Abstract
The identification of structural variant (SV) breakpoints plays a crucial role in understanding the genetic variants, mutagenic mechanisms, and functional consequences that drive various genetic diseases. While next-generation sequencing (NGS) has become a cornerstone in single nucleotide variant (SNP) discovery and characterization, short-read NGS technology faces significant challenges in resolving large genomic rearrangements such as duplications, deletions, inversions, and translocations. Nanopore sequencing offers a promising alternative by enabling precise mapping of chromosomal rearrangement breakpoints, and characterization of chromosomal alterations, thereby improving the genetic diagnosis of such conditions. Using long-read whole-genome sequencing, we examined the breakpoints of a cytogenetically balanced chromosomal translocation, t(8;22)(q13.3;q11.23), initially detected during prenatal diagnosis and later confirmed as de novo in a patient who developed NF2-associated schwannomatosis in late infancy. Nanopore sequencing revealed that the translocation disrupted the NF2 gene. This case highlights the power of nanopore long-read sequencing in detecting the exact consequences of de novo, apparently balanced translocations and in uncovering the genetic underpinnings of abnormal phenotypes. Given its ability to resolve complex SVs with high precision, nanopore sequencing might be considered a valuable complement to conventional genetic diagnostic methods, enhancing our understanding of genetic diseases and potentially improving diagnostic yield and risk assessment.
Similar content being viewed by others
Introduction
Balanced chromosomal abnormalities represent the most prevalent class of genomic SVs with an estimated population frequency ranging from 0.2 to 0.4%1,2. Notably, the occurrence of congenital abnormalities or intellectual disabilities in newborns harboring a de novo balanced chromosomal abnormality is approximately double that observed in the general population, specifically 6.1% for reciprocal translocation and 9.4% for inversions3. These findings strongly imply a causal relationship between balanced chromosomal abnormalities and the manifestation of clinical phenotypes.
The identification of de novo balanced chromosomal abnormalities during prenatal diagnosis often introduces significant uncertainty in genetic counseling, as their phenotypic consequences remain unpredictable3,4. These abnormalities typically involve structural rearrangements such as translocations or inversions, which can disrupt haploinsufficient genes or regulatory regions critical for normal development5,6,7. However, the precise mapping of these breakpoints has historically been challenging due to limitations in resolution and accuracy of conventional cytogenetic techniques. Recent advancements in long-read sequencing technologies markedly improved the ability to detect and characterize these SVs with high precision8 For instance, single-tube long fragment read (stLFR) whole-genome sequencing has demonstrated efficacy in identifying cryptic chromosomal rearrangements that were previously undetectable by standard methods9. This approach enables the resolution of complex SVs and the accurate delineation of breakpoint regions, facilitating a more comprehensive understanding of their potential impact on the phenotype.
Neurofibromatosis type 2 (NF2), also known as NF2-related schwannomatosis, is a hereditary tumor syndrome predisposing to the development of bilateral vestibular schwannomas, schwannomas in other central and peripheral locations, meningiomas and ependymomas10. This condition is caused by inactivating variants in the NF2 tumor-suppressor gene located at 22q12. These variants typically include single nucleotide variations or larger structural rearrangements, such as deletions affecting single or multiple exons, or the entire gene11,12,13. Several cases associated with chromosomal abnormalities have been reported, including the presence of a ring chromosome 2214,15,16,17,18,19, as well as translocations involving the long arm of chromosome 2215,20.
In NF2-related schwannomatosis, conventional methods such as Fluorescence In Situ Hybridization (FISH), Southern blotting, and targeted sequencing have been used to refine chromosome 22 translocation breakpoints21,22,23. but generally failed to achieve nucleotide-level resolution, especially in repetitive regions. This limitation underscores the need for long-read genome sequencing to precisely resolve balanced translocations and define their functional impact.
In the present case study, we employed long-read nanopore sequencing to investigate the genetic basis of NF2-related schwannomatosis in a patient with a de novo balanced translocation between chromosomes 8 and 22, t(8;22)(q13.3;q11.23), identified during prenatal diagnosis. This approach enabled precise breakpoint mapping, revealing that the translocation disrupted the NF2 gene, thereby elucidating the molecular etiology of the patient’s condition. Our findings underscore the utility of long-read sequencing technologies in resolving complex SVs and enhancing the accuracy of genetic diagnoses in the clinical setting.
Methods
Genomic DNA (gDNA) sequencing
The quality, quantity, and integrity of the extracted gDNA were assessed using the Qubit dsDNA BR Assay Kit (Invitrogen by Thermo Fisher Scientific, USA, Q32854/1) and the 4150 TapeStation System (Agilent, G2992AA) with genomic DNA ScreenTape Analysis kit (Genomic DNA Reagents 5067–5366 and Genomic DNA ScreenTape 5067–5365 Agilent).Before starting the library preparation, the gDNA were purified using 0.4–0.6X AMPure XP Beads (Beckman Coulter A63881, USA) to achieve an optimal fragment size distribution for long-read sequencing. Library preparation was performed using the Ligation Sequencing Kit V14 (SQK-LSK114, Oxford Nanopore Technologies, UK) following the manufacturer’s protocol. Briefly, 3 µg of fragmented gDNA were subjected to DNA repair and end-preparation using the NEBNext® FFPE DNA Repair Mix and Ultra II End Prep Enzyme Mix from the NEBNext® Companion Module v2 for Oxford Nanopore Technologies® Ligation Sequencing (NEB, Euroclone USA, E7672). Sequencing adapters were then ligated to the end-prepped DNA using the Ligation Adapter (LA) and Salt-T4 DNA Ligase (NEB). Adapter-ligated libraries were purified using AMPure XP Beads (AXP). The concentration of the adapter-ligated library was quantified using the Qubit dsDNA HS Assay Kit (Invitrogen- Thermo Fisher Scientific, USA, Q32854/1), which ensured a yield of 1200–1800 ng in a 96 µl volume. The libraries were loaded onto R10.4.1 flow cells (FLO-PRO114M) and sequenced on a PromethION 24/48 device (Oxford Nanopore Technologies). To maximize data output, the flow cells were washed and reloaded twice using the Flow Cell Wash Kit (EXP-WSH004, Oxford Nanopore Technologies, UK). The first wash was conducted approximately after 22 h, and the second approximately after 44 h of sequencing. MinKnow software (version 24.11.8) was used for data acquisition and super-high accuracy basecalling, with the quality score set to 10.
Data analysis
A quality report of the sequencing run was generated using NanoPlot (version 1.43.0,)24. Long reads were first filtered using Filtlong (version 0.2.1, https://github.com/rrwick/Filtlong), removing reads shorter than 1000 base pairs. The filtered data were then aligned to the human reference genome hg38 (GRCh38) using Minimap2 (version 2.28, https://github.com/lh3/minimap2) with the “-ax map-ont” argument and the FASTA reference file GCA_000001405.15. Subsequently, SAM files were converted to BAM format using Samtools (version 1.21, https://doi.org/10.1093/gigascience/giab008)25, and the resulting BAM files were sorted by chromosome order. Coverage of the sequencing run was calculated using Mosdepth (version 0.3.1026, . Variant calling was initially performed using Sniffles2 (version 2.6.127, and SVIM (version 2.0.0)28. Deletion variants in EXOC4, TOP3B, and GPHN were further confirmed using Nano-GLADIATOR29 using a window size of 10 kb and 100 kb. Variants were subsequently annotated using SnpEff (version 5.1d, https://pcingola.github.io/SnpEff/) based on the GRCh38 genome build. Finally, alignments and breakpoints detected by Sniffles2 were visualised using the Integrative Genomics Viewer (IGV, version 2.19.2, https://software.broadinstitute.org/software/igv/) and Ribbon (version 1.1, https://genomeribbon.com ).
Sanger sequencing of translocation breakpoints
To amplify the expected translocation breakpoints, we designed forward and reverse primer pairs, respectively: i) on chromosome 8 (chr8-F:5’- CAAGTGGAGTGATAGCGTGC − 3’) and NF2-intron 4 on chromosome 22 (chr22-R:5’- CCAGACACAAAAGGCCACAT- 3’), and ii) on chromosome 22 NF2-intron 4 (chr22-F:5’- GGAATCTCTGCACACACATCC-3’) and chromosome 8 (chr8-R:5’- CTCAAACACCAAGGGAGAAAGA-3’). PCR reactions were performed using the HotStarTaq DNA Polymerase (Qiagen, Hilden, Germany) and PCR products were sequenced by Sanger sequencing. Briefly, purified PCR products were sequenced on SeqStudio Genetic Analyzer sequencer (Applied Biosystems) using BigDye Terminator Chemistry (Thermo Fisher Scientific) according to the manufacturers recommendations. The results were analyzed with the Sequencing Analysis Software and Sequence Scanner Software v2.6 (Applied Biosystems).
Array-Comparative genomic hybridization
Chromosomal microarray (CMA) was performed using a SurePrint G3 Agilent Human Genome CGH Microarray 4 × 180k (Agilent Technologies, Santa Clara, California, USA). This platform is a high-resolution oligonucleotide-based microarray containing 180,000 CGH probes with a median spacing of 13 kb. Labelling and hybridization were performed following the protocols provided by Agilent: 800 ng of purified DNA of the patient and of a control DNA of the same sex (Agilent) were labelled for 2 h, minimizing light exposure, using the Agilent Genomic DNA Labelling Kit, using Cy5-dUTP for the patient DNA and Cy3-dUTP for the reference DNA. Labelled products were column purified (Agilent) and prepared combining test and control sample according to the Agilent protocol. After probe denaturation and pre-annealing with 50 µg of Human Cot-1 DNA (Agilent), hybridization was performed at 67 °C for 24 h in a rotating oven at 20 rpm. After two washing steps, the array slide was scanned with the SureScan DX Microarray Scanner (Agilent). The spot intensities were measured and the image files quantified using the Agilent Feature Extraction 12.1.1.1 software. Text outputs from the quantitative analyses were imported into Genomic Workbench Standard Edition 7.0.4.0 software (Agilent Technologies). Breakpoint positions were reported according to hg 19, build 37.
Results
Case presentation and preliminary analyses
A 15-year-old female was referred to the genetics clinic of the Careggi University Hospital, Florence, following a recent diagnosis of NF2-associated schwannomatosis. The patient presented with a two-month history of frontal headaches, nausea, and instability.
Magnetic resonance imaging (MRI) scans revealed multiple heterogeneously enhancing lesions, including bilateral masses in the cerebellopontine angles, a suspected ependymoma at the medulla oblongata-spinal cord junction, and a falx cerebri meningioma (Fig. 1a). Spinal MRI identified additional lesions at C3, C6, C7, C8, bilaterally in T1 and within the cauda equina. The patient underwent subtotal resection of the left vestibular schwannoma, with residual facial nerve paresis.
(a) MRI brain scan of the patient, showing multiple meningiomas (from left to right: axial, coronal and sagittal projections). (b) MLPA analysis of the NF2 gene: CNV analysis by Amplicon Suite software shows no evidence of rearrangements. (c) Array-CGH analysis of chromosomes 8 and 22 with SurePrint G3 Agilent Human Genome CGH Microarray 4 × 180k (Agilent Technologies, Santa Clara, California, USA). The 8q13.3 and 22q11.23 regions (highlighted in blue) do not display deletions or duplication.
Patient’s family history was unremarkable. At the time of her birth, the father was 50 and mother 39 years old. Due to advanced maternal age, an amniocentesis was performed at 16 weeks of gestation, revealing a fetal karyotype with an apparently balanced translocation: 46,XX, t(8;22)(q13.3;q11.23). Both parents showed normal karyotypes.
Genetic analysis of lymphocyte DNA for NF2 single-nucleotide variantswas performed using High Resolution Melting (HRM) of the coding regions and adjacent splice donor and acceptor sites, with no pathogenic variants identified. Multiplex Ligation-dependent Probe Amplification (MLPA) analysis also failed to detect any genomic rearrangements involving the NF2 gene (Fig. 1b).
The patient was subsequently lost to follow-up but re-presented at 31 years of age. Medical records indicated that, at 18 years old, she underwent sequential surgical resection of bilateral vestibular schwannomas and a foramen magnum meningioma over an 8-month period. These interventions resulted in bilateral hearing loss and facial nerve paresis. At 28 years of age, a treatment with bevacizumab was initiated and continued for two years. At 31, the brain and spinal MRI showed multiple intracranial meningiomas, several cervical spine ependymomas and numerous schwannomas affecting the cauda equina. Given the progressive worsening of instability, headaches, and visual symptoms, she underwent gamma knife stereotactic radiotherapy targeting the meningiomas located at the clinoid processes. Currently, the patient exhibits severely impaired visual acuity, bilateral deafness, and marked instability.
To confirm the absence of deletions involving the NF2 gene, an array-Comparative Genomic Hybridization analysis (array-CGH) with an average resolution of about 40 kb (Agilent Technologies 180 K) was performed. Although aCGH detected no deletion at 8q13.3 or 22q11.23 (Fig. 1c), it identified a heterozygous deletion of chromosome 22 involving TOP3B interpreted as a benign Copy Number Variation (CNV) given the high frequency in the Database of Genomic Variants (DGV: https://dgv.tcag.ca/dgv/app/home) and in gnomAD-Structural-variant database (https://gnomad.broadinstitute.org/gene/ENSG00000100038?dataset=gnomad_cnv_r4), of deletions involving the gene: 1.13% and 0.07%, respectively. Moreover, the aCGH identified two small deletions on chromosomes 7 and 14 involving, respectively, EXOC4 and GPHN that were evaluated as variant of uncertain significance. Parental DNA was not available to assess their possible de novo origin. The identified CNVs appeared to be incidental findings with no clear pathogenic relevance to the primary condition under investigation.
Nanopore long-reads sequencing identifies the translocation breakpoint in NF2.
To identify the breakpoints of chromosome 8 and chromosome 22, the gDNA library was constructed and sequenced on the nanopore PromethION platform. We performed a 72-hours MinKNOWN sequencing run following standard protocols. The run yielded a total of 3.63 million reads, of which 1,171,900 were discarded during Dorado basecalling due to low quality scores (< Q10). The remaining 2,458,100 reads had a median length of 11,296 base pairs, an N50 of 23,417 base pairs, and a total yield of 43.99 Gbases. Alignment to the GRCh38 (GCA_000001405.15) reference genome resulted in a mean coverage of 14.22X, with chromosome-specific coverages of 14.64X for chromosome 8, and 11.83X for chromosome 22.
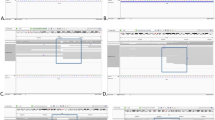
To investigate the previously reported translocation, we analyzed the aligned reads using long-read-specific SVs callers Sniffles2 and SVIM. Both tools identified a break end (BND) event involving positions 29,648,858 on chromosome 22 and 69,290,900 on chromosome 8 (Fig. 2a–c). Sniffles2 reported this BND with 8 supporting reads, and breakpoint coverage remained stable at around 15×, except for a drop to 9× at the start of the duplication. SVIM reported a similar result with 9 supporting reads (Fig. 2a–c). The coverage was consistent with previously reported thresholds for reliable detection of balanced SVs30.
(a) Integrated Genome Viewer (IGV) visualization of DNA long-read nanopore sequencing data showing genome breakpoints on chromosomes 8 and 22. (b) JBrowse2 Breakpoint Split View visualization of translocation on chromosomes 8 and 22. (c) Ribbon Genome Viewer visualization of long-read nanopore sequencing data identifies the translocation event. The chimeric reads between chromosome 8 and chromosome 22 are clearly evident. (d) PCR amplification shows a specific band of approximately 500 bp detected only in the proband’s DNA using specific primers for the derivative chromosomes 8 [der(8)] and 22 [der(22)]. Bands are absent in control DNA. (e) Schematic representation of chromosome structure variation according to nanopore sequencing results. Sanger sequencing of the 500 bp PCR products identifies the translocation breakpoints on derivative chromosomes 8 (top) and 22 (bottom). Dotted lines identify the junctions between the sequences of the two chromosomes.
To validate the translocation breakpoints of the proband, PCR amplification followed by Sanger sequencing was performed. We confirmed the breakpoints on chromosome 8 (GRCh38):g.69,290,900 and chromosome 22 (GRCh38):g.29,648,858 (Fig. 2c). The breakpoint on chromosome 8 was in an intergenic region between the LINC01592 and SULF1genes, whereas the breakpoint on chromosome 22 was mapped to intron 4 of the NF2 gene (Fig. 2d, Suppl Fig. 1a). These results confirm that, although detecting SVs, particularly in low-complexity31 or repetitive regions32, remains challenging, long-read nanopore sequencing effectively overcomes these limitations even with a low coverage.
Comparative detection of microdeletions by Array-CGH and Nanopore-Based approaches
Although array-CGH analysis did not detect deletions at 8q13.3 and 22q11.23, it revealed the presence of three small interstitial heterozygous deletions: (1) a ~ 69 Kb deletion involving the EXOC4 gene (arr[GRCh37] 7q33(133,483,238–133,552,629)x1); (2) a ~ 75 Kb deletion involving the GPHN gene (arr[GRCh37] 14q23.3(67,044,478–67,119,215)x1); and (3) a ~ 257 Kb deletion involving the TOP3B gene (arr[GRCh37] 22q11.22(22,313,128–22,569,881)x1) (Fig. 3a).
(a) Result of array-CGH analysis of chromosomes 8 and 22 with SurePrint G3 Agilent Human Genome CGH Microarray 4 × 180k (Agilent Technologies, Santa Clara, California, USA). Small deletions of 1.15 Mb, 460 Kb and 549 Kb have been identified respectively on chromosomes 7, 14 and 22. (b) Results of NanoGladiator CNV analysis in the genomic windows overlapping the deleted genes of interest.
To assess whether nanopore sequencing could also detect these small deletions identified by array-CGH, we applied multiple bioinformatic approaches. Sniffles2 successfully confirmed the deletions in the EXOC4 and GPHN. The EXOC4 deletion (chr7:133,797,907–133,871,892) was supported by 7 reads, with read depth across the deleted region dropping to roughly half the normal coverage (range ~ 4–19×) and a variant allele frequency (VAF) of 1.0. The GPHN deletion (chr14:66,577,693–66,658,868) was supported by 6 reads, with coverage reduced within the deleted interval (range ~ 5–17×) and a VAF of 0.857. SVIM provided consistent results for both deletions.
However, neither Sniffles2 nor SVIM detected the TOP3B deletion with sufficient read support. Only Sniffles2 reported this deletion, supported by a single read, which was not considered reliable. To further investigate and validate the deletion, we employed Nano-GLADIATOR, a CNV-specific analysis tool, using 10 kb and 100 kb window sizes. Nano-GLADIATOR successfully identified the TOP3B deletion, reporting a log2 ratio of − 1.14 and a copy number (CN) of 1.05 with the 100 kb window; similar results were obtained using the 10 kb window (log2 ratio: − 1.13, CN: 1.05). Nano-GLADIATOR also confirmed the deletions in EXOC4 and GPHN, thereby validating the array-CGH findings (Fig. 3b).
Discussion
In this study, we precisely mapped the breakpoints of a de novo balanced translocation, t(8;22)(q13.3;q11.23), using long-read nanopore sequencing in a patient diagnosed with NF2-associated schwannomatosis during adolescence, following the prenatal identification of the chromosomal rearrangement. To our knowledge, this is the first reported case of NF2-associated schwannomatosis in which a de novo balanced translocation has been molecularly characterized using nanopore sequencing.
Balanced de novo translocations identified in symptomatic individuals are frequently assumed to disrupt or delete disease-associated gene at the breakpoint regions. Following the clinical diagnosis of NF2-associated schwannomatosis in our proband, comprehensive molecular analyses, including HRM, MLPA, and high-resolution 180 K array-CGH failed to detect pathogenic variants or deletions within the NF2 gene. We hypothesized that the translocation breakpoint on chromosome 22 disrupted the NF2 gene, thereby contributing to the proband’s disease phenotype. This disruption likely caused heterozygous inactivation of the gene in all cells. As a consequence, the patient became predisposed to tumor development following the somatic loss of the second NF2 allele.
Despite several attempts to identify the translocation breakpoints involving the NF2 gene, including the use of fluorescence in situ hybridization (FISH), Southern analysis and DNA sequencing20,33,34, no significant progress has been made to date, even with the advent of short-read NGS. Long-read sequencing platforms, which offer superior resolution of SV breakpoints35,36, including translocations37,38, are increasingly recognized as the most effective approach for such analyses. However, their application in NF2-related schwannomatosis remains limited. To date, only a single study has reported the use of long-read whole-genome sequencing (WGS) in this context. Using PacBio sequencing, were identified SVs within the chromosome 22 region in a family affected by NF2-schwannomatosis previously undetected with short-read WGS. These included breakpoints involving NCF4 and PRR5, although their contribution to the disease phenotype remains uncertain39.
Microhomology was identified at translocation junctions: each breakpoint on chromosomes 8 and 22 is flanked by an identical 5‑bp motif, GGATT, strongly implicating microhomology-mediated mechanism. Such signatures are indicative of the involvement of microhomology-mediated end joining (MMEJ), homologous recombination or microhomology-mediated replication-based processes, such as microhomology-mediated break-induced replication (MMBIR) consistent with previously characterized mutational pathways that employ short homologous sequences to mediate double‑strand break repair and SV formation40,41,42.
This case highlights the value of integrating long-read sequencing into standard human genetic diagnostics, offering nucleotide-level resolution of structural breakpoints. Although traditional approaches, such as karyotyping combined with chromosomal microarray analysis, may detect balanced SVs, they do not allow nucleotide-level mapping of breakpoints and thereby fail to resolve whether rearrangements disrupt coding sequences, regulatory elements, or other genomic features. Long‑read sequencing overcomes this limitation by providing precise breakpoint definitions, which are essential for interpreting the functional impact of SVs43,44,45,46,47.
Although our analysis is retrospective, the detailed delineation of translocation breakpoints aligns with accumulating evidence that high-resolution breakpoint mapping can significantly enhance phenotype interpretation in balanced chromosomal rearrangements48,49,50,51 .In addition, defining breakpoints with base‑pair accuracy holds practical relevance for reproductive medicine, enabling targeted assessment in assisted reproductive technologies. Moreover, considering the ongoing improvements in accuracy, throughput, and cost-effectiveness of long-read sequencing technologies, it is increasingly plausible that clinical guidelines for evaluating de novo balanced chromosomal rearrangements will evolve to formally incorporate long-read sequencing into standard diagnostic workflows. Such integration promises to refine diagnostic precision and strengthen clinical decision-making in genomic medicine.
Data availability
The Fastq sequencing data generated in this study are available in the Sequence Read Archive (SRA https://www.ncbi.nlm.nih.gov/sra) under BioProject accession number (PRJNA1309293) and Biosample (SAMN50743668).
References
Jacobs, P. A., Browne, C., Gregson, N., Joyce, C. & White, H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J. Med. Genet. 29, 103–108 (1992).
Blake, J. et al. Sequencing of a patient with balanced chromosome abnormalities and neurodevelopmental disease identifies disruption of multiple high risk loci by structural variation. PLoS One. 9, e90894 (2014).
Warburton, D. De Novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am. J. Hum. Genet. 49, 995–1013 (1991).
Wallerstein, R. et al. Factors in decision making following genetic counseling for pre-natal diagnosis of de Novo chromosomal rearrangements. Clin. Genet. 69, 497–503 (2006).
Stankiewicz, P. & Lupski, J. R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61, 437–455 (2010).
Talkowski, M. E. et al. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N Engl. J. Med. 367, 2226–2232 (2012).
Redin, C. et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet. 49, 36–45 (2017).
Warburton, P. E., Sebra, R. P., Long-Read, D. N. A. & Sequencing Recent advances and remaining challenges. Annu. Rev. Genom Hum. Genet. 24, 109–132 (2023).
Jiang, L. et al. Identification of cryptic breakpoints through single-tube long fragment read whole genome sequencing based on preimplantation genetic testing. Npj Genom. Med. 10, 15 (2025).
Tamura, R. Current Understanding of neurofibromatosis type 1, 2, and schwannomatosis. Int. J. Mol. Sci. 22, 5850 (2021).
Moualed, D. et al. Prevalence and natural history of schwannomas in neurofibromatosis type 2 (NF2): the influence of pathogenic variants. Eur. J. Hum. Genet. 30, 458–464 (2022).
Ghalavand, M. A. et al. The genetic landscape and possible therapeutics of neurofibromatosis type 2. Cancer Cell. Int. 23, 99 (2023).
Kim, B. H. et al. NF2-Related schwannomatosis (NF2): molecular insights and therapeutic avenues. Int. J. Mol. Sci. 25, 6558 (2024).
Kehrer-Sawatzki, H. et al. Mutational analysis and expression studies of the neurofibromatosis type 2 (NF2) gene in a patient with a ring chromosome 22 and NF2. Hum. Genet. 100, 67–74 (1997).
Tsilchorozidou, T. et al. Constitutional rearrangements of chromosome 22 as a cause of neurofibromatosis 2. J. Med. Genet. 41, 529–534 (2004).
Denayer, E. et al. Pathogenesis of vestibular Schwannoma in ring chromosome 22. BMC Med. Genet. 10, 97 (2009).
Mahajan, S., Kaur, A. & Singh, J. Ring chromosome 22: a review of the literature and first report from India. Balkan J. Med. Genet. 15, 55–59 (2012).
Nussbaum, P. E., Patel, P. D., Nussbaum, L. A., Hilton, C. W. & Nussbaum, E. S. Bilateral vestibular schwannomas in a patient with ring chromosome 22: case report and review of the literature. Pediatr. Neurosurg. 56, 56–60 (2021).
Nunes, S. et al. Medulloblastoma development in a patient with a constitutional balanced t(5;22)(q35.1;q11.2) involving the NF2 gene. Case Rep. Oncol. 16, 36–44 (2023).
Arai, E. et al. Constitutional translocation t(4;22) (q12;q12.2) associated with neurofibromatosis type 2. Am. J. Med. Genet. 44, 163–167 (1992).
Kurahashi, H. et al. The constitutional t(11;22): implications for a novel mechanism responsible for gross chromosomal rearrangements. Clin. Genet. 78, 299–309 (2010).
Kurahashi, H. et al. Palindrome-mediated chromosomal translocations in humans. DNA Repair. (Amst). 5, 1136–1145 (2006).
Kehrer-Sawatzki, H. et al. The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum. Genet. 99, 237–247 (1997).
De Coster, W. & Rademakers, R. NanoPack2: population-scale evaluation of long-read sequencing data. Bioinformatics 39, btad311 (2023).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021).
Pedersen, B. S. & Quinlan, A. R. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2018).
Smolka, M. et al. Detection of mosaic and population-level structural variants with Sniffles2. Nat. Biotechnol. 42, 1571–1580 (2024).
Heller, D. & Vingron, M. SVIM: structural variant identification using mapped long reads. Bioinformatics 35, 2907–2915 (2019).
Magi, A. et al. Nano-GLADIATOR: real-time detection of copy number alterations from nanopore sequencing data. Bioinformatics 35, 4213–4221 (2019).
Hu, L. et al. Location of balanced Chromosome-Translocation breakpoints by Long-Read sequencing on the Oxford nanopore platform. Front. Genet. 10, 1313 (2020).
Sedlazeck, F. J., Lee, H., Darby, C. A. & Schatz, M. C. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 19, 329–346 (2018).
Carvalho, C. M. B. & Lupski, J. R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 17, 224–238 (2016).
Arai, E. et al. Mapping the breakpoint of a constitutional translocation on chromosome 22 in a patient with NF2. Genes Chromosomes Cancer. 6, 235–238 (1993).
Arai, E., Ikeuchi, T. & Nakamura, Y. Characterization of the translocation breakpoint on chromosome 22q12.2 in a patient with neurofibromatosis type 2 (NF2). Hum. Mol. Genet. 3, 937–939 (1994).
Leung, H. C. M. et al. Detecting structural variations with precise breakpoints using low-depth WGS data from a single Oxford nanopore minion flowcell. Sci. Rep. 12, 4519 (2022).
Norris, A. L., Workman, R. E., Fan, Y., Eshleman, J. R. & Timp, W. Nanopore sequencing detects structural variants in cancer. Cancer Biol. Ther. 17, 246–253 (2016).
Chow, J. F. C., Cheng, H. H. Y., Lau, E. Y. L., Yeung, W. S. B. & Ng, E. H. Y. Distinguishing between carrier and noncarrier embryos with the use of long-read sequencing in preimplantation genetic testing for reciprocal translocations. Genomics 112, 494–500 (2020).
Au, C. H. et al. Rapid detection of chromosomal translocation and precise breakpoint characterization in acute myeloid leukemia by nanopore long-read sequencing. Cancer Genet. 239, 22–25 (2019).
Perez-Becerril, C. et al. Improved sensitivity for detection of pathogenic variants in familialNF2-related schwannomatosis. J. Med. Genet. jmg-2023-109586 (2024). https://doi.org/10.1136/jmg-2023-109586
Chiang, C. et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and Transgenic integration. Nat. Genet. 44, 390–397 (2012).
Ottaviani, D., LeCain, M. & Sheer, D. The role of microhomology in genomic structural variation. Trends Genet. 30, 85–94 (2014).
Sfeir, A. & Symington, L. S. Microhomology-Mediated end joining: A Back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 40, 701–714 (2015).
Olivucci, G. et al. Long read sequencing on its way to the routine diagnostics of genetic diseases. Front. Genet. 15, 1374860 (2024).
Showpnil, I. A. et al. Long-read genome sequencing resolves complex genomic rearrangements in rare genetic syndromes. Npj Genom. Med. 9, 66 (2024).
De Clercq, G. et al. Full characterization of unresolved structural variation through long-read sequencing and optical genome mapping. Sci. Rep. 14, 29142 (2024).
Damián, A. et al. Long-read genome sequencing identifies cryptic structural variants in congenital aniridia cases. Hum. Genomics. 17, 45 (2023).
Yuan, J. et al. Performance of metagenomic Next-Generation sequencing and metagenomic nanopore sequencing for the diagnosis of tuberculosis in HIV-positive patients. Front. Cell. Infect. Microbiol. 14, 1423541 (2024).
Lei, M. et al. Long-read DNA sequencing fully characterized chromothripsis in a patient with Langer–Giedion syndrome and Cornelia de Lange syndrome-4. J. Hum. Genet. 65, 667–674 (2020).
Sanchis-Juan, A. et al. Complex structural variants in Mendelian disorders: identification and breakpoint resolution using short- and long-read genome sequencing. Genome Med. 10, 95 (2018).
Bestetti, I. et al. Long-read sequencing reveals chromothripsis in a molecularly unsolved case of Cornelia de Lange syndrome. Front. Genet. 15, 1358334 (2024).
Sinha, S. et al. Long read sequencing enhances pathogenic and novel variation discovery in patients with rare diseases. Nat. Commun. 16, 2500 (2025).
Acknowledgements
The authors would like to thank Dr. Ilaria Sani for assisting with and reviewing the chromosomal microarray data.
Funding
Supported by grants: University of Florence Grant for the research equipment within the framework of the National Research Program (PNR) 2021–2027—Year 2023 (F.D.L); Fondazione Italiana per la Ricerca sul Cancro (AIRC) under IG 2020 - ID 24503 (R.N.), European Union - Next Generation EU, National Recovery and Resilience Plan, Mission 4 Component 2—Investment 1.4 - National Center for Gene Therapy and Drugs based on RNA Technology - CUP B13C22001010001 (R.N. and L.P.) and Mission 4 Component 2-Investment 1.3—Mnesys A multiscale integrated approach to the study of the nervous system in health and disease – CUP B83C22004910002 (F.D.L.). Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them.
Author information
Authors and Affiliations
Contributions
R.N., F.D.L. and L.P. designed the study and drafted the manuscript; MM, DS, SC, SI, and MP performed the experiments; L.B. and A.P. analyzed the data; L.P. and F.D.L. wrote the manuscript. All authors revised and approved the final version of the article.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was based on data obtained as part of standard clinical care. According to the internal policy of Careggi University Hospital (Florence, Italy), written informed consent obtained during the genetic counseling process is considered sufficient for the use of anonymized patient data in retrospective observational reports involving individual patients. Therefore, ethical review and approval were not required, and no application was submitted to an ethics committee. The patient included in this study provided written informed consent for both the genetic analysis and the publication of the results.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Montini, M., Bonacchi, L., Sidoti, D. et al. Long-read genome sequencing resolves the breakpoints of a chromosome 8;22 balanced translocation in NF2-related schwannomatosis. Sci Rep 16, 549 (2026). https://doi.org/10.1038/s41598-025-30072-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-30072-8