Abstract
Colorectal cancer (CRC) manifests immunotherapy resistance. Therefore, research targeting the CRC immunotherapeutic strategies sheds light on the improvement of clinical treatment. KIAA1429 expression, clinical relevance, and immune correlations were analyzed using TCGA, GEPIA 2.0, and TIMER 2.0 databases. KIAA1429 expression and CD8+T cells infiltration were assessed by immunohistochemistry (IHC) in clinical specimens. Molecular functions were investigated through PPI network construction, along with KEGG and GO analyses. m6A binding sites of PD-L1 were predicted using SRAMP database. Expression correlations between KIAA1429 and PD-L1 were validated through cBioPortal analysis and experimental verification in CRC tissues/cell lines using IHC, Western Blot, and qRT-PCR. In CRC homozygous tumor-bearing mice, KIAA1429 was knockdown using siRNA, with treated with PD1 antibody. Tumor volume and weight of mice were observed. Flow cytometry and immunofluorescence were used to detect CD8+T cells infiltration, while immunohistochemistry was utilized to measure PD-L1 expression. Results: Bioinformatic analysis revealed that KIAA1429 was highly expressed in CRC, associated with poor prognosis and good diagnosis values, which is negatively correlated with CD8+T cells infiltration but positively correlated with PD-L1 expression. In vivo and in vitro experiments verified these results. Besides, knockdown of KIAA1429 in CRC cells significantly reduced PD-L1 mRNA and protein expression. Knockdown of KIAA1429 inhibits tumor growth by increasing CD8+T cell infiltration, decreasing PD-L1 expression and enhancing the efficacy of anti-PD1 immunotherapy in two mouse models. Conclusions: In summary, m6A methyltransferase KIAA1429 suppressed CRC tumor immunity by upregulating PD-L1 expression and reducing CD8+T cells infiltration. Targeting KIAA1429 might improve CRC anti-tumor immunity and enhance the efficacy of CRC immunotherapy.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is a malignant tumor that occurs in the colon or rectum and is one of the most common malignant tumors of the gastrointestinal tract1. According to the latest statistics, CRC ranks as the second most common cancer globally, with its incidence continuing to rise2. CRC patients are usually associated with poor prognosis and illustrated the resistant of various therapeutic strategies, including traditional surgery, radiotherapy, chemotherapy3.
Immune checkpoint inhibitors (ICIs) have emerged as a promising therapeutic strategy targeting CRC4. The most appealing immune checkpoint cascade is the programmed cell death protein-1/programmed death-ligand-1 (PD-1/PD-L1) pathway, which allows the evasion of tumor cells from immune surveillance of immune cells by regulating the binding of PD-L1 to PD-1 between tumor cells and immune cells5. Therefore, the disruption of PD-1/PD-L1 can restore anti-tumor immune activity of immune cells6. However, in most CRC cases, particularly those with proficient mismatch repair (pMMR), the immunotherapy targeting PD-1/PD-L1 is unsatisfied, and the underlying mechanisms remain unclear7. Therefore, it is important to unveiling the mechanism of CRC tumor immunity and to identify novel approaches to enhance immunotherapeutic efficacy.
In recent years, studies have pointed out that N6-methyladenosine(m6A) in tumor cells is associated with immune checkpoint expression, particularly in PD-L1 expression, which provides with novel clues to gain insights into the tumor immune microenvironment of CRC8. Current research has revealed that m6A modification can influence the stability, translation and localization of PD-L1 mRNA, thereby affecting PD-L1 protein expression and the subsequent immune evasion of tumor cells9. However, the specific mechanisms by which m6A regulates PD-L1 expression in CRC are still not fully understood.
KIAA1429 (Vir-like m6A Methyltransferase Associated, also known as VIRMA) is a crucial component of the m6A methyltransferase complex. Emerging evidence has shown that KIAA1429 plays an important role in various cancers, including CRC. In CRC, KIAA1429 has been reported to be involved in tumor progression10. However, the function and underlying mechanisms of KIAA1429 in CRC tumor immunity, especially its relationship with PD-L1 expression and immune cell infiltration, have not been well elucidated.
In this study, we investigated the role of the m6A methyltransferase KIAA1429 (Vir-like m6A Methyltransferase Associated, also known as VIRMA) in CRC tumor immunity, and discovered that KIAA1429 could impairs CRC tumor immunity by inhibiting CD8+T cells infiltration and facilitating PD-L1 expression, which provides a new strategy for improving CRC immunotherapy efficacy.
Results
High expression of KIAA1429 is associated with poor prognosis in CRC patients
We evaluated the expression of KIAA1429(Vir-like m6A Methyltransferase Associated, also known as VIRMA) in pan-cancer by using the TCGA database, which showed that KIAA1429 was significantly highly expressed in dozens of cancers, including colorectal cancer (CRC) (Fig. 1A). Similarly, we also applied TCGA database of CRC to assess the expression of KIAA1429 in CRC tissue samples compares to normal intestinal tissue, and found that KIAA1429 was highly expressed in CRC (Fig. 1B). In addition, we analyzed the expression of KIAA1429 in 367 CRC samples and 51 normal tissue samples using the GEPIA 2 database and similarly found that KIAA1429 was highly expressed in CRC (P < 0.05) (Fig. 1C). Consistent with this, the expression level of KIAA1429 in CRC tissues was also significantly higher than that in paracancerous normal tissues in 53 postoperative CRC tumor tissues and 33 paracancerous normal tissues we collected (P = 2.196e-07) (Fig. 1D).
Next, we further explored the relationship between KIAA1429 and the prognosis of CRC patients. Firstly, the Kaplan-Meier (KM) survival analysis showed a significant survival difference between the high KIAA1429 expression group and the low expression group. Survival was significantly lower in the high KIAA1429 expression group than in the low expression group (P = 0.027), and patients with high KIAA1429 expression had a higher risk of death compared to the low expression group (HR = 1.6, P = 0.028) (Fig. 1E). Next, we show the role of KIAA1429 in a multivariate prognostic column chart. This graph predicted the 1-, 3- and 5-year survival probability of patients based on clinical variables such as T-stage, N-stage, M-stage, age, gender, and KIAA1429 expression level. Patients with high KIAA1429 expression had a higher overall score, which predicted a lower probability of survival (Fig. 1F). To assess the accuracy of KIAA1429 expression in prognostic prediction, we plotted a calibration curve. The graph demonstrated the accuracy of 1-, 3- and 5-year survival prediction (Fig. 1G). Finally, the ROC curve further validates the diagnostic ability of KIAA1429 in predicting survival status. The AUC of 0.783 indicates that KIAA1429 has a good ability to differentiate between surviving and dying patients and performs well in terms of both sensitivity and specificity (Fig. 1H).
In summary, compared with normal tissue, these results indicate that KIAA1429 is highly expressed in CRC and is closely associated with poor patient prognosis. And KIAA1429 demonstrated high predictive accuracy for the survival status of CRC patients, suggesting its potential utility as a prognostic biomarker.
A: Elevated mRNA expression of KIAA1429 in pan-cancer (TCGA database); B: KIAA1429 is highly expressed in CRC tis sues compared to normal tissues (TCGA database, P < 0.001). C: Confirmation of the Figure B results through the GEPIA database(P < 0.05). D: Immunohistochemical staining confirmed higher KIAA1429 expression in CRC tissues compared to adjacent non-tumor tissues (Mann-Whitney U test, P = 2.196e-07); E: High expression of KIAA1429 correlates with poor prognosis (Kaplan-Meier analysis, Log-rank, P = 0.027); F: Nomogram for predicting 1-year survival probability based on clinical factors and KIAA1429 expression. G: Calibration curve assessing nomogram accuracy at 1-year, 3-year, and 5-year time points. H: ROC curve evaluating the diagnostic performance of KIAA1429 expression (AUC = 0.783).
KIAA1429 inhibits CD8+T cells infiltration in CRC patients
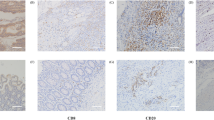
It has been shown that the infiltration of immune cells such as CD8+ T cells is closely related to cancer prognosis. To investigate whether KIAA1429 affects CRC tumor immunity, we first analyzed the infiltration of immune cells associated with the expression of KIAA1429 in CRC through the TCGA database, and preliminarily found that the infiltration of CD8+ T cells, CD4+ memory activated cells, Tregs, and M2 macrophages was associated with KIAA1429 expression (Fig. 2A). Next, we analyzed the infiltration of these four cells individually and found that the infiltration of CD8+T cells and Tregs was negatively correlated with the expression level of KIAA1429(Fig. 2B, C and D). To validate this finding, we assessed the relationship between KIAA1429 expression levels and immune cell infiltration in COAD and READ using the TCGA database. The analysis revealed a negative correlation between KIAA1429 expression and the infiltration of CD8+ T cells and Treg cells in both cancer types (P < 0.05, P < 0.01). (Fig. 2E).
Taken together, the results of the bioinformatics analysis above, we believe that the expression of KIAA1429 in CRC has a significant correlation with the infiltration of CD8+ T cells, so we also detected the infiltration of CD8+ T cells in the collected clinical samples, and the results obtained by Spearman correlation analysis showed that KIAA1429 was negatively correlated with the infiltration of CD8+T cells (P = 0.021) (Fig. 2F), which was consistent with the results of the bioinformatics analysis.
A: Correlation of KIAA1429 with tumor immune cell infiltration in CRC. B: CD8+ T cells and Treg cells are highly infiltrated with low KIAA1429 expression in CRC (TCGA database, P < 0.01, P < 0.001). C: Expression of KIAA1429 is negatively correlated with the level of CD8+ T cells infiltration in CRC (TCGA database, r=−0.015, P = 0.001). D: Expression of KIAA1429 is negatively correlated with the level of Treg cells infiltration in CRC (TCGA database, r=−0.233, P < 0.001). E: KIAA1429 was negatively correlated with the infiltration of CD8+ T cells and Tregs in both COAD and READ(TCGA database, P < 0.05, P < 0.01); F: Immunohistochemistry showed reduced CD8+ T-cell infiltration in CRC tissues compared to adjacent normal tissues (Spearman’s correlation analysis, r = 0.0995, P = 0.021).
m6A methyltransferase KIAA1429 facilitating PD-L1 expression in CRC
To further explore the role played by KIAA1429 in tumor immunity, we understood the function of KIAA1429 by constructing a protein-protein interaction (PPI) network as well as Genes and Genomes (KEGG) and Gene Ontology (GO) functional analyses. Through the KIAA1429 protein interaction network map, we found that KIAA1429 is closely related to a series of proteins involved in mRNA methylation and RNA splicing, suggesting that it may play a similar role (Fig. 3A). Functional enrichment analysis further supports the important role of KIAA1429 in key biological processes such as RNA modification, RNA splicing, and nucleoplasm transfer (Fig. 3B). KIAA1429 may regulate RNA splicing and stability through its role as a part of the methyltransferase complex, thereby affecting gene expression. It is well known that tumor cells can evade immune elimination by expressing PD-L1, which binds to PD-1 on immune cells such as CD8+ T cells. Given that KIAA1429 has a significant effect on CD8+T cells infiltration, we investigate the potential mechanisms by which KIAA1429 affects PD-L1. Firstly, we predicted the m6A binding site GAACU on PD-L1 mRNA by SRAMP database (Fig. 3C). This is a typical RRACH motif (R = purine A/G, H = non-guanine A/C/U) that can be recognized by methylases, suggesting that PD-L1 expression may be supervised in an m6A-dependent manner11. Secondly, we identified a positive correlation between KIAA1429 and PD-L1 in CRC patients through the cBioPortal database (P = 2.275e-3) (Fig. 3D). More importantly, we validated this result in tissue samples from CRC patients (P = 1.017e-07) (Fig. 3E and F).
To investigate the functional role of KIAA1429 in regulating PD-L1 expression, we knocked down KIAA1429 in SW620 cells using siRNA and measured PD-L1 mRNA and protein levels (Fig. 3G and H). The results showed that silencing KIAA1429 reduced PD-L1 mRNA levels compared with the normal group (P < 0.01), and its protein expression was also down-regulated compared with the normal group (P < 0.01) (Fig. 3I and J).
These results indicate that KIAA1429 plays a critical role in modulating PD-L1 mRNA and protein expression, potentially contributing to tumor immune evasion.
A: Protein-protein interaction (PPI) network of KIAA1429 with RNA-related proteins. B: Gene Ontology (GO) enrichment analysis of KIAA1429-related genes. C: SRAMP database predicts a significant m6A binding site on PD-L1 mRNA; D: KIAA1429 shows a positive correlation with PD-L1 expression in CRC tissues(cBioPortal data, P = 2.275e-03); E, F: Immunohistochemistry validated the positive correlation between KIAA1429 and PD-L1 expression in CRC samples (Spearman correlation analysis, P = 1.017e-07); G: Expression levels of KIAA1429 mRNA in various colorectal cancer cell lines, and KIAA1429 is highly expressed in SW620 cells (P < 0.001); H: siRNA-mediated silencing of KIAA1429 (siKIAA1429-1,−2,−3), with siKIAA1429-2 exhibiting the most effective knockdown(P < 0.01); I: Reduced PD-L1 mRNA expression following KIAA1429 knockdown in SW620 cells (P < 0.01); J: Western blot analysis confirmed downregulated PD-L1 protein expression in SW620 cells upon KIAA1429 silencing (P < 0.01).
Knockdown of KIAA1429 inhibits CRC tumorigenesis and promotes Anti-Tumor immunity in homozygous mice
To investigate the role of KIAA1429 in regulating anti-tumor immunity, we knocked down KIAA1429 (siKIAA1429) in MC38, an MSI-H type (Micro Satellite Instability-High) CRC cell line cells, using siRNA and injected the cells into homozygous C57BL/6 mice (Fig. 4A and B). Among them, we selected CT26-si-2 and MC38-si-3 for further experiments after verifying the knockdown efficiency via qPCR and determining that they exhibited better effects. Furthermore, the knockdown efficiency of KIAA1429 was further verified by qPCR in the collected tumor tissues (Supplementary Fig. 4 A). Tumor growth was monitored, and the results demonstrated that knockdown of KIAA1429 significantly reduced both tumor volume (P < 0.001) and weight (P < 0.001) compared to the control group (siNC) (Fig. 4C).
To assess the immune-modulatory effects of KIAA1429, we performed immunohistochemistry to examine PD-L1 expression and flow cytometry to evaluate CD8+T cells infiltration in tumor tissues. The siKIAA1429 group showed a corresponding reduction in PD-L1 expression and a significant increase in CD8+ T cells infiltration (P < 0.01) (Fig. 4E and Fig. 5).
To validate these findings, we extended the study to CT26 cells, an MSS-type (Micro Satellite Stability) CRC cell line, which were similarly subjected to KIAA1429 knockdown and injected into homozygous BALB/c mice (Fig. 4A and B). Consistent with the MC38 model, knockdown of KIAA1429 in CT26 cells resulted in reduced tumor volume and weight (Fig. 4D), increased infiltration of CD8+ T cells, and decreased PD-L1 expression in tumor tissues. Immunofluorescence staining further confirmed the enhanced CD8+ T cells infiltration in MC38 and CT26 tumors following KIAA1429 knockdown (Fig. 4E and Fig. 5).
These results were consistent with the results of biosignature analysis and pathological tissue testing of CRC patients, demonstrating that knockdown of KIAA1429 inhibits tumorigenesis and promotes anti-tumor immunity by enhancing CD8+T cells infiltration and reducing PD-L1 expression.
Knockdown of KIAA1429 enhances Anti-Tumor immunity in homozygous mice
Considering that increased CD8+ T infiltration has been reported to be associated with enhanced immunotherapeutic efficacy in different types of cancers, we investigated whether targeting KIAA1429 could enhance the effectiveness of anti-PD1 therapy in CRC. Using the MC38 tumor model, we treated homozygous mice with siKIAA1429 or siNC alongside anti-PD1 therapy once tumors reached a size of 50–100 mm³ (Fig. 4A and B). Compared with siNC, siKIAA1429 significantly inhibited the growth of MC38 tumors. Furthermore, the combination of siKIAA1429 and anti-PD1 exhibited the most pronounced tumor suppression, indicating a synergistic effect (Fig. 4C and D).
Based on the CT26 (MSS CRC) tumor model, we further explored whether targeting KIAA1429 could overcome anti-PD1 resistance in MSS CRC. Therefore, CT26 cells with KIAA1429 knocking down were injected into homozygous mice, followed by anti-PD1 treatment (Fig. 4A and B). Remarkably, knockdown of KIAA1429 enhanced the efficacy of anti-PD1 therapy in CT26 tumors, overcoming the typical resistance associated with MSS CRC (Fig. 4D).
The combination of KIAA1429 knockdown and anti-PD1 reduced the PD-L1 expression in CRC tumor tissues (Figs. 4E and 5A). In addition, flow cytometry analysis revealed that the combined therapy significantly increased CD8+T cells infiltration in both MC38 and CT26 homozygous models, as verified by immunofluorescence staining (Figs. 4E and 5A).
These findings suggest that targeting KIAA1429 not only enhances the efficacy of ICIs in MSI-H CRC, but also overcomes the resistance to ICIs in MSS CRC by increasing CD8+ T cells infiltration and inhibiting PD-L1 expression.
A: KIAA1429 was knocked down in MC38 and CT26 cells, and the knockdown efficiency was detected by qPCR. B: Representative tumors images in MC38 and CT26 homozygous mice; C: Body weight(left), tumor volume(middle), and tumor weight(right) in MC38 mice(n = 5); D: Body weight(left), tumor volume(middle), and tumor weight(right) in CT26 mice(n = 5); E: Immunohistochemical staining measures the expression of PD-L1 across different groups.
A: The results of immunohistochemical staining indicated that the expression of PD-L1 in the siKIAA1429 group was lower than that in the blank control group (siNC), and it was even lower in the combined treatment group (ANOVA). B: Flow cytometry detects CD8+T cells infiltration in tumor tissues of different subgroups of MC38 and CT26 homozygous mice; C: Immunofluorescence staining measures CD8+T cells infiltration in tumor tissues of different subgroups of MC38 and CT26 homozygous mice.
Discussion
KIAA1429 was first reported in 2014 as an indispensable molecule in the process of m6A modification12. KIAA1429 is a crucial member of the m6A methyltransferase complex, whose C-terminus is anchored to the nuclear speck by binding components such as WTAP and CBLL1, while the N-terminus recruits the METTL3/METTL14/WTAP catalytic core to mediate m6A modifications in the vicinity of the mRNA 3’UTR and termination codon13,14. Thus, KIAA1429 could modulate site-selectivity in RNA metabolism15. A study revealed that knockdown of KIAA1429 resulted in a decrease in the level of cellular m6A, which was more pronounced than knockdown of METTL3 or METTL14, indicating an important role of KIAA1429 in m6A modification12. Currently, it has been shown that KIAA1429 is commonly overexpressed in various cancers such as lung adenocarcinoma, gastric cancer, hepatocellular carcinoma, and is closely related to the poor prognosis of cancer16,17,18. In our study, we have noted that the m6A methyltransferase, KIAA1429, was highly expressed and correlated with poor prognosis in CRC by bioinformatics. These findings were validated in our collected pathological samples, where we observed high KIAA1429 expression levels consistent with the results of ZHOU et al., who similarly reported elevated KIAA1429 expression in CRC19.
In addition, anti-tumor immune function of the organism is critically dependent on the killing effect of immune cells on tumor cells20. Cytotoxic T lymphocytes (CTLs), also known as CD8+ T cells, are a key component of the adaptive immune response and play a major role in the immune defense against intracellular pathogens such as tumors21. Besides, Tumor cells can regulate the expression of immune checkpoints to evade the elimination of immune cells, thus weakening the body’s anti-tumor immunity22. Recent studies have shown that aberrant expression of m6A regulators has impaired anti-tumor immunity23. For instance, m6A methyltransferase METTL3 has been shown to induce immune escape of tumor cells by inhibiting immune cell infiltration such as CD8+T cells and enhancing PD-L1 expression in oral squamous cell carcinoma and non-small cell lung cancer, and targeting it can increase the effect of anti-PD-1 immunotherapy24,25. In addition, knockdown of the m6A methylated reading protein IGF2BP1 not only recruits the infiltration of immune cells in the immune microenvironment of hepatocellular carcinoma, but also down-regulates the expression of PD-L1, thus slowing down the progression of hepatocellular carcinoma26. Our findings analyzed by bioinformatics showed a significant correlation between KIAA1429 and PD-L1 and CD8+ T cells infiltration in CRC. Based on these results, our experiments in CRC validated for the first time that KIAA1429 correlates with PD-L1, CD8+ T cells infiltration and initially explored its regulatory mechanisms.
In our research, we firstly demonstrated that knockdown of KIAA1429 in SW620 cells reduced PD-L1 protein levels. This highlights KIAA1429’s role in suppressing CRC tumor immunity. Interestingly, our study found that the level of PD-L1 mRNA also decreased with the knockdown of KIAA1429. Based on the biological functions of KIAA1429 identified through bioinformatic analyses, which are primarily associated with m6A modification and RNA splicing, two potential explanations for this phenomenon can be proposed. First, according to the current study, some m6A regulators including KIAA1429 can execute function in m6A-dependent and m6A-independent manners19,27. For example, a study found that KIAA1429 regulates cell cycle protein-dependent kinase 1 (CDK1) expression in a m6A-independent manner in breast cancer28. Similarly, KIAA1429 might directly bind to and act as a splicer of PD-L1 pre-mRNA (Heterogeneous Nuclear RNA, hnRNA) to regulate the transcriptional level of PD-L1 mRNA in CRC, while the m6A binding site on PD-L1 may be other m6A regulators involved in binding. Secondly, m6A modification occurs almost throughout the entire process of mRNA generation and metabolism. m6A regulators have been shown to be involved in the regulation of PD-L1 mRNA stability29. The m6A methylation transferase METTL16 is able to bind and methylate the mRNA of MAT2A and rapidly dissociate to retain the intronic isoform, which is subsequently recognized and degraded by the methylation reading protein YTHDC130. Similarly, we hypothesized that KIAA1429 acts as an m6A methylation transferase regulating the level of m6A modification of PD-L1 mRNA by binding to the m6A site, thereby regulating mRNA splicing, which ultimately affects the expression of PD-L1 mRNA. Overall, our study indicates that KIAA1429 may promote immune evasion by regulating the expression of PD-L1 and CD8+ T cells infiltration. However, the precise molecular mechanisms underlying these effects warrant further investigation.
Numerous studies have explored targeting m6A regulators as a therapeutic strategy to enhance the efficacy of immunotherapy31. Targeted m6A regulators in tumor-bearing mice was found to improve the immune microenvironment of tumors and significantly enhance the efficacy of immunotherapy. For example, STM2457, an inhibitor of the m6A methyltransferase METTL3, has been shown to improve the immune microenvironment and significantly enhance anti-PD1 efficacy in CRC models32. Our results suggest that knockdown of KIAA1429 represents a promising strategy to improve CRC immunotherapy. In vivo experiments demonstrated that knockdown of KIAA1429 inhibited tumor growth, increased CD8+ T cells infiltration, and reduced PD-L1 expression in CRC-bearing mice. These findings are consistent with the bioinformatics analysis and pathological findings in clinical samples, further confirming the role of KIAA1429 in regulating tumor immunity for the first time.
Clinically, anti-PD1 therapy has shown variable efficacy in CRC patients. Despite its effectiveness, more than 50% of patients with MSI-H/dMMR CRC exhibit primary or secondary resistance, while MSS/pMMR CRC patients rarely respond to this treatment33,34. This resistance is often attributed to the formation of an immunosuppressive tumor microenvironment35. Given that knockdown of KIAA1429 in CRC downregulated PD-L1 expression and increased CD8+ T cells infiltration, we further explored the therapeutic significance of targeting KIAA1429 in CRC models constructed from MC38 (MSI-H) and CT26 (MSS) cells. By knockdown of KIAA1429 in MC38 (MSI-H) model and administering anti-PD1 treatment, we observed that PD-L1 expression levels was down-regulated and CD8+ T cells infiltration was increased in both anti-PD1 and combination therapy groups. In addition, the combination therapy showed better tumor growth inhibition than PD1 treatment alone. This is consistent with the fact that MSI-H CRC has a better immune response as a “hot tumor” in clinical practice. Meanwhile, the result also demonstrated that knockdown of KIAA1429 could enhance the immunotherapeutic effect of CRC. In the CT26(MSS) mouse model, we detected increased CD8+T cells infiltration and decreased PD-L1 expression in both anti-PD1 treatment group and combination treatment group. However, limited tumor shrinkage was observed in the anti-PD1 treatment group alone compared with the control group, which is also consistent with the clinical observation that MSS-type CRC patients show little response to ICIs. Meanwhile, the combination therapy group exhibited significant tumor suppression. Combined with the experimental results of Fang W and Bao Y et al. who also constructed a mouse subcutaneous tumor model using CT26, the mouse CRC subcutaneous tumor model did have a certain response to anti-PD1 treatment35,36. We hypothesized that although anti-PD1 monotherapy induced higher CD8+T cells and lower expression of PD-L1, its weak effects may be masked by the persistence of an immunosuppressive microenvironment. KIAA1429 is involved in this process by regulating PD-L1 expression and CD8+ T cell infiltration, so knockdown of KIAA1429 enhanced anti-PD1 immunotherapy effects in our study. In summary, these results indicate that knockdown of KIAA1429 not only enhances the immunotherapeutic efficacy of MSI-H CRC, but also breaks through the immunosuppression of MSS CRC, emphasizing the potential of KIAA1429 in improving the immunotherapeutic outcome of CRC.
In conclusion, our study demonstrated that KIAA1429 affects CRC prognosis by regulating PD-L1 expression and CD8+ T cells infiltration. Targeting KIAA1429 could improve the efficacy of CRC immunotherapy and helped overcome immunotherapeutic resistance in MSS CRC. However, there are also some limitations. For example, humanized mouse model of the immune system was not used to further validate the results of the experiments, and multicenter clinical samples were not collected. In addition, this study also exhibits certain methodological limitations. When assessing PD-L1 expression, we relied on immunohistochemistry (IHC) for quantification; however, the robustness of IHC-based quantification is less optimal than that of flow cytometry. Additionally, although we quantitatively analyzed the infiltration of CD8+ T cells in tumor tissues via immunofluorescence, we have not yet performed absolute quantification using flow cytometry with counting beads. In future research, beyond developing animal models that more closely mimic human tumor immunity and acquiring multi-institutional samples, we plan to further validate our findings by quantifying markers including PD-L1 and CD8+T cells through flow cytometry. Overall, these findings position KIAA1429 as a promising therapeutic target for CRC, offering new strategies for enhancing anti-tumor immunity and improving patient outcomes.
Materials and methods
Integrated analysis of KIAA1429 expression patterns, clinical significance, and function by databases
KIAA1429 expression profiles in CRC, and pan-CRC cohorts were systematically analyzed using TCGA transcriptomic data and GEPIA 2.0 validation. Visualization was performed through ggplot2, stats, and car packages in R (v4.2.1). PD-L1 co-expression were assessed via cBioPortal database.
Clinical prognostic evaluation incorporated Kaplan-Meier survival curves (GEPIA 2.0) and Cox regression-based nomogram construction (survival/rms packages), integrating pathological staging (TNM), age, gender, and KIAA1429(Also Known As VIRMA) expression levels. Diagnostic performance was quantified through ROC curve analysis (pROC package).
Functional characterization involved: (1) PPI network construction using STRING database (Cytoscape v3.7.1); (2) Pathway enrichment analysis via KEGG/GO annotations (MSigDB reference genes, FDR < 0.05 threshold)37,38,39.
Multi-dimensional immune infiltration profiling through CIBERSORT algorithm implementation (ggalluvial/ggplot2 visualization modules). Initially, we performed a preliminary screening by investigating the correlation between molecules and immune cell infiltration. We then delved deeper into the relationship between molecular expression levels and immune cell infiltration to elucidate the mechanistic link between KIAA1429 expression and immune cell dynamics. Furthermore, we validated the aforementioned results in COAD and READ via the TCGA database.
Methodological unification included standardized R package workflows (v4.2.1) for statistical computing and data visualization across all analytical phases.
Cell lines preparation
The human CRC-derived cell lines SW620, HCT8, HCT116, HT29, and the murine-derived CRC cell lines MC38 and CT26 were obtained from Shanghai Wen Ye Biotechnology Co. (Catalog numbers: MXC370, MXC153, MXC151, MXC183, MXC1130, MXC112). The cells were cultured in RPMI 1640 medium (10–040-CV, Corning) or DMEM medium (10–013-CV, Corning) with 10% fetal bovine serum (Sh30084.03, Hyclone) and 1% penicillin/streptomycin (C0222, Beyotime). Cultures were maintained at 37 °C in a humidified incubator with 5% CO2. These conditions ensured optimal cell viability and proliferation throughout the study.
Clinical samples acquisition
A total of 53 postoperative tumor tissue samples and 33 adjacent normal tissue samples were collected from CRC patients who underwent radical surgery at Taizhou People’s Hospital, affiliated with Nanjing Medical University, between 2020 and 2022. Clinicopathological data were gathered for all participants. (Patients inclusion criteria: Patients diagnosed with CRC through histopathological examination, Patients who underwent radical surgery for CRC at the hospital and No restrictions on age or gender. Patients exclusion criteria: Patients who had received prior anti-tumor treatments before surgery, Incomplete clinical records, Concurrent diagnosis of other malignancies, Presence of other severe systemic diseases.
The tissue specimens involved in this study were obtained from the Department of Pathology, Taizhou People’s Hospital, Nanjing Medical University. Clinical staging was referred to the American Joint Committee of Cancer (AJCC)/Union for International Cancer Control (UICC) TNM staging criteria for colorectal cancer (8th edition, 2017). The study was approved by the Ethics Committee of the Affiliated Taizhou People’s Hospital of Nanjing Medical University. Informed consent was waived by the Ethics Committee of the Affiliated Taizhou People’s Hospital of Nanjing Medical University (Approval Number: KY-2023-152-01), and all methods were carried out in accordance with relevant guidelines and regulations.
Animals
4–6 weeks of BALB/c, C57BL/6 female mice each 20 mice, each weighing 18 ± 2 g, each kind of mice were randomly divided into 4 groups, each group of 5 mice, The mice were housed under specific pathogen-free (SPF) conditions in a temperature-controlled environment at 25 °C, with a 12-hour light-dark cycle and ad libitum access to food and water. Mice subcutaneously injected CT26 and MC38 cells with or without KIAA1429 knockdown by siRNA. And the silencing effect was ensured by regular and quantitative intratumoral injection of the corresponding siRNA. Specifically, Injection of the corresponding siRNA was initiated at the same site (subcutaneously/intratumorally) 2 to 3 days after cell inoculation into the mice, with a frequency of twice a week. When the subcutaneous tumors grew to a size of 50–100 mm³, a group of mice of each species was randomly selected to receive intraperitoneal injections of anti-PD1(BioXCell, BE0146) twice a week. The specific groups and treatments are detailed in Table 1. Treatments were administered for 2–3 weeks. Tumor size and mouse body weight were recorded every three days, and tumor growth curves were plotted. At the end of treatment or when the tumor volume of mice ≥ 2000 mm3 the mice were executed by cervical dislocation method under Isoflurane (R510-22-10, RWD Life Science) inhalation anesthesia, and the subcutaneous tumor tissues were stripped and removed, and the weights were measured and photographed for recording.
The methods used to create animal models and dosage of drugs used refer to previous literature40,41. The animal study protocol was approved by the Animal Ethics Committee of the Collaborating Laboratory of Taizhou People’s Hospital affiliated with Nanjing Medical University (Approval Number: HJSW-24120101). All methods were conducted according to relevant guidelines and regulations which are reported under the ARRIVE guidelines.
Immunohistochemistry
Paraffin-embedded tissue specimens were sectioned into 4 μm slices. The water bath temperature was maintained at 45–50 °C, followed by natural flattening and baking in a thermostatic oven at 40 °C for 0.5–2 h. Deparaffinization and rehydration were carried out using 100% xylene and gradient ethanol. Endogenous peroxidase activity was sealed with 0.3% hydrogen peroxide for 15 min, followed by antigen repair. After blocking non-specific binding, the membrane was sealed with 5% Bovine Serum Albumin (BSA) for 30 min. Afterwards, primary antibodies, including KIAA1429 (25712-1-AP, Proteintech), PD-L1 (28076-1-AP, Proteintech), and CD8 (66868-1-IG, Proteintech), were applied and incubated overnight at 4 °C. After overnight incubation at 4 °C, the secondary antibodies were incubated for 30 min at room temperature. Immunostaining was visualized using the Envision System with diaminobenzidine as the chromogen. The result of CD8+T cell was evaluated by selecting three microscopic fields with the highest lymphocyte positivity. Quantitative analysis using Image Pro Plus 6.0 software determined the positive cell count, which was converted to cell density. The average density from the three fields was used for statistical evaluation. The result of KIAA1429 and PD-L1 was performed as follows: For each tissue section, three distinct microscopic fields were selected. For PD-L1, these fields were specifically chosen from areas showing positive expression. Using Image Pro Plus 6.0 software, the positive area and mean optical density were measured for each selected field. The integral optical density (IOD) was calculated by multiplying these two parameters, and the average IOD across the three fields was determined. This average served as the statistical indicator. Based on the median value of this indicator, samples were categorized into high - and low - expression groups.
Western blot
Add the sample buffer to the protein sample, adjust the final concentration of total protein to 1 mg/ml, boil the sample solution in boiling water at 100℃ for 5 min, put it on ice and centrifuge it at 3000 rpm/min for 1 min. 20 µl of sample solution was added to each well, and 10 µl of pre-stained marker was added to each well. 70v constant voltage electrophoresis was used for about 30 min, and the indicator bromophenol blue entered into the separator gel after it was changed to 90v constant voltage electrophoresis. Switch to 90v constant voltage electrophoresis, turn off the power when the indicator reaches about 0.5 cm from the lower end of the gel, and remove the gel plate. Configure the membrane transfer solution using Glycine (K41808066121, GE), Tris (2A210306310, Geneview) and methanol (10014118, Sinopharm Chemical Reagent Co.). After transferring the proteins in the gel to the PVDF membrane (ISEQ00010, Millipore) by the membrane transfer solution, the membrane was placed in 5% BSA closed at room temperature for 2 h, and then placed in the configured primary antibody solution β-actin (D191047, BBI), and at 4 °C overnight. The membrane was incubated with secondary antibody (D110087, BBI) for 2 h at room temperature. Protein bands were visualized with an enhanced chemiluminescence system. Antibodies were eluted using antibody stripping solution (SB-WB007z, share-bio) and then closed again for 2 h. Re-incubate the primary antibody PD-L1 (17952-1-AP, Proteintech), and secondary antibody (D110058, BBI) sequentially. Protein bands were visualized with an enhanced chemiluminescence system and quantified using Image J software, and each sample was tested 3 times.
qRT-PCR
Total RNA was extracted using Trizol reagent (9108, TAKARA), and the RNA content was detected using a spectrophotometer (NanoDrop 2000, Thermo Scientific). RNA was reverse transcribed into cDNA by reverse transcription kit, and then quantitative polymerase chain reaction was performed by SYBR Premix Ex Taq (20ul reaction system configuration: SYBR Premix Ex Taq 10 µl, cDNA 2 µl, forward primer (10 µM) 1 µl, reverse primer (10µM) 1 µl, and 6 µl of sterilized distilled water). Program steps: pre-denaturation: 95 ℃, 5 min, 1 cycle; PCR reaction: 95 ℃ 15 s, 60 ℃ 30 s, 40 cycles; extension reaction: 60 ℃ 2 min, 1 cycle). The relative mRNA levels were calculated using the 2−ΔΔCt method with GAPDH as the internal reference gene, and each sample was tested 3 times. Primer sequences are available at Table 2.
Flow cytometry
Tumor tissues were cut into fine pieces with ophthalmic surgical scissors. Add collagenase to digest at 37℃ for 10–30 min, remove the tissue pieces with a 200-mesh sieve, and collect the eluate. Add 1 ml of erythrocyte lysate separately, incubate at 4 °C for 10 min, centrifuge, 1200 rpm for 5 min. Discard the supernatant and shake. Add 2 ml of PBS wash solution, shake, centrifuge, 1200 rpm, 5 min. Add the corresponding antibody to each tube (CD3-FITC: 100204; Biolegend, CD4-PECY7: 100528; Biolegend, CD8-APC: 100712; Biolegend, CD45-pecy5: 103110; Biolegend) 5ul/each. Shake each tube after addition of antibody and keep away from light for 20 min at room temperature. Centrifuge and discard the supernatant, and shake. PBS 500 µl was added and the cell content was detected by flow cytometry (EXFLOW206; DAKEWE). Data analysis was performed with Flow Jo software (Version 10.8.1).
Immunofluorescence
Tumor tissues were dehydrated using gradient ethanol, embedded in paraffin, and sectioned into 4 μm slices. Sections were flattened in a 45–50 °C water bath, dried at room temperature, and baked at 40 °C for 0.5–2 h. The slices were deparaffinized using xylene and gradient alcohol and then rehydrated. After antigen repair using citric acid (PH6.0) antigen repair solution (G1202, Servicebio) the sections were closed with 5% BSA (G5001, Servicebio)/0.01mPBS (G0002, Servicebio) for 30 min. Primary antibodies, including CD3(17617-1-AP, Proteintech), CD8(66868-1-IG, Proteintech) were placed in the refrigerator at 4 °C overnight. and the next day it was left at room temperature for 15 min to rewarm, and fluorescent secondary antibody was added dropwise (FITC, D110061- 0100, BBI; CY3: D110088-0100, BBI), and incubate at room temperature for 30 min avoiding light. PBS washed for 5 min×5 times, PBS outside the specimen was wiped off with filter paper, and incubated for 2 min avoiding light by adding drops of DAPI. The specimen was washed with PBS for 1 min×3 times to remove the DAPI. Finally, the plate was sealed with glycerol and observed under the microscope immediately.
Statistical methods
The statistical analyses of the results generated by online interactive web servers using public databases [GEPIA2 database (http://gepia2.cancer-pku.cn), TIMER2.0 database (http://timer.cistrome.org), cBioPortal database (https://www.cbioportal.org), and SRAMP database (http://www.cuilab.cn/sramp)]. The results generated were calculated automatically. These results were considered statistically significant at *P < 0.05, **P < 0.01, and ***P < 0.001. The rest of the experimental results were plotted using Graphpad Prism 10 software for data analysis. The chi-square test was used for comparison of the count data and was considered statistically significant at P < 0.05. T-test and two-way analysis of variance (ANOVA) were used for comparison of measurement data, and P < 0.05 was considered statistically significant, and bar charts and scatter plots were plotted. Correlation analysis was performed using Spearman’s correlation analysis, and scatter plots were drawn.
Data availability
The datasets used and analysed during the current study available from the corresponding author on reasonable request.
References
Baidoun, F. et al. Colorectal cancer epidemiology: recent trends and impact on outcomes. Curr. Drug Targets[J]. 22 (9), 998–1009 (2021).
Filho, A. M. et al. The GLOBOCAN 2022 cancer estimates: Data sources, methods, and a snapshot of the cancer burden worldwide. Int. J. Cancer[J] 156(7), 1336–1346 (2024).
Ohishi, T. et al. Current Targeted Therapy for Metastatic Colorectal Cancer. Int. J. Mol. Sci[J] 24(2), 1702 (2023).
Yan, S. et al. Immune checkpoint inhibitors in colorectal cancer: limitation and challenges. Front. Immunol[J]. 15, 1403533 (2024).
Ghosh, C., Luong, G. & Sun, Y. A snapshot of the PD-1/PD-L1 pathway. J. Cancer[J]. 12 (9), 2735–2746 (2021).
Chamoto, K. et al. Insights from a 30-year journey: function, regulation and therapeutic modulation of PD1. Nat. Rev. Immunol[J]. 23 (10), 682–695 (2023).
Diaz, L. A. Jr. et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol[J]. 23 (5), 659–670 (2022).
An, Y. & Duan, H. The role of m6A RNA methylation in cancer metabolism. Mol. Cancer[J]. 21 (1), 14 (2022).
Zhou, X. et al. Targeting RNA N6-methyladenosine to synergize with immune checkpoint therapy. Mol. Cancer[J]. 22 (1), 36 (2023).
Zhu, W. et al. Role of m6A methyltransferase component VIRMA in multiple human cancers (Review). Cancer Cell. Int[J]. 21 (1), 172 (2021).
Wang, Y., Jin, P. & Wang, X. (6)-methyladenosine regulator YTHDF1 represses the CD8 + T cell-mediated antitumor immunity and ferroptosis in prostate cancer via m(6)A/PD-L1 manner. Apoptosis[J] 29 (1–2), 142–153 (2024).
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell. Rep[J]. 8 (1), 284–296 (2014).
Knuckles, P. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev[J]. 32 (5-6), 415–429 (2018).
Wen, J. et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell Self-Renewal. Mol. Cell[J]. 69 (6), 1028–1038e6 (2018).
Yue, Y. et al. VIRMA mediates Preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell. Discov[J]. 4, 10 (2018).
Guo, L. et al. Comprehensive analysis of the prognostic impact and immune implication of KIAA1429 in lung adenocarcinoma. Cancer Innov[J]. 1 (4), 328–343 (2022).
Miao, R. et al. KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer. J. Cell. Physiol[J]. 235 (10), 7420–7432 (2020).
Lan, T. et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer[J]. 18 (1), 186 (2019).
Zhou, Y. et al. m(6)A methyltransferase KIAA1429 acts as an oncogenic factor in colorectal cancer by regulating SIRT1 in an m(6)A-dependent manner. Cell. Death Discov[J]. 8 (1), 83 (2022).
Fu, C. & Jiang, A. Dendritic cells and CD8 T cell immunity in tumor microenvironment. Front. Immunol[J]. 9, 3059 (2018).
Hu, Y. et al. Predictive value of tumor-infiltrating lymphocytes detected by flow cytometry in colorectal cancer. Int. Immunopharmacol[J] 113(Pt A), 109286 (2022).
Li, L. et al. Dendrobine Suppresses Tumor Growth by Regulating the PD-1/PD-L1 Checkpoint Pathway in Lung Cancer (Curr Cancer Drug Targets[J], 2024).
Ma, C. et al. Pan-cancer analysis and experimental validation revealed the m6A methyltransferase KIAA1429 as a potential biomarker for diagnosis, prognosis, and immunotherapy. Aging (Albany NY)[J]. 15 (17), 8664–8691 (2023).
Ai, Y. et al. METTL3 Intensifies the Progress of Oral Squamous Cell Carcinoma via Modulating the m6A Amount of PRMT5 and PD-L1. J Immunol Res[J], 2021: p. 6149558. (2021).
Liu, Z. et al. N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol. Cancer[J]. 20 (1), 105 (2021).
Liu, Y. et al. Allosteric regulation of IGF2BP1 as a novel strategy for the activation of tumor immune microenvironment. ACS Cent. Sci[J]. 8 (8), 1102–1115 (2022).
Ma, L. et al. KIAA1429 is a potential prognostic marker in colorectal cancer by promoting the proliferation via downregulating WEE1 expression in an m6A-independent manner. Oncogene[J] 41 (5), 692–703 (2022).
Qian, J. Y. et al. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene[J] 38 (33), 6123–6141 (2019).
Wan, W. et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol. Cancer[J]. 21 (1), 60 (2022).
Shima, H. et al. S-Adenosylmethionine synthesis is regulated by selective N(6)-Adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell. Rep[J]. 21 (12), 3354–3363 (2017).
Pan, J. et al. Roles and therapeutic implications of m6A modification in cancer immunotherapy. Front. Immunol[J]. 14, 1132601 (2023).
Chen, H. et al. METTL3 inhibits antitumor immunity by targeting m(6)A-BHLHE41-CXCL1/CXCR2 axis to promote colorectal cancer. Gastroenterology[J] 163 (4), 891–907 (2022).
Taieb, J. et al. Deficient mismatch repair/microsatellite unstable colorectal cancer: Diagnosis, prognosis and treatment. Eur. J. Cancer[J]. 175, 136–157 (2022).
Fan, A. et al. Immunotherapy in colorectal cancer: current achievements and future perspective. Int. J. Biol. Sci[J]. 17 (14), 3837–3849 (2021).
Bao, Y. et al. Targeting m(6)A reader YTHDF1 augments antitumour immunity and boosts anti-PD-1 efficacy in colorectal cancer. Gut[J] 72 (8), 1497–1509 (2023).
Wang, F. et al. Targeting VCP potentiates immune checkpoint therapy for colorectal cancer. Cell. Rep[J]. 42 (11), 113318 (2023).
Kanehisa, M. et al. KEGG: biological systems database as a model of the real world. Nucleic Acids Res[J]. 53 (D1), D672–d677 (2025).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci[J]. 28 (11), 1947–1951 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res[J]. 28 (1), 27–30 (2000).
Lin, J. F. et al. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal. Transduct. Target. Ther[J]. 7 (1), 54 (2022).
Song, F. et al. Anlotinib potentiates anti-PD1 immunotherapy via transferrin receptor-dependent CD8(+) T-cell infiltration in hepatocellular carcinoma. Clin. Transl Med[J]. 14 (8), e1738 (2024).
Author information
Authors and Affiliations
Contributions
YD, YL, MW, and ML discussed the research question, derived the hypotheses and designed the study.YW and ML directed the study.YD, YL, MW, and YW Collected clinical specimens, performs tissue, cell and animal studies, and analyzes data.YD, YL, MW, and ML discussed the results,The first draft of the manuscript was written by YD, YL, and MW. All authors contributed and agreed to the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Du, Y., Liu, Y., Wang, M. et al. KIAA1429 impairs anti-tumor immunity via regulation of PD-L1 and CD8+ T cells in colorectal cancer. Sci Rep 15, 43619 (2025). https://doi.org/10.1038/s41598-025-27586-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-27586-6