Abstract
MamK, an actin-like protein conserved in magnetotactic bacteria, functions as a cytoskeletal element that positions the magnetosome, a bacterial geomagnetic sensor organelle. Specifically, MamK polymerizes into filaments associated with the magnetosome chain within each cell. The dynamics of these filaments are fundamental to magnetosome organelle positioning. However, the dynamic nature of the polymerized MamK filaments has not been characterized at the molecular level under physiological conditions. In this study, we used high-speed atomic force microscopy (AFM) to characterize the dynamic MamK polymerization process. MamK polymerized as double-helical filaments with a half-helical pitch distance of 41.3 ± 5.1 nm and a filament diameter of 6.3 ± 0.5 nm. The polymerizing MamK filaments elongated at average speeds of 12.4 ± 4.2 nm/min at the fast-growth ends (plus ends) and from − 4.4 to 8.0 nm/min at the slow-growth ends (minus ends) on the mica substrate in the solution containing 3 µM monomeric MamK. High-speed AFM demonstrated that MamK polymerized into dynamic double-helical filaments with polarity similar to that of eukaryotic actin filaments. Understanding the intrinsic dynamics of MamK, a well-conserved actin-like protein in magnetotactic bacteria, is key to elucidating the mechanism of magnetosome positioning in a bacterial cell.
Similar content being viewed by others
Introduction
Bacterial actin-like cytoskeletal proteins play several essential cellular roles, including mediating DNA segregation, maintaining cell shape, and participating in cell division1,2,3. MamK helps to maintain the position of the magnetosome (the magnetic organelle) of magnetotactic bacteria (MTB)4,5. A magnetosome is a membrane-enclosed organelle containing a single biomineralized magnetic crystal of magnetite or greigite. Magnetosomes are concatenated in a chain-like configuration located at the center of a cell, where they function as magnetic sensors that enable cells to navigate while swimming along the geomagnetic field6,7,8,9. The function of MamK mirrors that of eukaryotic actin, which regulates organelle dynamics and positioning.
The MamK cytoskeleton maintains the positioning of the magnetosome chain at the center of a cell along the cytoskeletal filament track. This linear positioning allows the magnetosome to function as an effective cellular magnetic sensor. In Magnetospirillum gryphiswaldense MSR-1, magnetosome chains are repositioned from the new cell poles to the middle of the daughter cells after cytokinesis10. Magnetosome repositioning is impaired by the expression of MamKD161A, which is defective in ATPase activity10. On the other hand, in M. magneticum AMB-1, MamK statically positions the magnetosome to help form an effective magnetic dipole and prevent magnetosome diffusion caused by thermal fluctuation11. The expression of ATPase-defective MamK also impairs magnetosome positioning in AMB-111. In the presence of ATP, the MamK monomers are polymerized to form filamentous structures12,13,14,15. ATPase activity is necessary for eukaryotic actin to form dynamic actin filaments with differential affinities for monomer addition at each filament plus or minus end. The actin filament dynamics (e.g., treadmilling) require various cytoskeletal functions, including cell migration and intracellular transportation. The indispensability of the MamK ATPase activity in magnetosome positioning suggests that MamK filament dynamics are required for bacterial organelle positioning.
The dynamic assembly of MamK has previously been investigated in vitro using light scattering to elucidate the kinetics of its polymerization13,14,15. Accordingly, the critical concentration in the presence of ATP was estimated to be approximately 0.7 µM for AMB-1 MamK13 and approximately 1.4 µM for MRS-1 MamK14. These values represent the minimum monomer concentrations required to form nucleation seeds capable of elongating into filaments. MamK dynamics have also been studied in vivo using fluorescence recovery after photobleaching (FRAP) and photoconversion assays involving fluorescent protein-tagged MamK. In MSR-1, both the photobleached and photoconversion areas shifted from the poles to the mid-cell, providing the first evidence of treadmilling MamK filaments10. The treadmilling speed was estimated to be approximately 300 nm/min.
While bulk MamK filament dynamics have been characterized using light scattering and FRAP, the behavior of individual filaments remains unexplored. In this study, we sought to determine whether MamK molecules polymerize into dynamic filaments with polarity (i.e., fast-growing plus-end and slow-growing minus-end filaments). To answer this question, we characterized the dynamic features of MamK filaments using high-speed atomic force microscopy (AFM). High-speed AFM was developed to study the structural dynamics of nanometer-sized soft biomolecules such as proteins, lipid bilayers, and living cells under physiological liquid conditions16,17,18,19. This method is well suited for characterizing the structural dynamics of filamentous proteinaceous polymers (e.g., actin and its binding proteins20,21, amyloid filamentous aggregates22, and bacterial cytoskeletal filaments of FtsZ)23,24. As a complementary technique for bridging the structural and biochemical characterizations, we used high-speed AFM to image the structural dynamics of the MamK filament in physiological buffer conditions.
Results and discussion
Imaging of the MamK filaments using atomic force microscopy (AFM)
Monomeric MamK was first purified to provide a suitable specimen for imaging the dynamic polymerization of MamK filaments using high-speed AFM. In previous studies of MamK biochemistry and structural biology13,25,26, the solutions used for polymerization contained glycerol (10%) to prevent aggregation. However, glycerol-containing solutions are unsuitable for achieving high spatial-temporal resolution using high-speed AFM. Moreover, we found that EDTA in a neutral pH buffer used for depolymerization in previously reported purification methods13,26 was ineffective under our purification conditions. Therefore, we modified the purification and sample storage methods to use an alkaline depolymerization buffer (DP buffer). An alkaline buffer with a pH higher than 9.5 effectively depolymerized MamK during purification (Figure S1).
The elution profile of the purified MamK (Figure S2A) obtained from size exclusion HPLC (the final step of the purification procedure) showed a single peak (Figure S2B). The pelleting assay of the purified MamK showed that polymerization was ATP-dependent (Figure S2C), with an ATPase activity of 0.23 min− 1 protein (Figure S2D). This ATPase activity is consistent with the value of approximately 0.2 min− 1 reported previously by Ozyamak et al.13. These assays were performed immediately after exchanging the alkaline DP buffer for neutral polymerization buffer (P buffer). These results indicate that the alkaline DP buffer did not affect the MamK ATPase and polymerization activities.
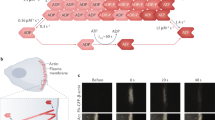
Ozyamak et al. reported that high potassium concentrations (> 75 mM) induce the formation of MamK filament bundles and inhibit filament disassembly, possibly due to inter-filament interactions within a bundle13. Therefore, we used low potassium (25 mM) and MamK (1, 1.5, or 3 µM) concentrations, with the latter being slightly above the critical concentration of 0.69 µM (AMB-1 MamK with ATP)13 for AFM imaging of individual dynamic MamK filaments. We observed the polymerized MamK filaments (1 µM) in P buffer with 1 mM ATP for 10 min, followed by loading on a bare mica surface. We observed double helical MamK filaments using the tapping mode of high-speed AFM in P buffer with 1 mM ATP (Fig. 1A). We could visualize the individual shapes of each MamK monomer along the filaments (Fig. 1B). Based on our observations, the average distance between the monomers was 5.5 nm (n = 18), the half-helical pitch distance of the helical configuration was 41.3 ± 5.1 nm (n = 100), and the diameter of filaments was 6.3 ± 0.5 nm (n = 100) (Fig. 1C-E). The half-helical pitch consisted of eight monomers. To our knowledge, this is the first description of the MamK filament structure under physiological conditions. The AFM MamK filament structure was consistent with the high-resolution images obtained using single-particle cryo-electron microscopy of the MamK filaments in a vitreous medium13,25,26.
MamK filaments as observed by AFM under physiological conditions. (A) An AFM image of the MamK filaments polymerized in the presence of 1 mM ATP. (B) A line profile of the height displacement along the black dashed line in the squared area in panel A. Triangles show the peaks of each MamK monomer comprising the protofilament (C, D). Histograms visualizing the length of the half-pitch (C) and diameters (D) of the double-helical filaments (n = 100). The red lines show the Gaussian fit curves. (E) Schematics of the MamK filament size based on the AFM observations. The AFM image was recorded at a rate of 1 s/frame in a 200 × 200 nm scanning area with a resolution of 200 × 200 pixels.
High-speed AFM imaging of elongating MamK filaments
High-speed AFM of the dynamic MamK polymerization processes was performed in P buffer containing monomeric MamK (3 µM or 1.5 µM) in the AFM imaging chamber. The AFM images shown in Figs. 2 and 3 were obtained under the 3 µM monomeric MamK condition. The bare mica surface was immersed in the sample solution for 3 min before generating images using AFM (Fig. 2A). We observed the monomeric MamK molecules as dynamic small globular structures densely but loosely attached to the mica surface before adding ATP (Movie S1), indicating that the MamK monomers are weakly associated with the bare mica surface in the P buffer. The mica substrate itself could not be imaged due to dense accumulation of MamK monomers, which appeared to saturate the surface in a two-dimensional fashion.
Dynamic elongation of the MamK filaments. (A) Schematics of the high-speed AFM experiments. Monomeric MamK was applied to the high-speed AFM sample chamber and became loosely associated with the mica surface. After incubation, MamK polymerization was triggered by addition of 1 mM ATP. (B) High-speed AFM images during the MamK polymerization in 3 µM MamK solution. The entire high-speed AFM video can be viewed in Movie S2. The AFM video was taken for about 30 min at 10 s/frames in a scanning area of 800 × 800 nm with a resolution of 400 × 400 pixels. (C) AFM images were taken 20 min after the addition of 1 mM ADP in place of ATP.
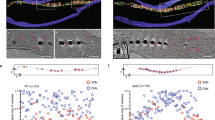
Dynamic polymerization properties of MamK filaments imaged using high-speed AFM. (A-C) Representative kymographs of (A) unidirectional, (B) bidirectional, and (C) treadmilling elongation modes. (D-F) Elongation time courses of fast-growing (solid lines) filament ends (plus ends) and slow-growing or shrinking (dashed lines) filament ends (minus ends) of (D) unidirectional (n = 4 individual filaments), (E) bidirectional (n = 3 individual filaments), and (F) treadmilling elongation (n = 4 individual filaments) patterns. Elongation speeds of the plus and minus ends in the three growth/elongation modes of the filaments in 3 µM (G) and 1.5 µM (H). Scale bars in panels A to C correspond to 20 nm.
We then added ATP to a final concentration of 1 mM in the AFM imaging chamber using a long gel-loading pipet tip. After adding ATP (Movie S2 and Fig. 2A), we observed straight elongating MamK filaments on the mica surface. The MamK filaments elongated dynamically over the mica surface (Fig. 2B and Movie S2). We frequently observed the sudden appearance of long filaments (Movie S2) that had polymerized in the fluid within the imaging chamber and become attached to the scanning area of the mica surface, with elongation after binding. The MamK filaments were of various lengths and were mostly straight. We did not observe any bent MamK filaments during the polymerization processes, indicating that MamK forms highly rigid straight filaments. Filament elongation stopped when the filament tips reached the sides of the adjacent filaments, possibly because contact with the adjoining filament blocked the monomer from binding to the filament tip. Figure 2C shows the mica surface 20 min after adding 1 mM ADP instead of ATP, which resulted in no polymerization of the MamK polymers.
The MamK cytoskeleton has intrinsic polarity
We analyzed the behavior of individual MamK filaments and classified filament growth into three modes: unidirectional, bidirectional, and treadmilling. We visualized the growing processes of the representative filaments as kymographs (Fig. 3A–C) with time courses of each pole position (Fig. 3D–F). In unidirectional growth, only one tip grew, while the other grew slower or not at all (Fig. 3A, D, and Movie S3). In bidirectional growth, both tips grew (Fig. 3B, E, and Movie S4). Treadmilling was defined as filament growth at one tip with regression at the other (Fig. 3C, F, and Movie S5).
AFM allows for the detection of materials that are fixed to a solid-liquid interface on a mica substrate. However, materials existing in a free state in the solution or sliding on the mica surface cannot be visualized in AFM observations. Therefore, the MamK filaments detected in our AFM observations were fixed to the mica surface due to interactions with the mica. Here, we observed the elongation and shrinking of these MamK filaments on the mica. Since the filaments are anchored in place, they cannot move or slide across the surface. The observed elongation and shrinking of the filaments result from the binding of new monomers to the filament tip (polymerization) and the release of monomers from the tip (depolymerization), respectively. The kymographs appear to show the stepwise growth of filaments (Fig. 3A-C); however, this is likely an artifact of the AFM imaging. The tips of the growing filaments became detached from the mica surface, and floating flexible structures unattached to the mica substrate could not be imaged using AFM. These growth dynamics are also illustrated in Supplementary Movies S3-5, which show representative filaments of each growth mode, with plus and minus ends indicated by red and white markers, respectively.
Unidirectional, bidirectional, and treadmilling growth occurred at rates of 70% (n = 66), 27% (n = 25), and 3% (n = 3) in the solution containing 3 µM monomeric MamK (Table 1). On the other hand, in the solution containing 1.5 µM monomeric MamK, unidirectional, bidirectional, and treadmilling growth occurred at 64% (n = 11), 18% (n = 3), and 18% (n = 3), respectively (Table 1). In both MamK concentrations, the three growth mode filaments were observed reproducibly.
We chose filaments that were not attached to the adjacent filaments for analysis of the growth modes and measurement of the tip elongation speeds of both tips (Fig. 3G, H; Table 2). Interestingly, we observed polarity in all three growth patterns, with fast-growing (plus end) and slow-growing or shrinking (minus end) ends. The elongation speeds of the plus ends were similar among the three growth modes. Specially, at 3 µM MamK, the speeds of plus ends were 12.4 ± 4.2, 18.9 ± 7.2, and 14.8 ± 6.6 nm/min in the unidirectional, bidirectional, and treadmilling modes, respectively, while at 1.5 µM MamK, they were 18.8 ± 8.4, 20.6 ± 7.2, and 19.6 ± 5.7 nm/min, respectively (Table 2). There are no large differences among growth speeds in each of the three growth modes and MamK concentrations. In contrast, the elongation speeds of the plus ends at the minus ends were variable among the growth modes. The elongation speeds of the minus ends in the unidirectional, bidirectional, and treadmilling modes were 0.8 ± 0.7, 8.0 ± 3.9, and − 4.4 ± 1.6 nm/min, respectively, at 3 µM MamK, and were 0.8 ± 1.4, 7.6 ± 1.1, and − 8.9 ± 1.9 nm/min respectively, at 1.5 µM MamK (Fig. 3G, H; Table 2). These estimates are based on data obtained from ten independent AFM experiments and suggest polarity in the polymerization of the MamK filaments, which, similar to eukaryotic actin filaments, can be attributed to differences in the critical concentrations at the plus and minus ends of the filaments.
In this AFM study, we simultaneously observed the three filament growth modes within the same imaging area. Given the uniform biochemical properties of MamK molecules, the coexistence of these distinct growth behaviors is likely attributable to the nanoscale heterogeneity in the observation conditions. Eukaryotic actin filaments vary in their polymerization modes depending on the monomer concentration in the solution (Figure S3). This suggests that the MamK monomer concentrations available at each filament tip were not uniform, despite the filaments being located on the same substrate. We observed dense MamK filaments exhibiting random orientations across the mica surface. Such dense packing may have hindered the supply of monomers to the filament ends. Moreover, because AFM only visualizes filaments attached to the mica surface, the solid-liquid interface between the buffer and substrate likely restricted the availability of MamK monomers at the filament tips. Alternatively, the heterogeneity of the growth modes might have resulted from the ATP hydrolysis states of MamK at the tips of individual filaments. The temporal exposure of ADP-form MamK in individual filaments might cause the heterogeneity of the growth modes.
We attempted to differentiate the concentrations of MamK used during AFM imaging to evaluate the impact of monomer concentration on filament growth modes. However, increasing the MamK concentration to 4 µM caused turbidity due to the formation of long MamK polymers or aggregates in the solution, which obstructed AFM imaging. On the other hand, lowering the concentration to 0.1 µM resulted in no detectable polymerized filaments on the mica surface. Moreover, reducing the concentration to 1 µM resulted in no dynamically polymerizing filaments being detected, but static filaments were observed. Given this narrow range of effective MamK concentrations for AFM imaging, we could not thoroughly assess how monomer concentration affects filament growth on the mica substrate.
There was no significant difference in the growth speeds of the filaments between the 1.5 µM and 3 µM conditions (Fig. 3G, H). This could be due to the similar concentration of monomeric MamK near the mica substrate, despite differences in MamK concentration in the solutions. The monomeric MamK might have an affinity for mica, causing it to accumulate on the mica surface. Conversely, the elongation speeds at the plus end were higher at 1.5 µM MamK compared to 3 µM (Table 2). This contradicts the expectation that a higher monomer concentration would result in faster elongation. Because the density of filaments observed on the mica substrates at 1.5 µM was lower than that at 3 µM (Fig. S4), we speculate that at 3 µM MamK, the increased filament density at the solid-liquid interface may structurally hinder filament elongation.
Another possible explanation for the different growth modes was the contamination of the truncated MamK monomer in the sample. A slightly degraded MamK monomer was present in the pellet fraction during the pelleting assay (Figure S2C), likely representing MamK peptides truncated at the N- or C-terminus. Although the abundance of the degraded form was low and unlikely to affect the polarity of most polymerizing filaments observed using AFM, it may have contributed to the different growth modes observed using AFM.
Only six treadmilling filaments were observed under our imaging conditions, indicating that filament disassembly was minimal. This may be due to the relatively high ATP concentration (1 mM) in the P buffer. This finding aligns with the in vitro bulk assembly using light scattering experiments, showing that filament disassembly was undetectable at 2 mM ATP13.
FRAP analyses showed that treadmilling is the primary dynamic mode of MamK filaments in Magnetospirillum cells, even under conditions of high intracellular ATP10. This suggests that additional factors are needed to regulate the dynamics of MamK filaments to maintain treadmilling filaments in MTB cells. Indeed, some magnetosome proteins have been reported to be involved in regulating MamK dynamics in AMB-1 cells. MamJ and LimJ promote the dynamic behavior of MamK filaments in AMB-1 cells as assessed by fluorescence recovery after photobleaching (FRAP) experiments27. McaA and McaB, which create space for new magnetosomes between pre-existing magnetosomes, also influence MamK dynamics28. Therefore, it is essential to understand the roles of these MamK-regulating proteins in the polymerization properties or filament structures of the MamK cytoskeleton at the molecular level. Additionally, MamY senses the curvature of the membrane and recruits the magnetosome chain to the geodetic cell axis29. As the next step, the dynamic interactions between MamK filaments and these magnetosome positioning proteins could be assessed using high-speed AFM imaging, using the method established in this study, to uncover how dynamic cytoskeletal filaments are regulated to achieve the precise positioning of magnetosomes in cells.
In this study, MamK filament polymerization was found to be inherently polar. This polarity contrasts with that of plasmid-encoded bacterial actin ParM, which polymerizes into non-polar filaments that elongate bidirectionally30. ParM filaments form a spindle that pushes plasmids toward opposite poles of the cell, facilitating equal plasmid segregation into daughter cells31. Conversely, unipolar cytoskeletal filaments, such as eukaryotic actin, support directional organelle transport and cellular movement. Therefore, the polarity of MamK filaments appears to be well suited for the directional translocation of magnetosomes. Reported magnetosome chain displacement speeds were approximately 18 and 20 nm/min in MamC-EGFP-expressing MSR-1 cells10 and in an Mms6-GFP-expressing mcaA deletion mutant of AMB-128, respectively. In comparison, MamK filaments observed using high-speed AFM in this study elongated from 12 to 21 nm/min at their fast-growing (plus) ends (Table 2). The velocities of magnetosome displacement in vivo differed insignificantly from that of the MamK elongation. However, FRAP analysis of mCherry-MamK in MSR-1 cells estimated a treadmilling speed of approximately 300 nm/min10, more than 20 times faster than the elongation speed measured using high-speed AFM. This discrepancy suggests that MamK polymerization dynamics are modulated in vivo by regulatory proteins (e.g., MamJ and McaA/B), influencing filament elongation speed, depolymerization, and nucleation. If this speculation is correct, this situation is analogous to how the polymerization properties of eukaryotic actin are regulated by actin-binding protein, which support the functions of the actin cytoskeleton32. Likewise, MamK-regulating proteins may be involved in fine-tuning MamK polymerization to facilitate precise magnetosome positioning in vivo.
In summary, we showed the dynamic polymerization process of the MamK cytoskeletal filaments at high spatial-temporal resolutions under physiological conditions. The MamK filament polymerization is notably characterized by its polarity. The intrinsic polarity of the polymerization of actin-like proteins drives the dynamic behavior of the actin-like cytoskeletal filaments in cells. Therefore, understanding the dynamic properties of the MamK cytoskeletal filaments is key to elucidating their function in magnetosome positioning. This work found conformable dynamic features between bacterial organelle-positioning MamK filaments and eukaryotic organelle-associated actin filaments. This insight provides a clue to understanding how bacteria have evolved actin-like cytoskeletal elements to support their organelle functions.
Methods
Bacterial strains and cultures
The Escherichia coli strains, XL-1 blue MRFʹ (Agilent Technologies, USA), and C41(DE3)33 were cultivated in LB broth at either 37℃ or 30℃, as described below. Whenever necessary, the antibiotic kanamycin was added at 20 µg/ml.
Purification of monomeric MamK protein
To generate the non-tagged MamK expression vector (pET-mamK_stop) for E. coli, the mamK gene, including its stop codon, was amplified from the AMB-1 genome using the primers mamK_inf_F (GAAGGAGATATACATATGAGTGAAGGTGAAGGCCAGGCC) and mamK_stop-inf_R (CACCAGGGTACCTCACGAGCCGGAGACGTCTCCAAGC). The amplified DNA fragment was inserted between the NdeI and KpnI sites of pET29b (Merck Millipore [Novagen], USA) using infusion cloning (Takara Bio, Japan). E. coli C41(DE3)(pET-mamK_stop) was grown at 30℃ to an A600nm of about 0.6, at which point MamK expression was induced with 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Nakarai Tesque, Japan) for 5 h. The cells were harvested by centrifugation at 8,000 × g for 15 min.
All purification steps were conducted at 4℃ unless indicated otherwise. The harvested E. coli cells (ca. 10 g in wet weight) were suspended in 100 ml of lysis buffer (10 mM Tris-HCl [pH 7.4], 25 mM KCl, 2 mM EDTA, and 1 mM DTT) and lysed by three passages through a French pressure cell (100 MPa). Cell debris was removed by centrifugation at 8,000 × g for 15 min, and the obtained supernatant was fractionated with ammonium sulfate at 20% saturation. The resulting precipitates were washed twice in lysis buffer containing 20% saturated ammonium sulfate. Then, the obtained precipitate was suspended in 50 ml of alkaline depolymerization (DP) buffer (10 mM CAPS [pH 9.6], 10 mM EDTA, and 2 mM DTT) and dialyzed thrice in 1 L of DP buffer for approximately 8 h. The sample solution was transparent during the dialysis due to MamK depolymerization. The resulting sample was ultracentrifuged at 100,000 × g for 1 h to remove the MamK polymers, aggregates, and other cellular compartments. The obtained supernatant was subjected to ion exchange chromatography (DEAE TOYOPEARL, 2.3 cm × 12 cm, Tosoh, Japan). The column was equilibrated with DP buffer. The MamK fraction was eluted using DP buffer containing 50 mM NaCl and concentrated in a dialysis tube using Aquacide III (Merck Millipore, USA). The concentrated MamK fraction was dialyzed in 1 L PB buffer for 12 h, ultracentrifuged at 100,000 × g for 1 h to remove aggregates, and then further purified by gel filtration (Superdex 200 Increase 10/300 GL, Cytiva, USA) using DP buffer for equilibration. The monomeric MamK fractions were pooled, and aliquots were stored at − 80℃ (Figures S2 and S5). Immediately before use in the polymerization experiments, the buffer was exchanged for a polymerization buffer (P buffer: 10 mM Tris-HCl [pH 7.5], 25 mM KCl, 2 mM MgCl2, and 1 mM DTT) using a desalting spin column (Zaba Spin Desalting Column, Thermo Fisher, USA). The desalted sample was used within a few hours.
Determination of the MamK depolymerization pH
Aliquots of the ammonium sulfate precipitation solution (20% saturation) of the cell-free extract from E. coli expressing MamK were suspended in 3 ml of each of the following buffers: 10 mM Tris-HCl (for pH 8.0, 8.5, and 9.0) or 10 mM CAPS-NaOH (for pH 9.5, 10.0, and 10.5), each containing 10 mM EDTA and 2 mM DTT. The suspension was dialyzed with 500 ml of each buffer for 12 h. The dialyzed samples were ultracentrifuged at 150,000 × g for 1 h. The MamK contents in the resulting precipitate and supernatant were quantified using SDS-PAGE (Figures S1 and S6). The proteins were stained with Coomassie brilliant blue (CBB), and the protein band intensities were quantified by image analysis using LAS3000 (Fujifilm, Japan) as implemented within the analysis software MultiGauge (GE Healthcare, USA).
High-speed AFM observations
Imaging was performed with a laboratory-built, tapping mode, high-speed AFM16. The tapping setup comprised a small silicon nitride cantilever (BL-AC10-DS, Olympus, Japan) with a spring constant k of ∼0.1 N/m and a resonance frequency f of ∼400–500 kHz in water. The free-oscillation peak-to-peak amplitude of the cantilever was set to 2 nm, and the amplitude set point was 1.6–1.8 nm. The tapping force was optimized to minimize damage to the MamK filaments. The AFM carbon styli were grown on each cantilever using an electron beam lithography system (ELS-7500UK, Elionix, Japan) with a deposition time of 60 s.
AFM observations of the MamK polymers were performed at room temperature on freshly prepared mica. The mica surface was silanized by depositing 3 µl of 0.2% 3-aminopropyltriethoxysilane (APTES) (Shin-Etsu Chemical, Japan) solution onto its surface. After silanization, the mica surface was washed three times with 20 µl ultrapure water, followed by three washes with 20 µl P buffer. MamK filaments were prepared by adding 1 mM ATP to a solution containing 1 µM monomeric MamK in polymerization buffer. The mixture was incubated for 10 min at room temperature. The resulting solution containing the MamK filaments was then applied to the APTES-treated mica and incubated for 5 min. After incubation, excess solution was removed by washing the mica three times with 20 µl P buffer containing 1 mM ATP. P buffer containing 1 mM ATP was used as the immersive imaging buffer. AFM images were recorded at 1 s/frame in a scanning area of 200 × 200 nm with a resolution of 200 × 200 pixels.
The MamK polymerization process was observed by loading 65 µl of 1.5 µM or 3 µM monomeric MamK in the polymerization buffer into a high-speed AFM sample chamber. The bare mica disc fitted on the AFM scanner was incubated in the corresponding samples. After confirming the accumulation of monomeric MamK on the mica using AFM imaging, we added to the sample chamber ATP to a final concentration of 1 mM. Successive AFM videos were taken for about 30 min at 10 s/frames in a scanning area of 800 × 800 nm with a resolution of 400 × 400 pixels.
Phosphate release assay
The production of inorganic phosphate by MamK polymerization was evaluated using a malachite green-sodium molybdate assay (Malachite Green Phosphate Assay Kit, BioAssay Systems, USA). The polymerization of MamK (3.03 µM) was initiated by adding 2 mM ATP at 25℃. The reaction was quenched with one volume of cold 0.6 M perchloric acid and stored on ice until all time points were collected34. The phosphate contents in the sample were quantified according to the manufacturer’s instructions. The absorbance was measured at 620 nm using a microplate photometer (Multiskan FC, Thermo Fisher Scientific, USA). The curves were normalized with a phosphate standard and a no-protein control.
Pelleting assay
To assess MamK polymerization, we mixed MamK with 1 mM ATP or ADP with P buffer and incubated the mixture at 28℃ for 10 min. The samples were then spun at 100,000 × g at room temperature for 1 h. The supernatant and pellet fractions were recovered and analyzed by SDS-PAGE (Figures S2 and S6). The MamK band intensities from both fractions were quantified using Multi Gauge v3.0 software (Fujifilm, Japan).
Physical and chemical measurements
The protein contents of MamK were assayed using Quick Start Bradford 1× Dye Reagent (Bio-Rad, USA). Spectrophotometric measurements were performed using a UV-2550 spectrophotometer (Shimadzu, Japan) with a 1-cm light path cuvette. SDS-PAGE was performed according to the methods of Laemmli35. Protein bands were stained with Coomassie brilliant blue G-250 (SERVA Blue G, Serva Electrophoresis GmbH, Germany).
Data availability
All data supporting this study’s findings are available in the paper and its supplementary information files.
Change history
26 January 2026
A Correction to this paper has been published: https://doi.org/10.1038/s41598-026-37236-0
References
Shaevitz, J. W. & Gitai, Z. The structure and function of bacterial actin homologs. Cold Spring Harb Perspect. Biol. 2, a000364. https://doi.org/10.1101/cshperspect.a000364 (2010).
Ozyamak, E., Kollman, J. M. & Komeili, A. Bacterial Actins their Divers. Biochemistry 52, 6928–6939 ; https://doi.org/10.1021/bi4010792 (2013).
Wagstaff, J. & Löwe, J. Prokaryotic cytoskeletons: protein filaments organizing small cells. Nat. Rev. Microbiol. 16, 187–201. https://doi.org/10.1038/nrmicro.2017.153 (2018).
Komeili, A., Li, Z., Newman, D. K. & Jensen, G. J. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311, 242–245. https://doi.org/10.1126/science.1123231 (2006).
Scheffel, A. et al. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440, 110–114. https://doi.org/10.1038/nature04382 (2006).
Uebe, R. & Schüler, D. Magnetosome biogenesis in magnetotactic bacteria. Nat. Rev. Microbiol. 14, 621–637. https://doi.org/10.1038/nrmicro.2016.99 (2016).
Müller, F. D., Schüler, D. & Pfeiffer, D. A compass to boost navigation: cell biology of bacterial magnetotaxis. J. Bacteriol. 202, 10–1128. 10.1128/ (2020). JB.00398 – 20.
Cornejo, E., Abreu, N. & Komeili, A. Compartmentalization and organelle formation in bacteria. Curr. Opin. Cell. Biol. 26, 132–138. https://doi.org/10.1016/j.ceb.2013.12.007 (2014).
Bazylinski, D. A. & Frankel, R. B. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2, 217–230. https://doi.org/10.1038/nrmicro842 (2004).
Toro-Nahuelpan, M. et al. Segregation of prokaryotic magnetosomes organelles is driven by treadmilling of a dynamic actin-like MamK filament. BMC Biol. 14, 88. https://doi.org/10.1186/s12915-016-0290-1 (2016).
Taoka, A. et al. Tethered magnets are the key to magnetotaxis: direct observations of magnetospirillum magneticum AMB-1 show that MamK distributes magnetosome organelles equally to daughter cells. mBio 8 (e00679- e00617). (2017).
Taoka, A., Asada, R., Wu, L. F. & Fukumori, Y. Polymerization of the actin-like protein MamK, which is associated with magnetosomes. J. Bacteriol. 189, 8737–8740. https://doi.org/10.1128/JB.00899-07 (2007).
Ozyamak, E., Kollman, J., Agard, D. A. & Komeili, A. The bacterial actin mamk: in vitro assembly behavior and filament architecture. J. Biol. Chem. 288, 4265–4277. https://doi.org/10.1074/jbc.M112.417030 (2013).
Sonkaria, S. et al. Insight into the assembly properties and functional organisation of the magnetotactic bacterial actin-like homolog, MamK. PLoS One. 7, e34189. https://doi.org/10.1371/journal.pone.0034189 (2012).
Deng, A. et al. In vitro assembly of the bacterial actin protein MamK from ‘Candidatus magnetobacterium casensis’ in the phylum Nitrospirae. Protein Cell. 7, 267–280. https://doi.org/10.1007/s13238-016-0253-x (2016).
Uchihashi, T., Kodera, N. & Ando, T. Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 7, 1193–1206. https://doi.org/10.1038/nprot.2012.047 (2012).
Ando, T., Uchihashi, T. & Scheuring, S. Filming biomolecular processes by high-speed atomic force microscopy. Chem. Rev. 114, 3120–3188. https://doi.org/10.1021/cr4003837 (2014).
Xiao, J. & Dufrêne, Y. F. Optical and force nanoscopy in microbiology. Nat. Microbiol. 1, 16186. https://doi.org/10.1038/nmicrobiol.2016.186 (2016).
Ando, T., Fukuda, S., Ngo, K. X. & Flechsig, H. High-speed atomic force microscopy for filming protein molecules in dynamic action. Annu. Rev. Biophys. 53, 19–39. https://doi.org/10.1146/annurev-biophys-030722-113353 (2024).
Kodera, N., Yamamoto, D., Ishikawa, R. & Ando, T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 468, 72–76. https://doi.org/10.1038/nature09450 (2010).
Ngo, K. X., Kodera, N., Katayama, E., Ando, T. & Uyeda, T. Q. Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife 4, e04806. https://doi.org/10.7554/eLife.04806 (2015).
Watanabe-Nakayama, T. et al. High-speed atomic force microscopy reveals structural dynamics of amyloid beta1-42 aggregates. Proc. Natl. Acad. Sci. U. S. A. 113, 5835–5840; (2016). https://doi.org/10.1073/pnas.1524807113
Hamon, L. et al. Mica surface promotes the assembly of cytoskeletal proteins. Langmuir 25, 3331–3335. https://doi.org/10.1021/la8035743 (2009).
Mateos-Gil, P. et al. Depolymerization dynamics of individual filaments of bacterial cytoskeletal protein FtsZ. Proc. Natl. Acad. Sci. U S A. 109, 8133–8138. https://doi.org/10.1073/pnas.1204844109 (2012).
Löwe, J., He, S., Scheres, S. H. & Savva, C. G. X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization. Proc. Natl. Acad. Sci. U. S. A. 113, 13396–13401; (2016). https://doi.org/10.1073/pnas.1612034113
Bergeron, J. R. et al. Structure of the magnetosome-associated actin-like MamK filament at subnanometer resolution. Protein Sci. 26, 93–102. https://doi.org/10.1002/pro.2979 (2017).
Draper, O. et al. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol. Microbiol. 82, 342–354. https://doi.org/10.1111/j.1365-2958.2011.07815.x (2011).
Wan, J. et al. McaA and McaB control the dynamic positioning of a bacterial magnetic organelle. Nat. Commun. 13, 5652. https://doi.org/10.1038/s41467-022-32914-9 (2022).
Toro-Nahuelpan, M. et al. MamY is a membrane-bound protein that aligns magnetosomes and the motility axis of helical magnetotactic bacteria. Nat. Microbiol. 4, 1978–1989. https://doi.org/10.1038/s41564-019-0512-8 (2019).
Garner, E. C., Campbell, C. S. & Mullins, R. D. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306, 1021–1025. https://doi.org/10.1126/science.1101313 (2004). (2004).
Garner, E. C., Campbell, C. S., Weibel, D. B. & Mullins, R. D. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315, 1270–1274. https://doi.org/10.1126/science.1138527 (2007).
Alberts, B. et al. The Cytoskeleton in Molecular Biology of The Cell (Senventh Edition) 949–1025 (W. W. Norton & Company, (2022).
Miroux, B. & Walker, J. E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298. https://doi.org/10.1006/jmbi.1996.0399 (1996).
Esue, O., Cordero, M., Wirtz, D. & Tseng, Y. The assembly of MreB, a prokaryotic homolog of actin. J. Biol. Chem. 280, 2628–2635. https://doi.org/10.1074/jbc.M410298200 (2005).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. https://doi.org/10.1038/227680a0 (1970).
Acknowledgements
This work was supported by JSPS KAKENHI 23K26816, 21K19071, 21KK0126, 19H02868, and 24117007. This work was supported by JST SPRING, Grant Number JPMJSP2135.
Author information
Authors and Affiliations
Contributions
Y.P., Y.K., Y.F., and A.T. designed the experiments; Y.P., Y.K., T.S., and A.T. performed the experiments; Y.K., T.S., and A.T. analyzed the data; Y.P., Y.K., and A.T. wrote the paper; and A.T. supervised the study. Y.P., and Y.K. contributed equally. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original PDF version of this Article contained errors in Figure 1. Full information regarding the corrections made can be found in the correction for this Article.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary Material 2
Supplementary Material 3
Supplementary Material 4
Supplementary Material 5
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pan, Y., Kikuchi, Y., Saito, T. et al. Visualizing the dynamic polymerization of the bacterial actin-like cytoskeleton for magnetic organelle positioning. Sci Rep 15, 44508 (2025). https://doi.org/10.1038/s41598-025-28026-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-28026-1