 proposed the DeepAVP model, a dual-channel deep neural network to analyze antiviral peptides (AVPs), consisting of an LSTM channel to capture long-term sequence dependencies and a CONV channel to analyze local evolutionary information. It integrates feature extraction directly into the neural network, optimizing it during training, unlike traditional methods that separate feature extraction. The PSSM layer in the CONV channel refines the BLOSUM matrix for better peptide prediction. Additionally, Xiao et al.14 developed iAMP-CA2L, a 2-level predictor designed to address the challenge of predicting the functions of AMPs, particularly when AMPs possess multiple functional classes. The model introduces CNN, BiLSTM, and SVM classifiers, utilizing cellular automata images to handle monofunctional and multifunctional AMPs. The first level identifies whether a peptide is an AMP or non-AMP, while the second level classifies the peptide into one or more functional types. DeepAVP-TPPred, introduced by Ullah et al.15, was created to address the limitations of existing methods by extracting two new transformed feature sets using image-based feature extraction algorithms and integrating them with evolutionary information-based features. These feature sets are then optimized using a novel feature selection technique called the binary tree growth algorithm. The final classification model is built using a DNN, enabling enhanced efficiency and generalization capabilities compared to existing predictors. Similarly, Deepstacked-AVPs, proposed by Akbar et al.16, utilize a combination of feature encoding methods, a Tri-PSSM-TS, word2vec-based semantic features, and CTDT descriptors, which represent the physiological properties of peptides. These features are fused into a single vector to overcome the limitations of individual encoding methods. Information gain (IG) is then applied to select the optimal feature set, and the final classification model is built using a stacked-ensemble classifier. Timmons et al.17 proposed the ENNAVIA model. This sequence-based deep neural network classifier predicts the antiviral activity of peptides by integrating cheminformatics techniques to model peptide sequences for classification. It is trained on a dataset of experimentally validated antiviral peptides, enabling an accurate in silico screening of candidates. The classifier aims to support large-scale analysis by leveraging the expanding volume of peptide sequence data.
Guan et al.18 proposed a two-stage DL framework for AVP identification. It consists of three modules: a contrastive learning module that encodes sequences using binary, BLOSUM62, and Z-scale representations and extracts features via multi-scale CNN and BiLSTM; a feature-enhanced transformer module that learns additional peptide characteristics; and a prediction module that fuses features from both modules to identify AVPs and their functional types. Yao et al.19 proposed ABPCaps, a capsule network-based model to identify antibacterial peptides (ABPs). ABPCaps integrates a CNN, LSTM, and capsule network, leveraging the capsule network’s ability to automatically extract critical features from both positive and negative samples. It employs a dynamic routing mechanism to assign appropriate weights to feature capsules, thereby enabling nuanced feature representation and improved handling of complex data patterns. The author also developed dbAMP-320, a comprehensive database of antimicrobial peptides (AMP) with extensive annotations on sequences, activity, properties, and structures. dbAMP-3 includes molecular docking data, tools for predicting toxicity and half-life (HemoFinder), a pipeline for designing stable, active AMPs, and links to over 20 AMP prediction tools and benchmark datasets, making it a key resource for AMP research and development. Akbar et al.21 proposed TargetAVP-DeepCaps, which employs a multi-faceted approach for AVP prediction. It utilizes the pre-trained ProtGPT2 model to generate contextual embeddings for peptide sequences. These sequences are transformed into 2D representations using SMR and RECM matrices, which are further processed with the CLBP technique to extract local features. A differential evolution mechanism constructs a weighted multiperspective feature vector, optimized through a hybrid MRMD + SFLA feature selection strategy. Finally, the model incorporates a self-normalized capsule network (Sn-CapsNet) to make predictions, providing a robust and interpretable framework for AVP identification. Singh et al.22 proposed the Deep-AVPiden model, which utilizes separable temporal convolutional networks, and its efficiency is further enhanced by a variation called Deep-AVPiden (DS), which employs point-wise separable convolutions. The models were used to identify potential AVPs in the natural defense proteins of plants, mammals, and fish. The identified peptides showed sequence similarity to experimentally validated antimicrobial peptides and could be chemically synthesized for antiviral testing. On the contrary, as described by Bai et al.23, training and fine-tuning bi-LSTM models is time-consuming due to their sequential, non-parallelizable nature and requires significant memory. In summary, a major challenge DL and deep neural networks face is the high computational cost associated with training and operation. Another key limitation is that many of these studies lack dedicated web servers, making it challenging for wet-lab researchers to efficiently discover and classify AVPs.
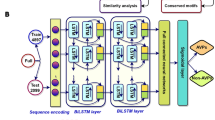
In this paper, we introduce an attention-based GRU model, which is widely used for sequence modeling due to its faster performance compared to LSTMs and its ability to capture long-range dependencies more effectively than CNNs. Firstly, we applied five feature extraction methods: one-hot encoding, GloVe, Word2Vec, FastText, and ProtBert, to extract meaningful patterns from the peptide sequence. Next, we combined these five base features to form a composite feature. To improve the predictive accuracy of the GRUATT-AVP model, we utilized the SHAP technique to eliminate irrelevant properties from the composite feature. Finally, we input the optimized features into our proposed model and trained it to identify antiviral peptides, aiming for precise predictions. Experimental results demonstrate that our model outperforms existing approaches and achieves state-of-the-art performance in AVP identification. The sequential workflow of this study is illustrated in Fig. 1.
Materials and methods
In this section, we provide a detailed description of the individual phases followed for GRUATT-AVP. The complete workflow is shown in Fig. 1. It consists of multiple steps, including data preparation, NLP-based feature encoding, optimal feature subset selection, selecting the best algorithm for model construction, and finally, model deployment and prediction.
This is a schematic representation of the proposed framework. At the top, the first section illustrates the data preprocessing steps, while the second section highlights feature extraction and selection using various NLP methods. The third section presents model construction, and the final section demonstrates model hosting and classification with the GRUATT-AVP model.
Dataset description
We collected a benchmark dataset from Stack-AVP24, where the data is gathered from multiple sources, including AVPdb25, HIPdb26, starPep27,28,29, DRAMP30, and SATPdb31, while non-AVPs were sourced from Swiss-Prot32 and AVPdb25. After collection, the data underwent cleansing; peptides containing non-standard amino acids (B, J, O, U, X, and Z) and those with fewer than five or more than fifty amino acids were excluded. Subsequently, the cluster database at high identity with tolerance (CD-HIT)33,34,35 was applied separately to AVPs and non-AVPs using a similarity threshold of 0.9 to filter out similar sequences. The CD-HIT and 0.9 threshold balances data diversity and redundancy, ensuring a wide variety of sequences to reduce overfitting while maintaining sufficient data for robust model training, which is crucial for tasks like predicting antiviral peptides where small sequence differences are significant. The final dataset comprised 5,413 peptides, divided into 2,707 AVPs and 2,706 non-AVPs. This dataset was further split into training (85% of the data) and test (15% of the data) sets for analysis.
Sequence encoding
This study examines various sequence encoding methods for extracting informative features by converting sequential data into numerical representations, ultimately developing a final prediction model. We employed NLP-based word embedding methods, GloVe, Word2Vec, and FastText, to capture semantic relationships between words. Different embedding dimensions based on the k-mer concept were explored, where a k-mer is a fixed-length (k) subsequence extracted from the original sequence. By varying k from 1 to 6, we aimed to capture the local context within the sequences. Furthermore, we employed a transformer-based method, known as ProtBert, with three different pooling mechanisms: Mean, Max, and CLS, to generate features.
K-mer
K-mers are consecutive segments of nucleotides or amino acids, each with a specific length k, contained within a sequence36. Although commonly used in DNA studies, k-mers can also be applied to RNA, proteins, and peptides. Any sequence of amino acids can be broken down into multiple successive k-mers, with the exact count depending on the sequence length (L) and the chosen k-mer size (k). For instance, in the sequence “AAGTCCAT” (L=8), there are seven 2-mers, six 3-mers, five 4-mers, four 5-mers, three 6-mers, and two 7-mers. The formula to determine the number of k-mers present in a sequence of length L is (L-k+1). This principle applies to sequences of any length or composition without exception.
One hot encoding
One-hot encoding37 is a method used to represent categorical data by converting each category into a binary vector. When applied to a protein sequence of length L, it transforms the sequence into an L \(\times\) n matrix, where n represents the number of different amino acids. Each row in this matrix contains n-1 zeros and a single 1, indicating the specific amino acid residue at that position in the protein sequence. This results in sparse, memory-inefficient, and high-dimensional data representations. In a one-hot encoding scheme, there is no inherent similarity between sequence or structural elements; each element is treated as either identical or different. For example, in a one-hot encoding of words, the words king, prince, and pot are equally dissimilar, despite king and prince sharing closer semantic meanings than king and pot or prince and pot. Similarly, biologically, an amino acid sequence of DDD is perceived to be more similar to EEE than to PPP or HHH. However, one-hot encoding does not inherently capture this biological similarity due to its binary nature.
GloVe
GloVe38 is an unsupervised learning algorithm that generates vector representations of words by leveraging local and global statistical information. It utilizes a log-bilinear regression model trained on the non-zero entries of a word-to-word co-occurrence matrix, which records the frequency of word co-occurrences in a corpus. Training the GloVe model involves a single pass through the entire corpus to gather co-occurrence statistics, which can be computationally intensive for large datasets. However, by focusing only on the non-zero entries in the co-occurrence matrix, GloVe reduces computational load, as these entries are far fewer than the total number of possible word pairs. Combined with a weighted least-squares regression model, this approach allows the loss function to converge more quickly. The resulting word vectors exhibit linear substructures that capture meaningful relationships between words.
Here, in Eq. (1), X denotes a word co-occurrence matrix. The probability of word j occurring in the context of word i is denoted as \(X_{(i,j)}\). v represents word embeddings with \(v_i\) and \(v_j\) being the respective word vectors for words i and j. \(b_i\) and \(b_j\) are constants, and f is a weight function. N indicates the size of the vocabulary.
FastText
FastText is a word embedding model designed for NLP tasks39. It encodes words as continuous-valued vectors, capturing both meaning and structure. FastText can handle unfamiliar words by breaking them down into smaller parts, such as character n-grams, which is especially useful for languages with complex word forms. The input layer breaks down text into n-gram character sequences to understand sub-word information. These sequences then move through the hidden layer, where a transformation generates feature representations. Finally, the output layer predicts the word’s context based on these representations. Key factors influencing fastText performance include the size of the word vectors (embedding dimension), the learning rate, the number of training rounds (epochs), and the context window size. fastText employs an architecture similar to the CBOW model, minimizing the softmax loss \(\ell\) over N documents.
In Eq. (2), \(x_n\) represents a bag of one-hot vectors, and \(y_n\) denotes the label of the n-th document. Unlike Word2Vec and GloVe, which rely on word-level representations, fastText utilizes character-level units to derive word representations.
Word2Vec
The Word2Vec model generates high-quality, dense word representations by processing large amounts of text. This unsupervised model builds a vocabulary from the text and creates word embeddings in a vector space40. This reduces the dimensionality compared to the traditional BOW method, which produces high-dimensional, sparse vectors. Word2Vec uses two main techniques to create word embeddings: the CBOW and the SGM. CBOW is faster and excels at representing common words, while the SGM is better at handling smaller datasets and provides superior representations for rare words. In the CBOW model, determining the target word \(W_t\) from n predictions is achieved through Eq. (3).
Here, \(W_t\) is the target word, and the sequence of words from \(W_{(t-n)}\) to \(W_{(t+n)}\) represents the context words. Equation (4) simplifies Eq. (3) by treating the hidden layer as equivalent to a SoftMax layer.
Here, \(W^{\prime }\) represents the output weight matrix between hidden layers. After performing matrix operations, \(h_t\) represents the average value of input vectors.
ProtBert
ProtBERT builds upon the BERT architecture41 and adapts a multi-layer bidirectional transformer encoder42, for protein sequence analysis, leveraging extensive datasets of protein sequences for training. Each layer in ProtBert consists of a multi-head self-attention sub-layer and a fully connected feed-forward sub-layer. The multi-head self-attention sub-layer allows the model to focus on different parts of a protein sequence simultaneously. Each sub-layer is wrapped with a residual connection, followed by layer normalization. The transformer architecture of ProtBert relies heavily on the attention mechanism. In the core function of the attention mechanism, a weighted sum of input values (V) is computed. The weights, which determine the amount of attention each input should receive, are derived from the dot product of queries (Q) and keys (K). A softmax operation ensures that these weights are normalized and sum to one. In ProtBert, pooling mechanisms summarize the information captured in the token representations into a fixed-size vector. This vector can then be used for various downstream tasks such as classification, regression, or other analyses. Our experiment used the three most popular pooling mechanisms: Mean, Max, and [CLS].
SHAP (Shapley additive exPlanations)
SHAP, developed by Lundberg and Lee43, is an additive feature attribution method that analyzes individual predictions by evaluating the contributions of features and ranking them by their importance. Using game theory principles, SHAP assigns an importance value to each feature for a specific prediction. SHAP values break down a prediction to reveal the effect of each feature. SHAP explains the prediction as follows:
In Eq. (5), g is the explanation model, while \(x' \in \{0,1\}^m\) is the coalition vector where 0 and 1 signify the absence or presence of a corresponding feature. M denotes the number of input features in the model, and \(\phi\) \(\in\) R (set of real numbers), \(\phi _i\) represents the attribution values for feature i. Based on game theory, Shapley values can be computed using the following equation:
In Eq. (6), N denotes the set of features in the model, and S represents all subsets derived from N. The function calculates the total contribution of a given feature set S. The notation \(S\subseteq N\setminus \{i\}\) indicates the value of the corresponding feature when i is known, compared to when the feature value i is unknown for all subsets.
Proposed model architecture
Our model, GRUATT-AVP, is an advanced neural network architecture that integrates two powerful mechanisms: the Attention Mechanism and the Gated Recurrent Unit (GRU). Combining these mechanisms enhances the model’s ability to process sequential data, focusing on the most relevant parts of the input sequence. Below, we describe the working principles of the Attention Mechanism, GRU, and GRUATT-AVP.
Attention mechanism
The attention mechanism44 is widely used in Seq2Seq (sequence-to-sequence)45 models, typically built using an Encoder-Decoder framework. The Encoder transforms the input data into a semantic vector, and the decoder generates the output data from this vector46. The attention mechanism helps by focusing on different parts of the input with varying levels of importance, improving data utilization efficiency. The following equation gives the mathematical formulation of the attention mechanism:
Here, \(u_w\) is a randomly initialized vector that gets updated during training. \(u_i\) represents the outcome of a fully connected operation applied to the hidden layer vector \(h_i\). \(W_i\) and \(b_i\) denote the weight matrix and bias term used in the attention computation, respectively. Lastly, \(\alpha _i\) signifies the attention score assigned to the i-th word in the sentence.
Gated recurrent unit (GRU)
GRU offers a simpler alternative to LSTM units while maintaining comparable performance47,48. The main architecture includes two gates: the update and reset gates. These gates work together to control the information flow, ensuring that relevant information is passed through the network to make accurate predictions. The update gate in a GRU determines how much of the previous memory and new information to retain. It is defined as:
In Eq. (10), \(x_t\) represents the current input vector, \(h_{t-1}\) is the value computed from the previous adjacent layer, \(W_z\) is the learnable weight matrix associated with the update gate, and \(\sigma\) denotes the sigmoid activation function. Then, the reset gate in GRU controls how much of the previous hidden state \(h_{t-1}\) should be reset or ignored based on the current input \(x_t\). It is defined as:
In Eq. (11), \(W_r\) is the learnable weight matrix for the reset gate. After that, the candidate’s hidden state is a new memory content that could be added to the network. It is computed using the reset gate to filter the previous hidden state.
In Eq. (12), W is the learnable weight matrix, and \(\odot\) denotes element-wise multiplication, tanh is a hyperbolic tangent function. The output range for tanh is (-1,1). The final hidden state combines the previous hidden state, \(h_{t-1}\), and the candidate’s hidden state, controlled by the update gate \(z_t\). Equation 13 describes the final hidden state.
GRUs have fewer parameters to train since they lack the output gate present in LSTMs, making them computationally more efficient. Their more straightforward structure often leads to faster training and inference times. Despite their simplicity, GRUs perform comparably to LSTMs on various tasks.
GRUATT-AVP model
The GRUATT-AVP takes feature vectors representing peptides created using various embedding methods. The GRU layer processes these sequences to capture patterns and dependencies over time. Next, an attention mechanism is applied to the GRU’s output, helping the model focus on the most critical parts of the input sequence by assigning different weights. This improves the model’s ability to identify key features and make more accurate predictions. The attention-weighted output is then passed through a dropout layer with a value of 0.1 to reduce overfitting, followed by a fully connected layer with a sigmoid activation function. This final layer produces a probability score ranging from 0 to 1, representing the model’s prediction for the peptide. The model is trained using a binary cross-entropy loss function, optimized with the Adam optimizer. A Step LR scheduler adjusts the learning rate periodically, enhancing convergence. Training involves multiple epochs, during which model parameters are updated based on gradients computed from the loss function. A new instance of the GRUATT-AVP model is created and initialized for each fold. During training, input features pass through the GRU layer, the attention mechanism, and the fully connected layer to generate predictions. The binary cross-entropy loss is calculated between the predictions and the target labels. Gradients are then backpropagated to adjust model parameters. The model’s performance is evaluated on the validation set, and the best parameters are saved based on validation accuracy.
Results and discussion
Evaluation metrics
In this study, we utilized various performance metrics49 to evaluate the efficiency of prediction methods and compare multiple classification models. In the context of classification problems, primary metrics include accuracy (ACC), Matthews’s correlation coefficient (MCC), sensitivity (Sen), specificity (Spe), precision (Pre), and so on. The equations of these metrics are as follows:
Here, True Positive (TP) refers to correctly identified positive instances, while True Negative (TN) refers to correctly identified negative cases. False Negative (FN) occurs when positive instances are mistakenly classified as negative, and False Positive (FP) occurs when negative instances are incorrectly classified as positive. Furthermore, we evaluated the model’s performance using the Area Under the Receiver Operating Characteristic Curve (AUROC). This is visualized by plotting the True Positive Rate (TPR) against the False Positive Rate (FPR) across different threshold values.
Hyperparameter tuning
Optimizing the performance of the proposed classification model requires a careful selection of hyperparameters50. These factors impact both training efficiency and the classification task. The critical hyperparameters for GRUATT-AVP include activation functions, batch size, dropout rate, learning rate, and maximum number of epochs. We performed hyperparameter optimization on the proposed GRUATT-AVP model to achieve the best results.
A summary of the chosen hyperparameter values is provided in Table 1. We did iterative manual tuning across multiple ranges. Each candidate was trained under 5-fold cross-validation with (shuffle=True) and a fixed seed \((random\_state=42)\) to maintain approximate label balance. Within each fold, we monitored validation accuracy of every epoch and checkpointed the best model. Among the candidates, the configuration that yielded the most stable and competitive validation results, with 2 GRU layers, a hidden size of 64, a batch size of 16, a learning rate of 0.01, and a dropout rate of 0.1, was selected for the final model. To limit overfitting in small datasets, we employed dropout, learning rate decay, and model selection based on cross-validation performance; we did not use early stopping or L2 weight decay.
Significance of pooling and K-mer in encoding more accurate motif information
A motif is a short peptide sequence that plays a key role in determining the function of AVPs. To better understand the motif information, we utilized k-mer embedding, which breaks sequences into smaller subsequences of length k, to facilitate the identification of motifs. We compared the effectiveness of k-mers (with k ranging from 1 to 6) for different feature extractors: GloVe, fastText, Word2Vec, and One-hot encoding. Additionally, selecting the right pooling strategy in ProtBert embeddings is crucial, as it affects the model’s ability to capture essential motif information in sequences, influencing prediction accuracy. To determine the best pooling strategy for our dataset, we tested three different pooling layers (Mean, Max, and CLS) when encoding peptide sequences. The model’s performance was then evaluated using various metrics. Table 2 compares the performance of different pooling strategies for ProtBert, and k-mer for fastText, One-hot encoding, GloVe, and Word2Vec.
The performance of ProtBert, One-hot encoding, fastText, and GloVe embeddings varies based on pooling strategies and k-mer values. In ProtBert, Mean Pooling outperforms the others across most metrics, achieving the highest AUC (0.955), accuracy (0.890), improving accuracy by 4.5% over the lowest CLS Pooling, MCC (0.780), sensitivity (0.879), F1 score (0.891), and AUPR (0.960). Mean pooling is more effective for peptide sequence databases because it averages the features across the entire sequence, thereby enhancing the accuracy of the predictions. It captures the holistic interactions and dependencies between amino acids, which are crucial for understanding the sequence’s overall biological function. Max Pooling, while not the top performer overall, excels in specificity (0.908) and AUPR (0.949). CLS Pooling generally underperforms, with the lowest values in AUC (0.932), MCC (0.691), and specificity (0.819). One-Hot Encoding using 3-mer shows the best performance among the k-mer strategies with an AUC (0.937), accuracy (0.864), improving accuracy by 2.7% over the lowest 2-mer, and F1 score (0.865). The 3-mer strategy performs best because it captures meaningful patterns at a mid-level of granularity, allowing the model to recognize both individual residues and common motifs that span three consecutive amino acids, thereby balancing the effects of excessive noise from shorter k-mers and overly large contexts from longer k-mers. Longer k-mers may capture more detailed patterns, but they also lead to overfitting and complexity that can reduce the model’s generalizability. However, the smaller k-mer sizes, particularly 1-mer and 2-mer, result in significantly weaker performance in comparison, likely because these smaller k-mers fail to capture relevant motif information and are too coarse to provide valuable insights. FastText, on the other hand, struggles with smaller k-mer lengths and more extensive k-mer lengths, such as 1-mer, 2-mer, 4-mer, 5-mer, and 6-mer. Where AUC values are as low as (0.758), (0.737), (0.724), (0.727), and (0.722), respectively. The main reason is that fastText relies on word embeddings that may not effectively represent individual amino acids or sequences when broken down into units that are too small or too large. FastText does not utilize specialized features or pre-trained embeddings designed explicitly for protein sequences. The best performance for fastText is observed with the 3-mer strategy, which achieves the highest AUC (0.920), accuracy (0.852), improving accuracy by 19.7% over the lowest 6-mer, and F1 score (0.848). For GloVe, the 3-mer strategy yields the best performance overall, with an AUC (0.926), accuracy (0.853), and an F1 score (0.852). The 1-mer and 2-mer strategies also perform relatively well with AUC (0.841) and AUC (0.839), respectively. The 4-mer, 5-mer, and 6-mer strategies show decreased performance across all metrics, likely due to overfitting with larger k-mer sizes and loss of essential sequence details. Word2Vec yields the worst result among all feature extractors across all k-mer values, indicating a low contribution of k-mer to Word2Vec. Word2Vec fails to effectively encode peptide sequences because it does not capture the complex biochemical properties, structural dependencies, and evolutionary relationships between amino acids, treating them as independent tokens. Its general-purpose design also lacks domain-specific knowledge of protein folding and molecular interactions, leading to suboptimal representations and poor performance in peptide-related tasks. Overall, the strong performance of 3-mers can be explained by their ability to capture short but biologically meaningful patterns of three amino acids that often form key structural regions, such as turns and binding sites. This length offers the right balance; shorter k-mers (1–2) lack enough context to represent functional motifs, while longer ones (4–6) tend to become too specific and lead to overfitting. On the other hand, Mean pooling works best for ProtBert because it averages information across the entire peptide sequence, allowing the model to consider overall relationships and dependencies between amino acids. This helps capture the global context needed to understand peptide function, unlike max pooling, which focuses only on the strongest signals, or CLS pooling, which relies on a single position in the sequence. By observing these details, it is clear that k-mer and pooling give weight to performance. Figure 2 shows the violin plot indicating the contribution of k-mer and pooling in different feature extractors.
Violin plots display the distribution of metric values. The width of each violin indicates how concentrated the values are. A wider area means a higher density of values at that performance level, while a narrow section suggests fewer values. The violin plots depict the performance of different encoding methods: One-Hot Encoding, Word2Vec, GloVe, fastText, and ProtBert, across various metrics (Accuracy, MCC, sensitivity, specificity, precision, F1-score) influenced by k-mer values (1-6) and pooling strategies for ProtBert. Figure 3 illustrates the ROC curves of fastText, GloVe, Word2Vec, and One-Hot Encoding on different k-mer values. The ROC curve of ProtBert is implemented on different pooling strategies.
The effects of different single and hybrid features
This section examines how various feature encoding strategies impact peptide sequence classification in the context of antiviral drug discovery. We investigated the outcomes based on single-view feature encoding (One Hot, Glove, fastText, Word2vec, ProtBert) and hybrid-view feature encoding, where multiple encoded features are paired to create new and potentially more effective encoding schemes. These feature sets were further passed to our proposed GRUATT-AVP model and tested with a 5-fold cross-validation. This study utilized five feature extractors: One-Hot Encoding, Word2vec, GloVe, fastText, and ProtBert, to determine their efficiency levels (Table 3).
ProtBert with mean pooling demonstrates the highest AUC (0.955), Accuracy (0.890), MCC (0.780), Sen (0.879), Spe (0.901), Pre (0.903), F1-score (0.891), and AUPR (0.960), making it the most powerful among all the feature extractors, and reflecting its ability to handle imbalanced data effectively. ProtBert’s accuracy is 2.6% higher than one-hot encoding, 3.7% higher than GloVe, 3.8% higher than fastText, and 19.1% higher than Word2Vec. ProtBert performs well in protein-related tasks because it is a transformer-based model pre-trained on large-scale protein sequence data, enabling it to capture complex, contextual relationships and dependencies within protein sequences that are critical for understanding their structure and function. The second-best feature extractor is One Hot Encoding with 3-mer, which also performs well, with AUC (0.937), Accuracy (0.864), MCC (0.730), Sensitivity (0.849), specificity (0.881), precision (0.883), F1-score (0.865), and AUPR (0.945). One-hot encoding also performs well in protein-related datasets because it provides a simple and effective way to represent each amino acid as a unique binary vector, capturing the presence or absence of each amino acid in a sequence without requiring complex relationships between them, and adding the k-mer concept further gives a better understanding of local patterns in the data. GloVe and fastText demonstrate competitive performance with similar accuracy (0.853) and (0.852), respectively. However, Word2Vec performs the worst among all the feature extractors with accuracy (0.699) and AUC (0.739). Word2Vec performs poorly in protein-related data because it relies on co-occurrence patterns between words, which do not translate effectively to the sequential and structural properties of amino acids in proteins, where local and global dependencies are more critical. At the transformer level, both ESM and ProtT5 are pretrained on large protein corpora, but ProtT5 achieves consistently better classification metrics: AUC 0.934, ACC 0.890, MCC 0.780, F1 0.893, and AUPR 0.918, in comparison with ESM. On the other hand, ESM remains competitive in terms of precision and specificity (0.867 and 0.869) and can be considered suitable, whereas ProtT5 is preferable when maximizing recall and balanced correctness is the priority. However, both ESM and ProtT5 underperform compared to ProtBert across most evaluation metrics.
A comprehensive list of feature sets was designed to obtain the best results by combining single-view features, including One-Hot Encoding, GloVe, fastText, Word2Vec, and ProtBert. These single-view features were combined to construct a robust hybrid feature space, thereby improving data representation. We adopted a straightforward concatenation strategy to combine different embeddings. We choose concatenation because it preserves the complete feature space of each representation without imposing assumptions on the relative importance of its components. The efficiency of these hybrid features was thoroughly assessed with the help of our proposed model, GRUATT-AVP. We generated 26 combinations using single-view feature sets and selected the five best-performing combinations. Table 4 compares the best five hybrid view features. The performance of the remaining 21 hybrid features are provided in the supplementary file (Supplementary Table 1).
Our best-performing five combinations align with theory and are empirically validated. We fuse four complementary embeddings: one-hot encoding (exact composition and position-specific motifs), fastText (local k-mer semantics and subword regularities), ProtBert (transformer-based, long-range dependencies and rich biological context), and GloVe (global co-occurrence structure and semantic relationships among residue patterns) to capture distinct sequence signals. This multi-scale representation enables the model to leverage both shallow, motif-level cues and deep, contextual information, thereby improving accuracy while preserving interpretability. The best-performing combination is (One Hot Encoding + fastText + ProtBert)(H1), achieving the highest AUC (0.986) and ACC (0.937) with strong results in other metrics like MCC (0.874), Sensitivity (0.935), Specificity (0.939), Precision (0.942), F1 (0.939), and AUPR (0.987). This is followed by (One Hot Encoding + GloVe + fastText + ProtBert)(H2), which performs similarly with AUC (0.985), ACC (0.935), and solid performance across all metrics, but a bit lower than H1. (One Hot Encoding + GloVe + ProtBert)(H3) performs well with AUC (0.983), ACC (0.933), and AUPR (0.984), but is slightly behind H1 and H2, as it misses the additional benefits of fastText sub-word modeling. (One Hot Encoding + ProtBert)(H4) shows a good performance, with AUC (0.978), ACC (0.928), AUPR (0.981), with a little lower precision (0.928) and F1 score (0.931), likely due to the absence of fastText and GloVe semantic and sub-word representation abilities. (GloVe + fastText + ProtBert)(H5) also performs comparably to H4 with AUC (0.979), ACC (0.918), and AUPR (0.981). Still, the lack of one-hot encoding reduces its ability to capture precise positional information, which in turn affects its overall classification ability. One noticeable thing is that the top five combinations all have ProtBert in common. The results of the top combinations stem from the synergy between ProtBert’s deep contextual understanding, fastText’s sub-word feature learning, and GloVe’s semantic embeddings, enabling the model to capture complex biological relationships in protein sequences more effectively. Combining these extractors allows a model to leverage their complementary strengths, improving performance on protein sequence classification tasks. In contrast, none of the top five combinations utilizes Word2Vec, which suggests the limited capability of Word2Vec in encoding antiviral peptides. Finally, based on the results in Table 4, we can conclude that the combined features yield better results than the single features for the experiment dataset. Figure 4 compares the performance of the single-view and hybrid-view features in a bar chart.
Finding the best feature selection technique
To determine the most effective feature selection strategy for our proposed model, we explored four different techniques: PCA (Principal Component Analysis)51, Lasso (Least Absolute Shrinkage and Selection Operator)52, ICA (Independent Component Analysis)53, and SHAP (Shapley Additive Explanations)43. Each technique was tested on five distinct feature portions: 200, 300, 400, 500, and 600 features, resulting in a total of 20 experiments. The goal was to identify the best feature selection technique with the optimal feature subset. After conducting all experiments, we determined the optimal feature subset for each method: PCA performed best with 400 features, Lasso with 600 features, ICA with 400 features, and SHAP with 500 features. Table 5 showcases the complete results of these feature selection techniques.
SHAP outperforms all other techniques, achieving the highest AUC (0.986), accuracy (0.948), AUPR (0.988), and MCC (0.896), which demonstrates superior performance in balancing positive and negative predictions. It also excels in sensitivity (0.942), specificity (0.954), precision (0.956), and F1 score (0.949). SHAP’s accuracy is 19% higher than PCA, 5.2% higher than Lasso, and 39.9% higher than ICA. PCA closely follows AUC (0.975) and accuracy (0.929), while Lasso achieves an AUC of 0.961 and an accuracy of 0.896, but lags behind SHAP and PCA in several key metrics. ICA performs the worst, with an AUC of 0.554 and an accuracy of 0.549, making it the least effective technique in this comparison. These impressive results make SHAP the most effective feature selection method for our proposed model, outperforming all other approaches in terms of predictive accuracy and overall performance. As a result, SHAP is considered the ideal feature selection strategy for this particular application, ensuring that the model can make the most accurate and reliable predictions. The rest of the 16 experiments based on other feature subsets are provided in the supplementary file (Supplementary Table 2)
SHAP utilization and performance study on traditional ML, DL, and our proposed model
At first, the best feature combination (One Hot Encoding + fastText + ProtBert) without applying any feature selection techniques was evaluated using both traditional ML and DL classifiers to compare their performance. The classifiers chosen for comparison were SVM54, XGBoost55, CNN56, and ResNet5057. Table 6 illustrates the comparison between all the classifiers mentioned.
Among all the models, the GRUATT-AVP model emerged as the top performer in the initial comparison. It achieved the highest AUC (0.986), accuracy (0.937), MCC (0.874), sensitivity (0.935), specificity (0.939), precision (0.942), F1 score (0.939), and AUPR (0.987), surpassing all other models in every metric. GRUATT-AVP’s accuracy is 4.7% higher than SVM, 7.4% higher than XGBoost, and 2.5% higher than CNN. SVM demonstrated a strong AUC (0.960), accuracy (0.890), and a good balance between sensitivity (0.882) and specificity (0.897). XGBoost performed decently with AUC and ACC scores of 0.944 and 0.863, respectively. CNN showed good performance in all metrics, highlighted by Sen (0.908), Spe (0.916), Pre (0.919), and F1-score (0.914). ResNet50 underperformed with the lowest AUC (0.836), accuracy (0.698), and F1 score (0.630), making it the least effective model in this comparison.
Subsequently, we employed SHAP to enhance the performance of our model by identifying the most critical data points from the best feature combination (One Hot Encoding + fastText + ProtBert). This approach, particularly beneficial for large-scale applications, allowed us to address the complexity of hybrid feature spaces and improve model performance by removing irrelevant and redundant information. We selected 200, 300, 400, 500, and 600 feature sets for SHAP, and each classifier was run five times using the different feature sets. The total number of experiments for this section is 25, and the best result for each classifier was chosen from the 200, 300, 400, 500, and 600 feature sets. Table 7 displays the performance evaluation after SHAP.
By focusing on significant features, we aimed to improve the performance and efficiency of classifiers by selecting the most suitable feature subset for each model. Among all the classifiers, our proposed GRUATT-AVP still performed best after SHAP utilization. It showed the highest result when using a subset of 500 features. The GRUATT-AVP model achieved an AUC (0.986), ACC (0.948), MCC (0.896), Sen (0.942), Spe (0.954), Pre (0.956), F1(0.949), and AUPR (0.988). The SVM classifier with a 200-feature subset achieved an excellent result, nearly close to GRUATT-AVP, with an AUC (0.979) and an ACC (0.925). The XGBoost classifier with a 200-feature subset gained an AUC (0.951) and ACC (0.881). The CNN classifier with 200 features also performs well, achieving a balanced AUC (0.925) and ACC (0.925). The ResNet50 classifier remains in the bottom position, with AUC (0.833) and ACC (0.764). Examining the values in Tables 6 and 7, we observe that all classifier results improved after using SHAP. The comparison clearly shows that SHAP improves the classifier’s performance. Figure 5 presents the ROC curve for before SHAP and after SHAP. Figure 6 displays the horizontal bar chart indicating the performance changes of different classifiers on different evaluation metrics before and after SHAP utilization. The remaining 20 experiments based on other feature subsets are provided in the supplementary file (Supplementary Table 3). SHAP analysis highlighted specific residues and short motifs that contributed most strongly to positive predictions, revealing a pronounced enrichment of arginine (R), lysine (K), and tryptophan (W) in high-importance k-mers. These residues are known for their cationic and aromatic properties, which facilitate electrostatic interactions with viral membranes and contribute to antiviral function.
Considering practical scenarios, we evaluated the GRUATT-AVP model on highly imbalanced datasets with ratios of 1:100 and 1:500. Despite the severe imbalance, the model maintained strong performance, demonstrating its robustness and applicability under real-world conditions. Table 8 shows the test results of these imbalanced datasets.
For rare-event settings (1:100 and 1:500), evaluation should center on precision–recall behavior. GRUATT-AVP achieves very high AUPR (0.978, 0.983), indicating that across thresholds it consistently ranks positives above negatives and sustains useful precision without sacrificing recall. At the selected operating point, recall is (0.940, 0.935), precision is (0.927, 0.903), and F1 is (0.933, 0.918), reflecting a balanced trade-off between detecting positives and limiting false positives. Overall, these results demonstrate that GRUATT-AVP is well-suited to imbalanced scenarios, as it preserves a high recall of rare positives, maintains precision at a practical level, and offers a strong PR curves that enables threshold tuning toward recall (screening) or precision (confirmation) without compromising reliability. Figure 7A shows the AUPR curves of GRUATT-AVP on imbalance datasets, and Fig. 7B shows the AUPR curves for different baseline models.
We evaluated pairwise differences on the same five cross-validation folds using paired t-tests and exact Wilcoxon signed-rank tests. Model-wise, t-test p-values were 0.0466 (SVM), 0.0296 (XGBoost), 0.0103 (CNN), 0.0177 (ResNet50), and 0.00181 (GRUATT-AVP), indicating statistically significant mean gains over all baselines. The corresponding Wilcoxon p-values were 0.3125, 0.3125, 0.1875, 0.1250, and 0.0625; with five folds, the two-sided exact Wilcoxon p-value is discrete and cannot be smaller than 0.0625, so only GRUATT-AVP is marginal by that criterion. Metric-wise, t-test p-values were 0.0252 (accuracy), 0.0306 (AUC), 0.00741 (precision), 0.0146 (sensitivity), and 0.00329 (F1), while Wilcoxon p-values were 0.3125, 0.4375, 0.1875, 0.1875, and 0.0625, respectively. Overall, the paired t-tests provide strong evidence of improved mean performance across models and metrics, and the Wilcoxon results are directionally consistent but conservative given the five-fold sample size. Table 9 displays all the values of metric-wise and model-wise comparisons.
Ablation study on our proposed model
In this study, we performed an ablation study, summarized in Table 10, to assess the importance of each component in our proposed model. The ablation study systematically examines the model to determine its effects on total performance.
-
Adding a Second GRU Layer (SGL): This experiment focuses on the impact of adding a new GRU layer to investigate the value of increasing the model’s temporal learning capacity.
-
Removing Both GRU Layers and Keeping One Attention Layer (RBGOA): This setup aims to assess the model’s performance when both GRU layers are omitted, and only one attention layer is implemented to evaluate the efficacy of the attention mechanism.
-
Adding a Second Attention Layer (ASA): This test evaluates the effectiveness of integrating a second attention layer to strengthen the connections between the encoder and decoder sections and increase the model’s attention to the necessary details.
-
Two GRU Layers Without Attention (TGA): This analysis establishes the significance of attention in the presence of two GRU layers, while no attention mechanism is used in the model.
-
One GRU Layer Without Attention (OGA): This assesses the model’s performance when it has only one GRU layer and no attention mechanism to allow a comparison of a single-layer GRU.
-
Removing the Fully Connected Layer (RFCL): Analyzing by excluding the final fully connected layer, vital for final predictions, is assessed to determine its usefulness in the model.
These ablations are crucial for understanding the significance of each component in evaluating the model’s efficacy and reliability. The proposed GRUATT-AVP model outperforms all six variants and yields the best results in all evaluation measures. Through cross-validation, GRUATT-AVP gained the best results. GRUATT-AVP is the top performer, achieving the highest AUC (0.986), ACC (0.948), F1 (0.949), and AUPR (0.988), along with strong results in MCC, Sensitivity, and Precision. ASA and OGA follow closely with AUC values of 0.955 and 0.957, and F1 scores of 0.893 and 0.887, respectively. SGL performs decently with AUC (0.908) and AUPR (0.925). TGA and RFCL show competitive performance, with RFCL slightly outperforming, while RBGGOA lags with the lowest AUC (0.589), MCC (0.160), and AUPR (0.582), indicating it is the weakest model. In conclusion, GRUATT-AVP outperforms the other models across most metrics. GRUATT-AVP achieved 6.7%, 36.8%, 5.5%, 7.1%, 6.2%, and 7.5% higher accuracy than SGL, RBGOA, ASA, TGA, OGA, and RFCL, respectively. This comparison highlights the importance of every component of the proposed model, as the performance difference is substantial and demonstrates that our model is well-constructed. We also examined the time and memory requirements of these models. Table 11 presents the computational costs associated with these models, where we can see that our model, GRUATT-AVP, demonstrated lower memory usage and shorter prediction time.
A web server for GRUATT-AVP is available at https://gruatt-avp.vercel.app/, offering a strong tool for AVP identification from sequence data. Figures 8 and 9 illustrate the workflow of our web application.
Comparison with state-of-the-art classifiers
This section compares our proposed model (GRUATT-AVP) with existing state-of-the-art models developed to identify AVPs. This test comparison was performed using an independent test dataset, which was consistently applied to all state-of-the-art models. Our proposed model has been compared with Deep-AVPiden22, iACVP8, AVPiden6, ENNAVIA17, iAMP-CA2L14, Meta-iAVP9, PreTP-Stack10, and DeepAVP13 . Table 12 displays a comparison with state-of-the-art classifiers.
The proposed model, GRUATT-AVP, stands out as the top performer in all aspects, achieving impressive scores with an accuracy of 0.948, a precision of 0.956, a recall of 0.942, and an F1 score of 0.949. This demonstrates its superior ability to make accurate predictions while balancing precision (minimizing false positives) and recall (minimizing false negatives), which is crucial for many real-world applications. Deep-AVPiden follows closely behind, recording an accuracy of 0.898, precision of 0.902, and recall of 0.900. While it performs well, it still lags behind GRUATT-AVP in overall effectiveness. iACVP has a considerably lower F1 score (0.581) and exhibits a notable weakness in recall (0.465), indicating that while its precision is decent (0.773), it fails to identify positive cases effectively. AVPIden performs even worse, with low accuracy (0.599) and precision (0.572), though it compensates somewhat with a higher recall of 0.737. Models such as Meta-iAVP and DeepAVP demonstrate a generally poor performance across all metrics. The iAMP-CA2L, having a good precision (0.888), has a very low accuracy (0.523) and a relatively weak recall (0.662), leading to an underwhelming F1 score of 0.759. PreTP-Stack and ENNAVIA perform the worst, with both models consistently showing low scores across accuracy, precision, recall, and F1, particularly in recall, which limits their effectiveness. Overall, GRUATT-AVP is the most effective in this comparison. It demonstrates superior predictive accuracy, balance between precision and recall, and an exceptional F1 score, making it the most effective model in this group. This underscores the strength and robustness of GRUATT-AVP in tackling complex tasks compared to other state-of-the-art models.
Conclusions
This paper emphasizes the importance of identifying antiviral peptides as a crucial step in developing novel therapeutic approaches for viral diseases and accelerating the discovery of antiviral drugs. Our proposed model, GRUATT-AVP, is a robust and innovative DL framework designed to predict antiviral peptides. It leverages the power of a hybrid architecture, combining an Attention mechanism with a Gated Recurrent Unit (GRU), to effectively capture the sequential and contextual dependencies within peptide sequences. By incorporating the Attention mechanism, the model focuses on the most relevant parts of peptide sequences, thereby improving prediction accuracy and interpretability. Meanwhile, the GRU component enhances its ability to process long-range dependencies and complex relationships within biological sequences. In GRUATT-AVP, we utilized an optimized combination of NLP-based word embeddings to effectively represent peptide sequences, capturing both semantic and syntactic information within them. To enhance the interpretability of the model and understand how specific features influence its predictions, we employed SHAP, a powerful tool for feature attribution and explanation. SHAP provides a detailed breakdown of feature contributions by calculating the Shapley values, allowing us to pinpoint which features play the most significant roles in the model’s decision-making process. This step ensures transparency and provides valuable insights into the mechanisms underlying the model’s ability, thereby improving the overall model accuracy and increasing trust in the model’s application for antiviral research. A web server for GRUATT-AVP is available at https://gruatt-avp.vercel.app/, offering a strong tool for AVP identification from sequence data.
Despite GRUATT-AVP’s strong performance on our benchmark, it is essential to note that the model was trained on a relatively small, balanced dataset of 5,413 peptides (2,707 AVPs and 2,706 non-AVPs). These sequences were curated from a handful of public databases and de-redundified with CD-HIT, so they do not reflect the diversity and class imbalance found in natural proteomes. Consequently, our current results may not generalize to peptide types not present in the training set, such as sequences from understudied organisms or different viral families. All predictions remain computational and require wet-lab experimental validation to confirm antiviral activity and eliminate false positives. To address this, future work will expand the dataset with more heterogeneous positive and negative examples from additional sources and evaluate the model on highly imbalanced test sets. Future work will explore advanced protein language models to learn richer contextual representations and combine them in hybrid architectures, as well as Explainable AI (XAI) techniques, such as LIME, for improved interpretability. The goal is to enhance AVP prediction through a hybrid deep learning model. Upcoming versions will integrate additional peptide sequence data to refine predictions and advance protein-based analysis, integrating wet-lab facilities.
Data availability
The datasets for this research can be directly accessed from our web application site at: https://gruatt-avp.vercel.app/dataset.
Abbreviations
- AVP:
-
Antiviral peptide
- AMP:
-
Antimicrobial peptides
- GRU:
-
Gated recurrent unit
- RNN:
-
Recurrent neural network
- TCN:
-
Temporal convolutional network
- LSTM:
-
Long short term memory
- CNN:
-
Convolutional neural network
- RF:
-
Random forest
- BiLSTM:
-
Bidirectional long short-term memory
- SVM:
-
Support vector machine
- PCA:
-
Principal component analysis
- LASSO:
-
Least absolute shrinkage and selection operator
- ICA:
-
Independent component analysis
- ROC:
-
Receiver-operating characteristic curve
- NLP:
-
Natural language processing
- W2V:
-
Word2Vec
- GloVe:
-
Global vector
- SHAP:
-
SHapley additive explanations
- ML:
-
Machine learning
- DL:
-
Deep learning
- MCC:
-
Matthew’s correlation coefficient
- PSSM-TS:
-
Tri-segmentation-based position-specific scoring matrix
- CTDT:
-
Composition/transition/distribution-transition
- IG:
-
Information gain
- LDA:
-
Linear discriminant analysis
- CBOW:
-
Continuous bag of words
- SGM:
-
Skip-Gram model
- CD-HIT:
-
Cluster database at high identity with tolerance
References
Rampersad, S., & Tennant, P. Replication and expression strategies of viruses. Viruses 55 (2018).
Mothes, W., Sherer, N. M., Jin, J. & Zhong, P. Virus cell-to-cell transmission. J. Virol. 84(17), 8360–8368 (2010).
Vilas Boas, L. C. P., Campos, M. L., Berlanda, R. L. A., de Carvalho Neves, N. & Franco, O. L. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 76, 3525–3542 (2019).
Ali, F., Kumar, H., Alghamdi, W., Kateb, F. A. & Alarfaj, F. K. Recent advances in machine learning-based models for prediction of antiviral peptides. Arch. Comput. Methods Eng. 30(7), 4033–4044 (2023).
Thakur, N., Qureshi, A. & Kumar, M. AVPpred: collection and prediction of highly effective antiviral peptides. Nucleic Acids Res. 40(W1), W199–W204 (2012).
Pang, Y., Yao, L., Jhong, J. H., Wang, Z. & Lee, T. Y. AVPIden: a new scheme for identification and functional prediction of antiviral peptides based on machine learning approaches. Brief. Bioinform. 22(6), bbab263 (2021).
Qureshi, A., Tandon, H. & Kumar, M. AVP?IC50Pred: multiple machine learning techniques?based prediction of peptide antiviral activity in terms of half maximal inhibitory concentration (IC50). Pept. Sci. 104(6), 753–763 (2015).
Kurata, H., Tsukiyama, S. & Manavalan, B. iACVP: markedly enhanced identification of anti-coronavirus peptides using a dataset-specific word2vec model. Brief. Bioinform. 23(4), bbac265 (2022).
Schaduangrat, N., Nantasenamat, C., Prachayasittikul, V. & Shoombuatong, W. Meta-iAVP: a sequence-based meta-predictor for improving the prediction of antiviral peptides using effective feature representation. Int. J. Mol. Sci. 20(22), 5743 (2019).
Yan, K. et al. PreTP-Stack: prediction of therapeutic peptide based on the stacked ensemble learning. IEEE/ACM Trans. Comput. Biol. Bioinf. 20(2), 1337–1344 (2022).
Chowdhury, A. S., Reehl, S. M., Kehn-Hall, K., Bishop, B. & Webb-Robertson, B. J. M. Better understanding and prediction of antiviral peptides through primary and secondary structure feature importance. Sci. Rep. 10(1), 19260 (2020).
Sharma, R. et al. Deep-AVPpred: Artificial intelligence driven discovery of peptide drugs for viral infections. IEEE J. Biomed. Health Inform. 26(10), 5067–5074 (2021).
Li, J., Pu, Y., Tang, J., Zou, Q. & Guo, F. DeepAVP: a dual-channel deep neural network for identifying variable-length antiviral peptides. IEEE J. Biomed. Health Inform. 24(10), 3012–3019 (2020).
Xiao, X., Shao, Y. T., Cheng, X. & Stamatovic, B. iAMP-CA2L: a new CNN-BiLSTM-SVM classifier based on cellular automata image for identifying antimicrobial peptides and their functional types. Brief. Bioinform. 22(6), bbab209 (2021).
Ullah, M., Akbar, S., Raza, A. & Zou, Q. DeepAVP-TPPred: identification of antiviral peptides using transformed image-based localized descriptors and binary tree growth algorithm. Bioinformatics 40(5), btae305 (2024).
Akbar, S., Raza, A. & Zou, Q. Deepstacked-AVPs: predicting antiviral peptides using tri-segment evolutionary profile and word embedding based multi-perspective features with deep stacking model. BMC Bioinformatics 25(1), 102 (2024).
Timmons, P. B. & Hewage, C. M. ENNAVIA is a novel method which employs neural networks for antiviral and anti-coronavirus activity prediction for therapeutic peptides. Brief. Bioinform. 22(6), bbab258 (2021).
Guan, J. et al. A two-stage computational framework for identifying antiviral peptides and their functional types based on contrastive learning and multi-feature fusion strategy. Brief. Bioinform. 25(3), bbae208 (2024).
Yao, L. et al. Abpcaps: a novel capsule network-based method for the prediction of antibacterial peptides. Appl. Sci. 13(12), 6965 (2023).
Yao, L. et al. dbAMP 3.0: updated resource of antimicrobial activity and structural annotation of peptides in the post-pandemic era. Nucleic Acids Res. 53(D1), D364–D376 (2025).
Akbar, S., Raza, A., Zou, Q., Alghamdi, W., Kang, X., Ali, H. & Luo, X. Accelerating Prediction of Antiviral Peptides Using Genetic Algorithm-Based Weighted Multiperspective Descriptors with Self-Normalized Deep Networks. J. Chem. Inf. Model. (2025).
Singh, V. & Singh, S. K. A separable temporal convolutional networks based deep learning technique for discovering antiviral medicines. Sci. Rep. 13(1), 13722 (2023).
Bai, S., Kolter, J. Z., & Koltun, V. An empirical evaluation of generic convolutional and recurrent networks for sequence modeling. arXiv preprint arXiv:1803.01271 (2018).
Charoenkwan, P., Chumnanpuen, P., Schaduangrat, N. & Shoombuatong, W. Stack-AVP: a stacked ensemble predictor based on multi-view information for fast and accurate discovery of antiviral peptides. J. Mol. Biol. 437(6), 168853 (2025).
Qureshi, A., Thakur, N., Tandon, H. & Kumar, M. AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res. 42(D1), D1147–D1153 (2014).
Qureshi, A., Thakur, N. & Kumar, M. HIPdb: a database of experimentally validated HIV inhibiting peptides. PLoS ONE 8(1), e54908 (2013).
Aguilera-Mendoza, L. et al. Overlap and diversity in antimicrobial peptide databases: compiling a non-redundant set of sequences. Bioinformatics 31(15), 2553–2559 (2015).
Aguilera-Mendoza, L. et al. Graph-based data integration from bioactive peptide databases of pharmaceutical interest: toward an organized collection enabling visual network analysis. Bioinformatics 35(22), 4739–4747 (2019).
Aguilera-Mendoza, L. et al. Automatic construction of molecular similarity networks for visual graph mining in chemical space of bioactive peptides: an unsupervised learning approach. Sci. Rep. 10(1), 18074 (2020).
Kang, X. et al. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 6(1), 148 (2019).
Singh, S. et al. SATPdb: a database of structurally annotated therapeutic peptides. Nucleic Acids Res. 44(D1), D1119–D1126 (2016).
UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res.47(D1), D506–D515 (2019).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23), 3150–3152 (2012).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13), 1658–1659 (2006).
Huang, Y., Niu, B., Gao, Y., Fu, L. & Li, W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26(5), 680–682 (2010).
Jenike, K. M., Campos-Domínguez, L., Boddé, M., Cerca, J., Hodson, C. N., Schatz, M. C. & Jaron, K. S. Guide to k-mer approaches for genomics across the tree of life. arXiv preprint arXiv:2404.01519 (2024).
Yang, K. K., Wu, Z., Bedbrook, C. N. & Arnold, F. H. Learned protein embeddings for machine learning. Bioinformatics 34(15), 2642–2648 (2018).
Pennington, J., Socher, R., & Manning, C. D. Glove: Global vectors for word representation. In Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP), pp. 1532–1543 (2014).
Bojanowski, P., Grave, E., Joulin, A. & Mikolov, T. Enriching word vectors with subword information. Trans. Assoc. Comput. Ling. 5, 135–146 (2017).
Mikolov, T., Sutskever, I., Chen, K., Corrado, G. S. & Dean, J. Distributed representations of words and phrases and their compositionality. Adv. Neural Inf. Process. Syst. 26 (2013).
Elnaggar, A. et al. Prottrans: Toward understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44(10), 7112–7127 (2021).
Devlin, J., Chang, M. W., Lee, K., & Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 conference of the North American chapter of the association for computational linguistics: human language technologies, volume 1 (long and short papers), pp. 4171-4186, (2019).
Lundberg, S. M., & Lee, S. I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 30 (2017).
Luong, M. T., Pham, H., & Manning, C. D. Effective approaches to attention-based neural machine translation. arXiv preprint arXiv:1508.04025 (2015).
Sutskever, I., Vinyals, O., & Le, Q. V. Sequence to sequence learning with neural networks. Adv. Neural Inf. Process. Syst. 27 (2014).
Niu, Z., Zhong, G. & Yu, H. A review on the attention mechanism of deep learning. Neurocomputing 452, 48–62 (2021).
Cho, K., Van Merriënboer, B., Gulcehre, C., Bahdanau, D., Bougares, F., Schwenk, H., & Bengio, Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv preprint arXiv:1406.1078 (2014).
Chung, J., Gulcehre, C., Cho, K., & Bengio, Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv preprint arXiv:1412.3555 (2014).
Rainio, O., Teuho, J. & Klén, R. Evaluation metrics and statistical tests for machine learning. Sci. Rep. 14(1), 6086 (2024).
Feurer, M. & Hutter, F. Hyperparameter optimization 3–33 (Springer International Publishing, 2019).
Abdi, H. & Williams, L. J. Principal component analysis. Wiley interdisciplinary reviews: computational statistics 2(4), 433–459 (2010).
Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Stat Methodol. 58(1), 267–288 (1996).
Hyvärinen, A. & Oja, E. Independent component analysis: algorithms and applications. Neural Netw. 13(4–5), 411–430 (2000).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
Chen, T. & Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, pp. 785–794 (2016).
O’shea, K. & Nash, R. An introduction to convolutional neural networks. arXiv preprint arXiv:1511.08458 (2015).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778 (2016).
Funding
The project is partially supported by Multimedia University (MMU) IR Fund (Project ID MMUI/220041).
Author information
Authors and Affiliations
Contributions
Md. Tarek Aziz: Writing-original draft, Methodology. Abu Sufian Rupok: Formal analysis and Writing. S M Hasan Mahmud: Conceptualization, Supervision, Project administration, Writing -review & editing. Kah Ong Michael Goh: Funding acquisition, review & editing. Md. Faruk Hosen: Formal analysis, Visualization. Watshara Shoombuatong: Draft validation. Dip Nandi: Writing - review & editing, Draft validation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Aziz, M.T., Rupok, A.S., Mahmud, S.M.H. et al. GRUATT-AVP: leveraging a novel attention-based gated recurrent unit to advance the accuracy of antiviral peptide prediction. Sci Rep 15, 42509 (2025). https://doi.org/10.1038/s41598-025-26565-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-26565-1
Keywords
This article is cited by
-
Exploring gallbladder cancer prognosis using machine learning and explainable AI
Discover Computing (2026)