Abstract
Tie1, an orphan receptor, is a receptor tyrosine kinase that is expressed in endothelial cells. It can form a polymer with Tie2, thereby regulating the Ang/Tie2 signaling pathway, which is crucial for angiogenesis and plays a significant role in tumor progression. However, the specific role of Tie1, particularly in tumor processes, remains poorly understood. In this study, we investigated the functional effects of Tie1 knockdown in cervical cancer both in vitro and in vivo. We used CCK-8, wound healing, and Transwell assays to evaluate cervical cancer cell proliferation and migration in vitro. Additionally, we established subcutaneous xenograft tumor and lung metastasis mouse models to examine tumor growth and metastasis. The impact of Tie1 knockdown cervical cancer cell-conditioned medium on human umbilical vein endothelial cell (HUVEC) angiogenesis was assessed using an angiogenesis assay. Tie1 knockdown inhibited activation of the Tie2/PI3K/Akt signaling axis and weakened the migration and invasion abilities of cervical cancer cells in vitro and in vivo. Addition of Ang1 partially reversed the effects of Tie1 knockdown. Knockdown of Tie1 also reduces CD31 protein expression in vitro and in vivo. Tie1 derived from cervical cancer cells exerts an oncogenic role by promoting progression through activation of the Ang1/Tie2/PI3K/Akt signaling axis. These findings may provide new biomarkers and identify potential therapeutic targets for cervical cancer.
Similar content being viewed by others
Introduction
Cervical cancer (CC) is a significant gynecological health issue and is ranked as the fourth most prevalent tumor in women1. Approximately 600,000 women worldwide are diagnosed with cervical cancer each year, and more than 300,000 deaths have been attributed to this disease. Most CC cases are associated with high-risk human papillomavirus (HPV) infections2. Cervical cancer is the second most commonly diagnosed and fatal cancer in women in developing countries. Patients with early or locally advanced disease usually have a favorable prognosis. However, the five-year survival rate for metastatic or recurrent cervical cancer is less than 20%3,4. Therefore, it is crucial to identify key molecular mechanisms underlying cervical cancer progression.
Tie is a tyrosine kinase receptor widely expressed in vascular endothelial cells5,6. The family of receptor tyrosine kinases includes two members: Tie1 and Tie2 (also termed TEK)7. Angiopoietins (Ang1, Ang2, and Ang4, also termed Angpt) bind to Tie28. The Ang/Tie2 signaling pathway plays a pivotal role in regulating angiogenesis, inflammatory vascular remodelling, and vascular leakage9,10. The Ang/Tie2 signaling pathway is utilized not only by endothelial cells for angiogenesis but also by cancer cells to sustain their own growth, invasion, and metastasis11,12,13,14. The orphan receptor Tie1 does not bind to any Ang but forms a polymer with Tie2, regulates the Ang/Tie2 pathway, and plays an important role in pathological angiogenesis and tumor progression15,16. The current understanding of how Tie1 regulates the Ang/Tie2 signaling axis and related downstream molecules to promote the malignant progression of cervical cancer remains limited, and further studies are required.
Therefore, to elucidate the role of Tie1 in cervical cancer invasion and metastasis, we established a cervical cancer xenograft model in nude mice and conducted a series of in vivo and in vitro experiments. Our study identifies a novel molecular mechanism by which Tie1 facilitates cervical cancer progression. These results indicate that Tie1 may serve as a critical regulator of the Ang1/Tie2/PI3K/Akt pathway, thereby promoting the invasive metastasis of cervical cancer.
Materials and methods
Cell culture
The cell lines used in this study, namely the human cervical cancer cell lines HeLa (RRID: CVCL_0030) and SiHa (RRID: CVCL_0032), the immortalized human cervical epithelial cell line H8 (RRID: CVCL_9389), and human umbilical vein endothelial cells (HUVEC, were sourced from the Pathology Laboratory of Xinjiang Medical University. Prior to experimentation, all cell lines were confirmed to be free of mycoplasma contamination, and their identities were authenticated by STR profiling. The culturing conditions for All cells involved Dulbecco’s modified Eagle’s medium (DMEM; HyClone, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco, USA), 100 U/mL penicillin, and 100 µg/mL streptomycin (Gibco), maintained at 37 °C in an atmosphere of 5% CO2.
Western blot analysis
Cells were lysed on ice using radioimmunoprecipitation assay (RIPA, from Solarbio, China) supplemented with protease and phosphatase inhibitors. Subsequently, equal volumes of protein extracts were boiled for 5 min and separated by SDS-PAGE. The proteins were then transferred onto polyvinylidene fluoride (PVDF; Sigma-Aldrich LLC, 3010040001) membranes. The membranes were incubated overnight at 4 °C with a series of primary antibodies, including Tie1 (#DF4582, Affinity, China), GAPDH (#AF7021, Affinity, China), Phospho-PI3Kinase p85α (Y607) (#ab182651, Abcam, UK), PI3Kinase p85α (#AF6241, Affinity, China), phospho - Akt (Ser473) (#9271s, Cell Signaling Technology, USA), Akt (#4691s, Cell Signaling Technology, USA), mouse anti - β - actin (#66009–1 - Ig, Proteintech, China), and Angiopoietin − 1 (#HY - P70061, MedChemExpress, China). The following day, the membranes were washed five times with TBST and incubated with an HRP-conjugated secondary antibody for 1 h. Finally, the membranes were rinsed five times with TBST and visualized using the Western Bright ECL detection system (Bio-Rad, USA).
Cell migration (scratch wound) assay
For the cell migration assay, HeLa cells were seeded into 6 - well plates at a density of 2 × 105 cells per well and incubated for 6 h. Subsequently, a sterile pipette tip was used to create a scratch in the middle of each well and the detached cells within the scratches were removed. At the initial time point (0 h), scratch images of each cell group were obtained using an optical microscope, and their widths were analyzed and quantified using Image J (Fiji Is Just ImageJ, version 1.54f; http://imagej.org). After an additional 24 h of incubation, the width of the scratches in each group was measured (24 h), and the cell migration rates were calculated based on the change in scratch width.
Cell invasion assay
HeLa cells were resuspended in DMEM medium without fetal bovine serum, and about 1 × 105 cells were inoculated in the upper chamber of the Transwell culture plate coated with substrate gel (the lower chamber was added with an appropriate amount of DMEM medium containing 10% fetal bovine serum), After 24 h of incubation, the cells on the upper surface of the membrane were removed by swabbing. The cells that had invaded to the lower surface were then washed, fixed, and stained. After staining, images of the cells in the lower chamber were collected using a light microscope and analyzed using the ImageJ software.
Angiogenesis formation experiment
Thawed BD Matrigel Matrix (40 µL; Corning, 356234) was placed in each well of a 96-well plate and allowed to stand for 30 min. To replicate the process of tube formation, transfected cancer cell cultures for culturing human umbilical vein endothelial cells (HUVEC) were seeded onto pre-coated plates at a density of 5 × 104 cells/well and incubated at 37 °C with 5% CO2. After 4 h, tube formation was observed under an inverted microscope (Olympus IX73P1F). The tube length was measured using the Angiogenesis Analyzer plugin in Fiji (ImageJ).
CCK8 assay
Cell viability was analyzed using the Cell Counting Kit-8 (CCK8, Biotek, Shanghai, China) according to the method provided by the manufacturer’s instructions. Cells were inoculated and cultured in 96-well microtiter plates (Corning, USA) at a density of 5 × 103 cells/well in 100 µL culture medium. After 24 h of treatment, 10 µL of the CCK-8 reagent was added to each well and incubated for 2 h. All experiments were performed in triplicate. Absorbance was measured at 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA), and wells without cells were used as blank wells. Cell proliferation was expressed as the absorbance.
Cell transfection
For the transfection of Tie1 knockdown, a lentiviral vector harboring the puromycin sequence was employed to construct Tie1shRNA. Subsequently, Tie1shRNA was encapsulated in lentiviral envelopes and transfected into HeLa cells for 16 h. Puromycin was used for screening purposes. Real-time fluorescence quantitative PCR or protein blotting assays were performed to assess the expression levels of target genes.
Gene expression by qRT-PCR
Total RNA was extracted from HeLa cells using an RNeasy Plus Mini Kit (TransGen, China). Subsequently, 1 µg of the obtained RNA was used for cDNA synthesis using multi-transcript reverse transcriptase and random primers (Applied Biosystems). The synthesized cDNA was then subjected to duplicate analysis by qRT-PCR (7500 Rapid Real-Time Fluorescent Quantitative PCR System, Bio-Rad, USA) using Sybr Green Chemistry reagents and corresponding forward and reverse primer pairs. To ensure reliable quantification, the expression of the target genes was normalized to that of genotype-independent GAPDH. The 2 - ΔΔCT method was used for quantification.
Xenografts and tumor growth analysis in vivo
BALB/C nude mice (female, 3 to 5 weeks old, sourced from Slaccas, Shanghai, China) were maintained under sterile conditions with a constant humidity ranging from 60% to 70% and a room temperature of 18 to 20 °C. Nude mice were housed in a clean and quiet environment. Optimal temperature and humidity levels were provided and adequate food and water were supplied. Special housing conditions involve the use of individually ventilated cages and the regular replacement of bedding to ensure a hygienic environment. The mice were used in accordance with the protocol approved by the Animal Care and Use Committee of the Xinjiang Medical University. For the subcutaneous xenograft model, 4 × 106 human HeLa cells were prepared in 200 µL of phosphate-buffered saline (PBS) and injected subcutaneously into the right axillary tissue of immunocompromised BALB/C female nude mice aged 3–5 weeks. Once the tumor size reached approximately 60 mm3, the mice were randomly divided into five groups (five mice per group): control, NC group, NC + Ang1 group, Tie1, and Tie1 knockout + Ang1 groups. According to the schedule, Ang1 (5 ng/g) or PBS was administered by intraperitoneal injection (ip) once daily for 14 days. Tumor growth was measured five times per week using a Vernier caliper. After 15 days, once necrosis manifested in the xenografts and the nude mice exhibited visible emaciation, the mice were sacrificed by cervical dislocation ( 25 mice in total). Mice were euthanized by cervical dislocation. This procedure was performed only by experienced researchers to ensure instantaneous death and minimize any potential pain or distress. Subsequently, the tumors were dissected using surgical scissors. Tumor volume was calculated using the following formula: tumor volume = [L × W2]/2, where W represents the tumor width and is the tumor length.
For the metastatic xenograft model, cells (1 × 106) were prepared in 200 µL PBS and injected into the tail vein of immunocompromised BALB/C female nude mice aged 3–5 weeks. Mice were randomly divided into five groups (five mice per group): control, NC, Tie1 knockout, and Tie1 knockout + Ang1 groups. One week after injecting shRNA-transfected HeLa cells through the tail vein, Ang1 (5 ng/g) or PBS was administered by intraperitoneal injection (ip) once daily. The health status and behavior of nude mice were observed every other day, including diet, activity level, mental state, and body weight changes. After three weeks, the nude mice were sacrificed by cervical dislocation when bioluminescence image signals were detected in their lungs (20 mice in total). The specific method of euthanasia is as described above. The liver and lung tissues were dissected using surgical scissors for subsequent experiments. Animal experimental equipment was provided by the Animal Experiment Center of Xinjiang Medical University. We strictly adhered to animal welfare principles. All the researchers involved in the experiment received professional training in animal care and handling before they started.
Small animal in vivo imaging
One day before the experiment, the mice were fasted with access to water. On the day of imaging, after appropriate treatment, the mice were mildly anesthetized using 1.5 ~ 2.5% isoflurane in oxygen delivered via a nose cone, with anesthesia depth monitored by lack of response to toe pinch and maintained throughout the procedure. Body temperature was maintained at 37 °C using a heating pad and placed on the heating platform of the imaging dark box. After setting the parameters of the imaging system, a fluorescent substrate (d-luciferin potassium) (#HY-12591B, MedChemExpress, China) was used for bioluminescence imaging. Ten to 15 min before imaging, d-Luciferin was intraperitoneally injected at a dose of 150 mg/kg (or 10 µL/g of luciferin stock solution) based on the animal’s body weight. After waiting for a uniform distribution, images were collected. The software accompanying the imaging system was used to analyze the images, enabling measurement of the light signal intensity, delineation of regions of interest, and calculation of the average light intensity. Quality control was implemented using equipment calibration, control experiments, and repeatability experiments. The experimenters wore protective equipment and strictly adhered to the animal experimental ethics and norms. After the experiment, the animals were properly disposed.
Histopathology and immunostaining
For histological assessment, the dissected cervical tumors were fixed in 10% neutral buffered formalin for approximately one week. Subsequently, the sections were embedded in paraffin and sectioned. The obtained samples were stained with hematoxylin and eosin (H&E). For immunohistochemical (IHC) staining, the tissue slides were first dewaxed using xylene (Zsbio, China) and then rehydrated with ethanol. To inhibit endogenous peroxidase activity, 3% H₂O₂ in methanol was used. After rinsing the sections with PBS, the slides were incubated with 10% normal goat serum (Zsbio, China) at 20–25 °C for 30 min. Subsequently, the were incubated with primary antibodies (p - Tie2, 1:100; p - p85α, 1:100) overnight at 4 °C, followed by incubation with secondary antibodies for 30 min at room temperature. After DAB staining, hematoxylin re-staining, dehydration, and clarification in xylene, slides were mounted. The staining degree was evaluated using Image Pro Plus 6.0 by calculating the staining intensity and positive area fraction. Staining intensity was determined by calculating the cumulative optical density values (integral optimal density, IOD). The degree of staining was expressed as the average density value (mean density), which is equivalent to the average optical density (AOD), calculated as AOD = integrated optical density (IOD)/positive area fraction.
Co-immunoprecipitation (Co-IP)
After equalizing the protein concentration of the admixture, the lysates were subjected to immunoprecipitation with the relevant antibodies (Tie1, Tie2, and PI3K) or IgG for 2 h. Subsequently, the were incubated overnight at 4 °C with Protein A/G agarose beads (Thermo Fisher, USA). Thereafter, the immunoprecipitated proteins were washed with lysis buffer and eluted from agarose beads with 4× upsampling buffer. Finally, the bound proteins were denatured and separated using protein blot analysis.
Protein–protein docking method
First, we obtained the structures of key target proteins from the PDB database (https://www.rcsb.org). The PyMOL software (PyMOL Molecular Graphics System, version 3.1; https://pymol.org) was used to remove water molecules and extract the target protein structure. The obtained protein structure was then input into GRAMM (https://gramm.compbio.ku.edu/) to perform the protein-protein docking simulation.
LigPlot+ (LigPlot + version 4.5; https://www.ebi.ac.uk/thornton-srv/software/LigPlus/) was used to analyze the chemical bonds in the results and PyMOL was used for visualization. Typically, a binding energy lower than − 7 kcal/mol indicates a strong intermolecular interaction, making it more valuable for research.
Bioinformatic analysis
Data processing and download of the cervical cancer dataset: The GSE9750 dataset was obtained from the GEO database (Gene Expression Omnibus; https://www.ncbi.nlm.nih.gov/geo/). The supplementary material provides information regarding this dataset. The ‘Limma’ software (Linear Models for Microarray and Omics Data, version 4.5; https://bioconductor.org/packages/release/bioc/html/limma.html) was utilized to analyze mRNA expression differences. The significance threshold for the analysis was set at an adjusted P < 0.05.
Pan-cancer of Tie1 analysis: Using the TIMER2.0 database (Tumor IMmune Estimation Resource; http://timer.cistrome.org/ ), we obtained the differential expression profile of TIE1 across pan-cancer and normal tissues. The relationship between TIE1 expression and clinical prognosis in pan-cancer was further analyzed via GEPIA2 (Gene Expression Profiling Interactive Analysis; http://gepia2.cancer-pku.cn/#index).
Statistical analysis
Data are expressed as means ± standard deviation (SD). Statistical significance was set at P < 0.05. Differences were analyzed using Student’s t-test or one-way ANOVA. All statistical analyses were performed using the SPSS 26 software ( Statistical Package for the Social Sciences; version 26.0; https://www.ibm.com/cn-zh/spss ).
Results
Tie1 is highly expressed in cervical cancer tissues and cells
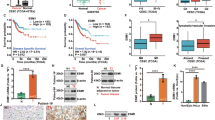
To investigate the expression of Tie1 in cervical cancer, we first found that it was highly expressed in cervical cancer and other tumor tissues such as head and neck squamous cell carcinoma, liver hepatocellular carcinoma, stomach adenocarcinoma, using bioinformatics analysis (Fig. 1A, Fig. S1A). This was also demonstrated in cervical cancer cell lines by western blotting when compared to the normal cervical cell line, H8 (Fig. 1B). To investigate the role of Tie1 in cervical cancer, we used lentiviral shRNA to knock down Tie1 in HeLa and SiHa cells. This resulted in a reduction in protein and mRNA levels compared with those in the controls (Fig. 1C,D). Among the treatments, Tie1-shRNA3 demonstrated the highest knockdown efficiency, and was therefore selected for further experiments.
Tie1 is highly expressed in cervical cancer tissues and cells. (A) Bioassay analysis of Tie1 expression in the cervical (n = 33) and paracancerous tissues(n = 20). (B) The Efficiency of Tie1 overexpression was verified by protein blotting in HeLa, SiHa, and H8 cells. (C) Western blotting was used to detect lentiviral control (sh-scramble) or Tie1-shRNA (sh-Tie1)-infected HeLa and SiHa cells for Tie1 protein expression. β-Actin was used as a loading control. (D) Real-time fluorescence quantitative PCR was used to detect Tie1 mRNA levels in lentiviral control (sh-scramble) and Tie1-shRNA (sh-Tie1)-infected HeLa and SiHa cells. β-Actin was used as a loading control. Data are shown as mean ± SD of 3 independent experiments. Statistical significance was assessed by one-way ANOVA with Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001.
Tie1 knockdown inhibited migration, invasion and proliferation of cervical cancer cells which was partly reversed by Ang1
To demonstrate the effect of Tie1 on cervical cancer, proliferation, migration, and invasion assays were performed. CCK8 experiments showed that the knockdown of Tie1 inhibited the proliferation of HeLa and SiHa cells (Fig. 2A). Knockdown of Tie1 also markedly impeded the migratory and invasive capacity of HeLa and SiHa cells (Fig. 2B,C, Fig. S1B,C). Addition of Ang1 (200 ng/ml) partially reversed the inhibitory effects of Tie1 knockdown on proliferation and invasion; however, its effect on restoring cell migration was minimal. Angiogenesis experiments were performed to investigate the effects of Tie1 expression on angiogenesis in cervical cancer and vascular endothelial cells. The results showed that HUVEC angiogenesis was reduced in the conditioned medium of cervical cancer cells with Tie1 knockdown but increased by the addition of Ang1 (Fig. 2D). These data indicate that Tie1 not only promotes the proliferation, migration, and invasion of cervical cancer cells but also serves as a paracrine effector medium to regulate angiogenesis of endothelial cells.
Tie1 knockdown inhibited the migration, invasion, and proliferation of cervical cancer cells, which was partially reversed by Ang1. (A) Effect of Tie1 knockdown on the proliferation of HeLa and SiHa cells was evaluated. (B) The effect of Tie1 knockdown on cell migration was assessed using a wound healing assay in HeLa and SiHa cells. Scale bar = 200 μm. (C) The impact of Tie1 knockdown on the invasion capacity of HeLa and SiHa cells was evaluated using a Transwell assay. Scale bar = 50 μm. (D) Effect of Tie1 knockdown on the tube-forming capacity of Huvec cells was investigated. Data are shown as mean ± SD of 3 independent experiments. Statistical significance was assessed by one-way ANOVA with Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001.
Tie1 regulates Tie2 phosphorylation, followed by activation of PI3K/Akt signaling pathway in cervical cancer cells, and Ang1 enhances phosphorylation of PI3K p85α and Akt
To investigate the molecular mechanism of Tie1 in cervical carcinogenesis, we first performed co-immunoprecipitation (Co-IP) assays. The results showed that Tie1 interacts with Tie2, and Tie2 in turn interacts with PI3K p85α (Fig. 3A). Additionally, molecular docking technology indicated that the binding energy of Tie1 and Tie2 was − 12.5 kcal/mol, while the binding energy of Tie2 and PIK3R1, also known as PI3K p85α, was − 7.8 kcal/mol (Table 1). These two significant negative values suggest strong binding affinity between the respective pairs. Using the PyMOL software, the complexes formed by Tie1, Tie2, Tie2, and PIK3R1 were visualized, clearly illustrating their unique binding modes. The Tie1-Tie2 binding interface is stabilized by a fundamental hydrogen-bond network, with hydrogen bond donors from Tie1 (Gln989, Arg849, and Asn822) and acceptors from Tie2 (Arg780, Gln795, and His866). These donor–acceptor pairs specifically interact in a “Tie1-donor to Tie2-acceptor” manner, forming three core hydrogen bonds that constitute the structural backbone of the complex. Further analysis revealed the presence of unconventional sulfur-mediated hydrogen bonds: the thiol groups of Cys892 and Cys1118 in Tie1 form “S–N” and “S–O” type hydrogen bonds with Lys863 and Glu508 in Tie2, respectively. Due to the involvement of sulfur atoms, these hydrogen bonds exhibit higher bond energy than conventional ones, significantly enhancing binding specificity. In terms of bond length, the hydrogen bond between Tie1 Arg849 and Tie2 Gln795 measures 2.72 Å, representing a moderate-strength hydrogen bond, whereas the hydrogen bond between Tie1 Cys1118 and Tie2 Glu508 is only 1.87 Å—significantly shorter than typical hydrogen bonds—with bond energy approaching that of a covalent interaction. This ultra-short hydrogen bond serves as a key structural feature contributing to the stability of the Tie1–Tie2 complex, enabling it to withstand intracellular environmental fluctuations and ensuring persistent functionality under physiological conditions (Fig. 3B, Fig. S2). Additionally, salt bridges and hydrophobic interactions within the binding interface act synergistically with the hydrogen-bond network, filling spatial gaps and further enhancing the overall stability of the complex.
Analysis of protein-protein interactions between TIE1 and TIE2 and between TIE2 and PIK3R1 in cervical cancer. (A) Collection of HeLa cells loaded, CO-IP detection analyzes the interaction of Tie1 and Tie2, Tie2 and PI3K p85α in HeLa cells. (B) Molecular docking technology indicated that Tie1 interacts with Tie2, Tie2 interacts with PIK3R1.
In the Tie2–PIK3R1 binding interface, interactions are similarly governed by a “hydrogen bond + salt bridge” framework, with clearly defined roles for participating residues. Key hydrogen bond–forming residues include Lys363 and Asp923 from Tie2, and Ser255 and Glu81 from PIK3R1. These residues engage in bidirectional hydrogen bonding (“Tie2 donor to PIK3R1 acceptor” and “PIK3R1 donor to Tie2 acceptor”), forming multiple cross-interacting hydrogen bonds that create a flexible yet stable binding interface. Salt bridges—electrostatic interactions between oppositely charged side chains—are represented by one pair between Tie2 Asp178 and PIK3R1 Lys423, and another between Tie2 Lys442 and PIK3R1 Asp60. These two salt bridges are symmetrically distributed on either side of the binding interface, not only strengthening electrostatic interactions but also serving to “lock” the spatial conformation of the interface, preventing dissociation due to protein conformational fluctuations. Furthermore, a hydrogen bond between Tie2 Pro170 and PIK3R1 Ser1091 measures 2.99 Å in length. Although slightly longer than the ultra-short hydrogen bonds observed in the Tie1–Tie2 complex, this bond is still shorter than the conventional upper limit of 3.5 Å, classifying it as a moderate-strong hydrogen bond. Its role involves “filling” the spatial gaps between salt bridges, thereby reducing water molecule penetration into the binding interface and lowering the dissociation energy of the complex. The combination of strong hydrogen bonds, salt bridges, and hydrophobic interactions among these amino acid residues collectively enhanced the stability of the Tie2-PIK3R1 interaction, promoting the formation of a stable complex (Fig. 3B, Fig. S2).
We further investigated whether the expression of Tie1 influenced the PI3K/Akt signaling pathway. The results revealed that knockdown of Tie1 in cervical cancer cells resulted in a reduction in the levels of phosphorylated Tie2, PI3K p85α, and Akt (Fig. 4A-C). The phosphorylation levels of PI3K p85α and Akt were elevated upon addition of Ang1(Fig. 4B-C). We next investigated whether insulin-like growth factor-1 (IGF-1), an agonist of PI3K, could reverse the effects of Tie1 knockdown on phosphorylation of the PI3K/Akt pathway. After 100 ng/ml IGF-1 stimulation was given to Tie1 knocked down cells, western blotting was performed. The results showed that IGF-1 significantly increased PI3K p85α and Akt phosphorylation in Tie1-shRNA3 HeLa cells, while the total protein was relatively unchanged (Fig. 4D). Previous studies have proposed that knockdown of Tie1 can suppress the activation of the PI3K/Akt signaling pathway in HeLa cervical cancer cells, and this inhibitory effect can be partially alleviated by PI3K agonists. Subsequently, we investigated whether IGF-1 induced migratory and invasive behaviors. The experimental data demonstrated that IGF-1 enhanced wound healing and invasion (as shown in Fig. 4E). Moreover, the PI3K agonist IGF-1 partially reversed the inhibition of the PI3K/Akt pathway induced by Tie1 knockdown, which in turn led to the activation of this pathway and contributed to the malignant progression of cervical cancer17. These results suggest that Ang1 may have a promoting role in cancer, and that Tie1 may enhance its tumorigenic function in cervical cancer by regulating the Ang1/Tie2/PI3K/Akt signaling pathway.
Tie1 regulates Tie2 phosphorylation, followed by activation of PI3K/Akt signaling pathway in cervical cancer cells, and Ang1 enhances phosphorylation of PI3K p85α and Akt. (A) Tie1-shRNA3 HeLa cells were subjected to western blotting to assess phosphorylation levels of p-Tie1 and p-Tie2. (B) Western blot analysis of PI3K p85α, and Akt phosphorylation in control and Tie1-shRNA3 200 ng/ml Ang1-treated HeLa cells, with β-actin as the loading control. (C) Western blot analysis of PI3K p85α, and Akt phosphorylation in control and Tie1-shRNA3 200 ng/ml Ang1-treated SiHa cells, with β-actin as the loading control. (D) Western blotting was used to analyze changes in the expression of PI3K/Akt signaling pathway proteins in Tie1-shRNA3 HeLa cells treated with IGF-1 (100 ng/ml). (E) The difference in cell migration distance under the action of IGF-1 was detected by the scratch test. Differences in invasive ability following IGF-1 treatment. Data are shown as mean ± SD of 3 independent experiments. Statistical significance was assessed by one-way ANOVA with Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001.
Knockdown of Tie1 inhibited the formation of subcutaneous xenograft tumors in nude mice
HeLa cells with Tie1 knockdown or control were injected into nude mice to establish a subcutaneous xenograft model (Fig. 5A), which was treated with phosphate-buffered saline (PBS). The Tie1 knockdown and negative control groups were treated with Ang1 (5 ng/g, intraperitoneal injection, 15 days). Knockdown of Tie1 led to the deceleration of subcutaneous tumor growth in nude mice without a notable impact on body weight. In addition, the addition of Ang1 resulted in accelerated tumor growth without significant weight loss compared with the controls (Fig. 5B, Fig. S3A). Live imaging of small animals demonstrated a schematic representation of the tumor size for each group (Fig. 5C). Hematoxylin and eosin-stained (H.E.) sections of the tumors revealed that the necrotic area was larger in the Tie1 knockdown plus Ang1, PBS (vehicle), and control groups than in the Tie1 knockdown group. This suggests that the knockdown of Tie1 can inhibit the growth of cervical cancer in vivo, and Ang1 can accelerate its growth. (Fig. 5D, top panel). Given the propensity of advanced cervical cancer to metastasize to lymph nodes, lymph node metastasis was examined in nude mice. The results demonstrated a decreased lymph node metastasis rate in nude mice with Tie1 knockdown and an increased rate in nude mice with Tie1 knockdown plus Ang1 (Fig. 5D, bottom, Fig. S3B). Subsequently, immunohistochemistry (IHC) was conducted on xenograft tumors to evaluate the expression of p-Tie2 and p-PI3K p85α in vivo. These results demonstrated that the knockdown of Tie1 markedly suppressed the protein expression of p-Tie2 and p-PI3K p85α. Ang1 addition elevated the protein expression of p-Tie2 and p-PI3K p85α in Tie1 knockdown cells (Fig. 5E, Fig. S3C). The effect of Tie1 on the expression of these proteins was evaluated using western blotting. This analysis revealed that the levels of p-Akt, p-PI3K p85α, and CD31 were diminished in the Tie1 knockdown group. Ang1 upregulated the phosphorylation of these proteins in the Tie1 knockdown group (Fig. 5F).
Knockdown of Tie1 inhibited the formation of subcutaneous xenograft tumors in nude mice. (A) Pattern diagram of subcutaneous xenograft tumors in nude mice. (B) Comparison of the volume of transplanted tumors and the effect of Ang1 (5ng/g) on the growth of subcutaneous tumors in control and Tie1-shRNA groups (n = 5 mice per group). (C) Small animal live imaging luminescence showing the subcutaneous graft tumor size in nude mice (n = 5 mice per group). (D) Histological examination of tumor tissue (40x) and lymph node metastasis (100x and 200x) in different groups. (E) After Ang1 treatment, the expression of p-Tie2, and p-PI3K p85α was detected using immunohistochemistry (IHC) (400 ×). (F) Western blot analysis of the effects of Ang1 on the expression of key proteins in nude mice and the changes in key proteins in the Tie1-shRNA3 group. Data are shown as mean ± SD. Statistical significance was assessed by one-way ANOVA with Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001.
Tie1 enhances in vivo metastasis in nude mice
To investigate the effects of Tie1 and Ang1 in vivo, a lung metastasis model was constructed using tail vein injections (Fig. 6A). For all tail vein-injected nude mice, no significant difference in body weight was observed before treatment ( Fig. S3D). After the mice were anesthetized, in vivo imaging was performed and metastases in the lungs were observed (Fig. 6B). H&E staining demonstrated that lung metastases increased in the control group, whereas it was reduced in Tie1 knockout mice and increased in Tie1 knockdown plus Ang1 knockdown mice(Fig. 6C, Fig. S3E ). Subsequently, IHC was conducted on lung metastases to evaluate the expression of p-Tie2 and p-PI3K p85α in vivo. These results demonstrated that the knockdown of Tie1 markedly suppressed the protein expression of p-Tie2 and p-PI3K p85α(Fig. S4A-B). Perhaps, because of the relatively short intervention time, liver metastasis was not observed. Our data confirm the role of Tie1 in promoting HeLa cell metastasis in vivo. Additionally, we performed a comprehensive bioinformatic analysis of public datasets (including TCGA) via TIMER and GEPIA2 platforms. The results demonstrated that Tie1 expression was significantly dysregulated in multiple human cancers. Most importantly, survival analysis revealed that high Tie1 expression was significantly associated with poorer overall survival (OS) in several cancer types, including lung squamous cell carcinoma (LUSC) and stomach adenocarcinoma (STAD). High Tie1 expression was also linked to worse disease-free survival (DFS) in colorectal cancers (COAD and READ) (Figure S5). In summary, the knockdown of Tie1 led to a decrease in the lung metastasis of cervical cancer cells. Ang1 can increase cervical cancer cell metastasis to the lungs, and Ang1 may play a role in promoting cervical cancer.
Tie1 enhances in vivo metastasis in nude mice. (A) Modal pattern of tail vein injections into nude mice. (B,C) Representative bioluminescence images, H&E staining of pulmonary metastatic foci, and rates of lung metastasis in the NC, Tie1-shRNA3, and Tie1-shRNA3 + Ang1 groups (n = 3 mice per group). Scale bars: 100–20 μm. Data are shown as mean ± SD. Statistical significance was assessed by one-way ANOVA with Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
The Ang/Tie signaling pathway is a crucial regulator of blood and lymphatic vessel growth6, as well as of endothelial homeostasis during acute injury, inflammation, and pathological angiogenesis18,19,20. Most previous studies have focused on their role in vascular endothelial cells, while only a few have focused on the function in tumor cells. Recent studies have reported that Ang/Tie can be secreted by cancer cells21,22. The orphan receptor tyrosine kinase Tie1 has received increasing attention as a potential cancer-related gene23. However, the role of Tie1 in tumors, particularly the underlying mechanisms, remains insufficiently detailed. In this study, we provide the evidence that Tie1 derived from cervical cancer cells promotes invasion and metastasis by activating the Ang1/Tie2/PI3K/Akt pathway.
In our study, bioinformatics analysis revealed that Tie1 was highly expressed in human cervical cancer tissues and this was confirmed by western blotting in HeLa and SiHa cell lines. Therefore, these cell lines were selected for subsequent experiments. Our results demonstrated that Tie1 knockdown inhibited the proliferation, migration, and invasion of HeLa and SiHa cells in vitro. In vivo assays further revealed that Tie1 knockdown reduced tumor size, lymph node metastasis, and lung metastasis in nude mice. These results indicate that cervical cancer cells express functional Tie1 receptors which promote proliferation, invasion, and metastasis. This finding extends previous reports of Tie1 function in Lewis lung carcinoma to cervical cancer.
Angiogenesis formation assays in this study revealed that conditioned medium from Tie1-knockdown cells inhibited HUVEC tube formation, indicating a paracrine regulatory role for Tie1 in cervical carcinoma. This finding aligns with the reports in other cancers, such as Lewis lung carcinoma, where Tie1 derived from cancer cells mediated angiogenesis to promote progression and metastasis14,24.
Tie1 and Tie2 interact with each other. Tie1 regulates Tie2, and is critical for downstream signal transmission25,26. The PI3K/Akt/mTOR pathway mediated by Ang/Tie2 is a main channel involved in tumor angiogenesis and the immunosuppressive microenvironment27. To explore the mechanism by which Tie1 promotes the progression of cervical cancer, we first identified interactions between Tie1 and Tie2, as well as between Tie2 and PI3K p85α, using Co-IP. Additionally, molecular docking analysis indicated strong binding affinity between Tie1 and Tie2 (binding energy = -12.5 kcal/mol) and between Tie2 and PIK3R1 (PI3K p85α) (binding energy = -7.8 kcal/mol). These results provide evidence for physical interactions in cervical cancer cells. Subsequently, we demonstrated that Tie1 knockdown inhibited the phosphorylation of Tie2, as assessed by western blotting. It has been reported that Tie1 derived from cervical cancer cells promotes tumor progression via the PI3K/AKT signaling pathway28. In our study, Tie1 knockdown also reduced phosphorylation levels of Akt and PI3K p85. A xenograft mouse model confirmed these results. In vivo assays also showed that CD31 expression was decreased in Tie1-knockdown groups, indicating that Tie1 knockdown inhibits angiogenesis in cervical cancer. We further conducted a pan-cancer analysis of Tie1 using the TCGA database. The results revealed that high Tie1 expression is significantly associated with poorer OS in several cancer types, including LUSC and STAD. Additionally, high Tie1 expression was also linked to worse DFS in COAD and READ, which hinted Tie1 pro-cancer role in some types of carcinoma. In conclusion, Tie1 may act as a signaling molecule that regulates the Tie2/PI3K/Akt signaling axis, thereby promoting the malignant growth of cervical cancer cells. Ang1 treatment partly restored PI3K/Akt phosphorylation and functional effects.
The constitutive expression of Ang1, the natural ligand of Tie2, in perivascular cells suggests its involvement in endothelial cell survival, potentially contributing to the maintenance of endothelial tissue integrity11,29. The role of Ang1 in tumorigenesis remains controversial, with conflicting reports in the literature. Some researchers have reported that overexpression of Ang1 improved tumor perfusion and growth, whereas others observed that Ang1 impaired angiogenesis and subsequently inhibited tumor growth30,31,32,33. To date, the function of Ang1 in tumors remains unclear. In this study, we probed the effects of Ang1 on cervical cancer. The results showed that proliferation and invasion of cancer cells were significantly enhanced following Ang1 treatment, with no significant change in migration in either control or Tie1-knockdown groups. Additionally, tube-forming assays showed that vascular network formation was increased by Ang1, suggesting a pro-tumorigenic role of Ang1 in cervical cancer. In vivo experiments in nude mice further confirmed the pro-proliferative, pro-invasive, and pro-metastatic effects of Ang1, positioning it in the upstream of the Tie1/Tie2 receptor complex. This pro-tumor effect of Ang1 has been confirmed by other researchers. For example, Ang1 can increase vascular density in glioblastoma multiforme (GBM)34. Additionally, some studies have shown that Ang1 promotes triple-negative breast cancer cell proliferation by upregulating carboxypeptidase A435. Collectively, our findings demonstrate that Tie1 promotes invasion, metastasis, and angiogenesis by regulating the Ang1/Tie2/PI3K/Akt signaling axis in cervical cancer. These findings suggest Tie1 may serve as a potential therapeutic target. Several limitations of this study should be noted, including the lack of HPV status detection and validation in patient-derived xenograft model.
In summary, based on in vitro and in vivo data from cervical cancer cell lines and nude mice, we provide the compelling evidence that Tie1 promotes angiogenesis, invasion, and metastasis by activating the Ang1/Tie2/PI3K/Akt signaling pathway, and that Ang1 exerts a pro-tumorigenic role in cervical cancer. Tie1 may be a promising target for therapeutic intervention in cervical cancer patients.
Data availability
If necessary, all raw data can be obtained from the corresponding author.
Abbreviations
- GEO:
-
Gene expression omnibus
- Tie1:
-
Kinase with immunoglobulin like and EGF like domains 1
- Tie2:
-
TEK receptor tyrosine kinase
- ANG:
-
Angiopoietin
- RT-qPCR:
-
reverse transcription-quantitative
- HUVEC:
-
human umbilical vein endothelial cells
- Co-IP:
-
Co-immunoprecipitation
- IGF-1:
-
Insulin-like growth factor 1
- PI3K:
-
Phosphoinositide 3-kinase
- IHC:
-
Immunohistochemistry
- H.E.:
-
Hematoxylin and eosin staining
- Akt:
-
Protein kinase B
- DPI:
-
diphenyleneiodonium
References
Di Fiore, R. et al. Cancer stem cells and their possible implications in cervical cancer: A short review. Int. J. Mol. Sci. 23 https://doi.org/10.3390/ijms23095167 (2022).
Hu, Z. & Ma, D. The precision prevention and therapy of HPV-related cervical cancer: new concepts and clinical implications. Cancer Med. 7, 5217–5236. https://doi.org/10.1002/cam4.1501 (2018).
Sharma, S., Deep, A. & Sharma, A. K. Current treatment for cervical cancer: an update. Anti-cancer Agents Med. Chem. 20, 1768–1779. https://doi.org/10.2174/1871520620666200224093301 (2020).
Tewari, K. S. et al. Bevacizumab for advanced cervical cancer: final overall survival and adverse event analysis of a randomised, controlled, open-label, phase 3 trial (Gynecologic oncology group 240). Lancet (London England). 390, 1654–1663. https://doi.org/10.1016/s0140-6736(17)31607-0 (2017).
Partanen, J. et al. A novel endothelial cell surface receptor tyrosine kinase with extracellular epidermal growth factor homology domains. Mol. Cell. Biol. 12, 1698–1707. https://doi.org/10.1128/mcb.12.4.1698-1707.1992 (1992).
Wu, X. & Liu, N. The role of Ang/Tie signaling in lymphangiogenesis. Lymphology 43, 59–72 (2010).
Sato, T. N. et al. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature 376, 70–74. https://doi.org/10.1038/376070a0 (1995).
Davis, S. et al. Isolation of angiopoietin-1, a ligand for the TIE2 receptor, by secretion-trap expression cloning. Cell 87, 1161–1169. https://doi.org/10.1016/s0092-8674(00)81812-7 (1996).
Augustin, H. G., Koh, G. Y., Thurston, G. & Alitalo, K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat. Rev. Mol. Cell Biol. 10, 165–177. https://doi.org/10.1038/nrm2639 (2009).
Chan, B., Yuan, H. T., Ananth Karumanchi, S. & Sukhatme, V. P. Receptor tyrosine kinase Tie-1 overexpression in endothelial cells upregulates adhesion molecules. Biochem. Biophys. Res. Commun. 371, 475–479. https://doi.org/10.1016/j.bbrc.2008.04.091 (2008).
Li, Y. et al. Activation of Angiopoietin-Tie2 signaling protects the kidney from ischemic injury by modulation of Endothelial-Specific pathways. J. Am. Soc. Nephrology: JASN. 34, 969–987. https://doi.org/10.1681/asn.0000000000000098 (2023).
Reikvam, H. et al. Targeting the angiopoietin (Ang)/Tie-2 pathway in the crosstalk between acute myeloid leukaemia and endothelial cells: studies of Tie-2 blocking antibodies, exogenous Ang-2 and Inhibition of constitutive agonistic Ang-1 release. Expert Opin. Investig. Drugs. 19, 169–183. https://doi.org/10.1517/13543780903485659 (2010).
Xie, H. et al. Endothelin-1/Endothelin receptor type A-Angiopoietins/Tie-2 pathway in regulating the cross talk between glomerular endothelial cells and podocytes in Trichloroethylene-Induced renal immune injury. J. Inflamm. Res. 14, 761–776. https://doi.org/10.2147/jir.S301104 (2021).
La Porta, S. et al. Endothelial Tie1-mediated angiogenesis and vascular abnormalization promote tumor progression and metastasis. J. Clin. Investig. 128, 834–845. https://doi.org/10.1172/jci94674 (2018).
Song, S. H., Kim, K. L., Lee, K. A. & Suh, W. Tie1 regulates the Tie2 agonistic role of angiopoietin-2 in human lymphatic endothelial cells. Biochem. Biophys. Res. Commun. 419, 281–286. https://doi.org/10.1016/j.bbrc.2012.02.009 (2012).
Savant, S. et al. The orphan receptor Tie1 controls angiogenesis and vascular remodeling by differentially regulating Tie2 in tip and stalk cells. Cell. Rep. 12, 1761–1773. https://doi.org/10.1016/j.celrep.2015.08.024 (2015).
Wang, Q. et al. AQP3 promotes the invasion and metastasis in cervical cancer by regulating NOX4-derived H(2)O(2) activation of Syk/PI3K/Akt signaling axis. J. Cancer. 15, 1124–1137. https://doi.org/10.7150/jca.91360 (2024).
Gamble, J. R. et al. Angiopoietin-1 is an antipermeability and anti-inflammatory agent in vitro and targets cell junctions. Circul. Res. 87, 603–607. https://doi.org/10.1161/01.res.87.7.603 (2000).
Papapetropoulos, A. et al. Angiopoietin-1 inhibits endothelial cell apoptosis via the Akt/survivin pathway. J. Biol. Chem. 275, 9102–9105. https://doi.org/10.1074/jbc.275.13.9102 (2000).
Kim, I. et al. Angiopoietin-1 regulates endothelial cell survival through the phosphatidylinositol 3’-Kinase/Akt signal transduction pathway. Circul. Res. 86, 24–29. https://doi.org/10.1161/01.res.86.1.24 (2000).
Yang, P. et al. Cervical cancer cell-derived angiopoietins promote tumor progression. Tumour Biology: J. Int. Soc. Oncodevelopmental Biology Med. 39, 1010428317711658. https://doi.org/10.1177/1010428317711658 (2017).
Oliveira, S., Pereira, S. S., Costa, M. M., Monteiro, M. P. & Pignatelli, D. Ang-Tie angiogenic pathway is distinctively expressed in benign and malignant adrenocortical tumors. Int. J. Mol. Sci. 23 https://doi.org/10.3390/ijms23105579 (2022).
Kontos, C. D., Cha, E. H., York, J. D. & Peters, K. G. The endothelial receptor tyrosine kinase Tie1 activates phosphatidylinositol 3-kinase and Akt to inhibit apoptosis. Mol. Cell. Biol. 22, 1704–1713. https://doi.org/10.1128/mcb.22.6.1704-1713.2002 (2002).
D’Amico, G. et al. Tie1 deletion inhibits tumor growth and improves angiopoietin antagonist therapy. J. Clin. Investig. 124, 824–834. https://doi.org/10.1172/jci68897 (2014).
Xu, M. et al. LECT2, a ligand for Tie1, plays a crucial role in liver fibrogenesis. Cell 178, 1478–1492e1420. https://doi.org/10.1016/j.cell.2019.07.021 (2019).
Zhang, Y., Kontos, C. D., Annex, B. H. & Popel, A. S. A systems biology model of junctional localization and downstream signaling of the Ang-Tie signaling pathway. NPJ Syst. Biology Appl. 7, 34. https://doi.org/10.1038/s41540-021-00194-6 (2021).
Wu, X. et al. Macrophage-derived SHP-2 inhibits the metastasis of colorectal cancer via Tie2-PI3K signals. Oncol. Res. 31, 125–139. https://doi.org/10.32604/or.2023.028657 (2023).
Wei, Y. et al. Cervical cancer cell-derived Tie1 expression via PI3K/AKT signaling pathway promotes tumor progression. Exp. Cell Res. 439. https://doi.org/10.1016/j.yexcr.2024.114060 (2024).
Pan, L., Liu, Z., Chen, Y., Yang, B. & Cheng, B. Angiopoietin-1: can be produced by endothelial cells and act in an autocrine agonistic manner? Clin. Hemorheol. Microcirc. 74, 341–345. https://doi.org/10.3233/ch-190731 (2020).
Hayes, A. J. et al. Expression and function of angiopoietin-1 in breast cancer. Br. J. Cancer. 83, 1154–1160. https://doi.org/10.1054/bjoc.2000.1437 (2000).
Cai, E. et al. Angiopoietin-1 is associated with a decreased risk of lymph node metastasis in early stage cervical cancer. Histol. Histopathol. 35, 1029–1034. https://doi.org/10.14670/hh-18-234 (2020).
Thapa, K., Khan, H., Kaur, G., Kumar, P. & Singh, T. G. Therapeutic targeting of angiopoietins in tumor angiogenesis and cancer development. Biochem. Biophys. Res. Commun. 687, 149130. https://doi.org/10.1016/j.bbrc.2023.149130 (2023).
Hu, A., Roberts, C., Moscalu, A., Redston, M. & Yoo, J. Ang1 and Ang4 differentially affect colitis and carcinogenesis in an AOM-DSS mouse model. PloS One. 18, e0281529. https://doi.org/10.1371/journal.pone.0281529 (2023).
Zadeh, G., Koushan, K., Pillo, L., Shannon, P. & Guha, A. Role of Ang1 and its interaction with VEGF-A in Astrocytomas. J. Neuropathol. Exp. Neurol. 63, 978–989. https://doi.org/10.1093/jnen/63.9.978 (2004).
Liu, X. et al. Angiopoietin-1 promotes triple-negative breast cancer cell proliferation by upregulating carboxypeptidase A4. Acta Biochim. Biophys. Sin. 55, 1487–1495. https://doi.org/10.3724/abbs.2023082 (2023).
Acknowledgements
We appreciate Dr. Shayahati Bieerkehazhi (UT Health Science Center at Houston, University of Texas, USA) for language edition of the manuscript.
Funding
This work was supported by the open project of the Key Laboratory of the Ministry of Education for Research on Highly Prevalent Diseases in Xinjiang (#2023A02, to Yonghua Shi).
Author information
Authors and Affiliations
Contributions
YH-S developed the study concept and design. HJ-W, X-W, BJ-L, and QX-W, XY-L performed the experiments and collected data. HJ-W, YH-S analyzed the data. HJ-W and YH-S drafted the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical statement
This work adheres to the ARRIVE 2.0 guidelines for reporting animal research. All experimental protocols were approved by the Ethics Committee of Xinjiang Medical University (Approval No. IACUC-JT-20240228-39), and efforts were made to minimize animal suffering in accordance with the 3R principles (Replacement, Reduction, Refinement).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wei, H., Wang, X., Wang, Q. et al. Tie1 derived from cervical cancer promotes the invasion and metastasis by Ang1 mediating Tie2/PI3K/Akt signaling axis and angiogenesis. Sci Rep 15, 43979 (2025). https://doi.org/10.1038/s41598-025-27665-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-27665-8