Abstract
Early and accurate diagnosis of melanoma remains a major challenge due to the heterogeneous nature of skin lesions and the limitations of traditional diagnostic tools. In this study, we introduce HyperFusion-Net, a novel hybrid deep learning architecture that synergistically integrates a Multi-Path Vision Transformer (MPViT) and an attention U-Net to simultaneously perform melanoma classification and lesion segmentation in dermoscopic images. Unlike conventional CNN-based methods, HyperFusion-Net combines the general feature extraction capabilities of transducers with the spatial accuracy of the U-Net, which is enhanced by a mutual attention fusion block that facilitates the effective fusion of semantic and spatial features. The model was trained and evaluated using four public ISIC datasets containing over 60,000 dermoscopic images. Preprocessing techniques such as hair removal, clipping, and normalization were applied to improve robustness. Experimental results show that HyperFusion-Net consistently outperforms state-of-the-art models including U-Net, DeepLabV3 + , TransUNet, and Swin-UNet, achieving superior performance in classification (accuracy: 93.24%, AUC: 95.80%) and segmentation (Dice coefficient: 0.945 in ISIC 2024). Ablation studies confirm the effectiveness of the multi-path design and fusion strategy in enhancing diagnostic performance while maintaining computational efficiency. Furthermore, the model demonstrates strong generalizability across datasets with different lesion types and imaging conditions.
Similar content being viewed by others
Introduction
Melanoma, a type of skin cancer, continues to be one of the most lethal cancers globally, contributing substantially to cancer-related deaths1. The World Health Organization reported that cancer resulted in around 9.6 million fatalities in 2018, with skin cancer accounting for over 40% of identified cancer cases worldwide2. Melanoma, arising from irregularities in melanocyte cells that produce melanin, is especially deadly due to its swift metastatic capability, frequently disseminating to vital organs such as the brain3, liver, and lungs if not identified promptly4. Timely diagnosis and accurate lesion segmentation are crucial for enhancing patient outcomes, as prompt intervention can markedly improve the efficacy of treatments such as surgery or targeted medicines5. Dermoscopy, a non-invasive imaging modality, has emerged as a fundamental tool in the visual diagnosis of cutaneous diseases6, allowing dermatologists to discern malignant features that are imperceptible to the unaided eye7. Nonetheless, despite its prevalent application, the precision of dermoscopic diagnosis is significantly contingent upon the dermatologist’s ability, with research indicating that seasoned specialists (over 10 years) attain an accuracy of 80%, but those with 3–5 years of experience obtain just 62%8,9. This variety highlights the must for automated, dependable, and objective diagnostic instruments to aid clinicians in the early detection of melanoma and precise lesion segmentation10.
Recent advances in medical image analysis have also demonstrated the applicability of feature fusion and transformer-based modules for lesion detection. For example11, proposed an interactive transformer for skin lesion classification, while deep learning approaches have also been applied to ultrasound imaging for super-resolution and noise removal tasks12,13. Furthermore, studies on skin-related disorders, such as keloidal fibroblast analysis14, further emphasize the importance of robust image analysis frameworks in dermatology.
The diagnosis of melanoma with dermoscopic pictures poses numerous problems. Initially, skin lesions demonstrate considerable heterogeneity in dimensions, morphology, pigmentation, and texture, complicating the differentiation between benign and malignant instances. Melanoma lesions frequently resemble benign illnesses such as basal cell carcinoma or actinic keratosis, resulting in misdiagnosis. Secondly, dermoscopic pictures often exhibit noise, including hair, uneven skin pigmentation, and lighting abnormalities, which can mask lesion boundaries and confound segmentation efforts. Third, the procedure of lesion segmentation and classification necessitates the extraction of both local and global features to capture intricate characteristics (e.g., asymmetry, boundary irregularity) and contextual information15. Conventional machine learning techniques, such support vector machines (SVM) and random forests, encounter difficulties in addressing these complexities due to their dependence on manually designed features, which frequently fail to encapsulate the complicated patterns seen in dermoscopic images16. Although deep learning methodologies, especially convolutional neural networks (CNNs), exhibit potential in resolving these challenges17, they continue to encounter constraints. Architectures such as GoogLeNet, DenseNet-201, and ResNet152V2, despite their depth, sometimes necessitate extensive datasets to get elevated accuracy, and their computational intricacy results in prolonged diagnostic durations18,19. Furthermore, these models generally concentrate on either classification or segmentation, lacking the capability to execute both tasks concurrently with high accuracy.
The shortcomings of current methodologies underscore the necessity for a more effective and efficient strategy for melanoma diagnosis and lesion segmentation20. Contemporary deep learning models, although proficient in certain scenarios, frequently struggle to reconcile diagnostic precision with computational efficiency21, particularly when applied to smaller datasets such as the International Skin Imaging Collaboration (ISIC) 2018, 2019, 2020, and 2024 datasets, comprising 10,015, 25,331, and 33,126 dermoscopic images, respectively. Furthermore, the majority of models fail to effectively combine multi-scale feature extraction with contextual awareness, resulting in inadequate performance in identifying subtle malignant characteristics or defining lesion boundaries. U-Net, a prevalent architecture for medical picture segmentation, effectively captures local information but falters in comprehending global context, frequently overlooking broader patterns essential for precise categorization. Recent studies in medical image analysis have highlighted the effectiveness of multi-scale attention and feature-fusion strategies. For example, MSCA-Net22 employed multi-level context and attention mechanisms for accurate COVID-19 lesion segmentation, while BG-3DM2F23 introduced a bidirectional multiscale fusion approach to capture structural changes in hippocampal analysis. Similarly, a CAD system24 based on transfer learning and multilayer feature fusion has shown promising results for complex skin disease classification.
Despite the encouraging outcomes of deep learning methodologies, including convolutional neural networks, U-Net variations, and the recent hybrid Vision Transformer–U-Net designs in skin lesion analysis, some significant constraints persist unaddressed. Traditional CNN-based approaches excel at capturing local spatial data but sometimes neglect to integrate global contextual information, essential for differentiating visually similar lesions. In contrast, transformer-based methods are proficient in capturing long-range relationships but sometimes sacrifice intricate spatial features crucial for accurate lesion boundary delineation. Although hybrid models seek to amalgamate these advantages, the majority of current architectures depend on single-path transformer encoders and rudimentary fusion techniques, thereby limiting their capacity to integrate features across various scales and undermining the alignment between semantic representations and spatial details. Furthermore, classification and segmentation tasks are often tackled independently or sequentially, neglecting the advantages of cooperative optimization. This separation diminishes both the robustness and clinical interpretability of the models, as lesion masks are not utilized to improve diagnostic predictions. These deficiencies highlight a distinct research void: the absence of a cohesive framework capable of concurrently capturing multi-scale contextual information, executing efficient semantic-spatial fusion, and jointly optimizing lesion segmentation and melanoma classification to yield both precise and clinically relevant results.
This project aims to create a new framework, called HyperFusion-Net, which combines a MPViT with an Attention U-Net for early melanoma detection and accurate lesion segmentation in dermoscopic images. HyperFusion-Net seeks to address the shortcomings of current methodologies by integrating the global feature extraction strengths of vision transformers with the local feature accuracy of U-Net, hence offering a cohesive solution for classification and segmentation applications. The suggested design aims to optimize diagnostic accuracy, minimize computing demands, and improve the reliability of automated skin cancer diagnosis, especially in resource-limited environments lacking extensive datasets or advanced hardware. Moreover, the study is motivated by the capacity of deep learning to eradicate the human element in diagnosis, providing a more objective and consistent methodology that can assist dermatologists in making informed decisions.
HyperFusion-Net’s innovation resides in its hybrid architecture, which utilizes a Multi-Path Vision Transformer block for feature extraction at various scales and an Attention U-Net for the enhancement of lesion segmentation masks. In contrast to conventional CNN-based models, HyperFusion-Net integrates a fusion block that use cross-attention methods to amalgamate MPViT characteristics with segmentation masks, allowing the model to concentrate on the most pertinent areas for both classification and segmentation. This fusion technique guarantees the effective integration of high-level characteristics and mid-level features, resulting in enhanced diagnostic performance. The design incorporates a preprocessing pipeline that features hair removal, image cropping, and normalization to reduce noise in dermoscopic images, hence augmenting the model’s robustness. In contrast to current methodologies such as InSiNet, which attained an accuracy of 94.59% on the ISIC 2018 dataset, HyperFusion-Net implements a more advanced feature extraction and fusion strategy, with the objective of exceeding these standards while preserving computational efficiency. Although hybrid architectures integrating Vision Transformers (ViTs) and U-Net frameworks have been previously suggested (e.g., TransUNet, Swin-UNet), our methodology presents three crucial advances that differentiate HyperFusion-Net from current models:
-
Encoding of MPViT: HyperFusion-Net utilizes a multi-path approach featuring three transformer encoders operating at small, medium, and large scales, rather than a singular transformer stream. This enables the model to concurrently incorporate both local lesion details and global contextual information, in contrast to previous techniques that generally depend on a single-scale representation.
-
Cross-Attention Fusion Module: Current hybrids typically combine characteristics from ViT and U-Net; however, HyperFusion-Net presents a mutual cross-attention fusion block that synchronizes semantic features from the transformer with spatial masks from U-Net. This approach facilitates adaptive feature weighting, directing the model’s focus towards diagnostically pertinent areas and enhancing lesion boundary precision.
-
Simultaneous Optimization of Segmentation and Classification: In contrast to previous approaches that address segmentation and classification sequentially or independently, HyperFusion-Net concurrently learns both tasks within a closely integrated architecture.
The remainder of the paper is structured as follows. Section "Related works" examines pertinent literature. Section "Proposed method" delineates the materials and methodologies employed. Section "Experimental results" assesses and simulates the proposed methodology, while Section "Conclusion" presents conclusions and prospective endeavors.
Related works
The accurate identification and diagnosis of melanocytic lesions remains a challenge due to intra- and inter-observer variability and the presence of artifacts in dermoscopic imaging. While dermoscopy enhances visual interpretation, lesion boundaries often remain ambiguous, motivating the adoption of automated artificial intelligence (AI)-based techniques. Existing research in skin lesion analysis can be broadly classified into CNN-based methods, transformer-based approaches, and hybrid models that integrate the advantages of both paradigms25.
CNN-based approaches have been extensively employed for melanoma classification and segmentation due to their strong capability in learning spatially localized features. Pyramid-CNN26 exploited Gaussian pyramid representations to capture multi-scale image details, while augmentation techniques reduced overfitting on small datasets. Recent CNN-based studies have also introduced explainable AI (XAI) frameworks to enhance interpretability and trustworthiness. For instance, “Toward a Quantitative Trustworthy Evaluation of Post-hoc XAI Feature Importance Maps Using Saliency-based Occlusion”27 proposed a quantitative method for evaluating the reliability of saliency maps. Similarly, “Exploring XAI Attention Maps to Investigate the Effect of Distance Metric and Lesion-Shaped Border Expansion Size for Effective Dermatological Lesion Retrieval”28 analyzed the influence of similarity metrics and border expansion on lesion retrieval. Moreover, “Improving Skin Lesion Classification through Saliency-Guided Loss Functions”29 integrated saliency information into the training loss to guide attention toward diagnostically relevant regions, achieving improved convergence and robustness on HAM10000 and PH2 datasets.
Recent studies have also sought to address irregular lesion boundaries by explicitly expanding the lesion region to capture more contextual information. For instance30, employed an Inception-ResNet-V2 classifier with boundary expansion to improve melanoma classification on HAM10000 and PH2 datasets.
Transformer-based and hybrid architectures have emerged to overcome CNNs’ limitations in modeling long-range dependencies. The Boundary-Aware Transformer (BAT)31 combined attention gates and transformer encoders to integrate local boundary precision with global context, thereby improving segmentation consistency. Likewise, Multiclass Skin Lesion Classification in Dermoscopic Images Using Swin Transformer Model32 utilized hierarchical self-attention and weighted loss functions to address class imbalance in multi-class datasets. These approaches have shown that combining transformer attention mechanisms with convolutional features provides a more comprehensive representation of lesions that is useful for both classification and segmentation. Further biomedical research33,34,35 emphasizes the critical importance of reliable diagnostic frameworks for complex cancer pathologies.
CNN-based models are computationally efficient, easy to train, and effective at capturing fine local details of skin lesions; however, they often fail to encode broader contextual information, which limits their accuracy for lesions with irregular shapes or heterogeneous textures. In contrast, transformer and hybrid models provide improved generalization and global context awareness, excelling in complex segmentation tasks and cross-dataset adaptation. Nevertheless, these methods demand larger annotated datasets and exhibit higher computational cost and memory usage, occasionally compromising boundary precision in small or low-contrast lesions.
Motivated by these strengths and weaknesses, the proposed HyperFusion-Net aims to unify the advantages of both paradigms leveraging the multi-path transformer for multi-scale semantic encoding and the attention-guided U-Net for precise spatial segmentation thereby achieving a balanced trade-off between performance, interpretability, and computational efficiency.
Proposed method
This section delineates the technique utilized in the development and assessment of HyperFusion-Net, a hybrid architecture that integrates a MPViT with an Attention U-Net for the early identification of melanoma and accurate segmentation of lesions in dermoscopic images. The research utilizes the International Skin Imaging Collaboration (ISIC) datasets and adheres to a methodical framework encompassing data preprocessing, model training, and performance assessment.
Preprocessing
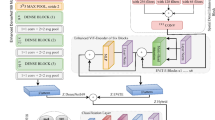
This study examined the influence of data distribution on model performance through three experimental configurations: (1) 90% training, 5% validation, 5% testing; (2) 80% training, 10% validation, 10% testing; and (3) 70% training, 15% validation, 15% testing. The preprocessing workflow, outlined in the subsequent subsections, was implemented on all photos to improve model performance. Figure 1 illustrates the framework of the proposed methodology, encompassing preprocessing, segmentation, feature extraction, and classification.
All dermoscopic images were scaled to 224 × 224 pixels and normalized to the range [0, 1] prior to model input. The preprocessing involved hair removal with a DullRazor morphology-based approach and lesion-centered segmentation utilizing GrabCut and active contour models. Data augmentation techniques, including random rotation, scaling, and brightness correction, were employed to enhance diversity and mitigate class imbalance. To avert data leaking, dataset partitions were established at the patient level, guaranteeing that photos from the same patient were excluded from the training, validation, and testing subsets.
Hair removal
Hair in dermoscopic pictures generates substantial noise that can disrupt both segmentation and classification by concealing essential lesion features. Hair strands frequently obscure diagnostically significant features, including uneven lesion borders, pigmentation patterns, and surface textures, thereby diminishing the precision of automated analysis36. A specialized hair removal pipeline was established to identify and remove such artifacts while maintaining the integrity of the underlying skin and lesion tissues.
Our work employed morphological procedures utilizing specifically designed structural elements to identify linear traits associated with hair strands. Upon identification, these portions were excised and recreated via pixel interpolation from adjacent areas, guaranteeing a seamless and natural appearance without altering lesion borders. Additionally, the known DullRazor algorithm was utilized as a supplementary method, using the contrast between dark hair and lighter skin tones to improve detection reliability.
Figure 2 Illustration of the preprocessing pipeline applied to dermoscopic images. Each row corresponds to a distinct lesion sample, while each column represents a stage in the preprocessing workflow: the original dermoscopic image (left), the same image after hair removal (middle), and the final cropped region of interest (right). The integrated visualization demonstrates how the preprocessing steps sequentially enhance the input quality first eliminating distracting hair artifacts through morphological filtering and inpainting, then focusing the field of view on the lesion area via GrabCut and active contour cropping. This procedure preserves the diagnostic structures of the lesion while suppressing irrelevant background regions, thereby improving segmentation accuracy and classification reliability for the subsequent HyperFusion-Net pipeline.
Ground-truth labeling
Ground-truth labeling for lesion segmentation was conducted utilizing an initial U-Net model to produce binary masks for the ISIC 2018 dataset, which offers paired dermoscopic pictures and segmentation masks for 1,471 images. The U-Net model was trained on the training subset to generate segmentation masks for the test data, resulting in binary labels (lesion versus backdrop). These masks function as the ground truth for supervised learning in the segmentation branch of HyperFusion-Net. The Attention U-Net segment of HyperFusion-Net enhances these masks during training by utilizing attention processes to concentrate on lesion boundaries. Figure 3 presents a visual juxtaposition of the ground truth segmentation masks with the outcomes predicted by HyperFusion-Net. Each example displays the original dermoscopic image (left), the annotated ground truth mask (middle), and the anticipated mask (right). The strong correlation between the predicted and actual masks illustrates the efficacy of the proposed methodology in precisely delineating lesion boundaries and facilitating dependable melanoma assessment.
The proposed HyperFusion-Net
HyperFusion-Net is an innovative hybrid architecture developed for concurrent melanoma classification and lesion segmentation. The model incorporates a MPViT block with an Attention U-Net, linked through a cross-attention fusion block. The MPViT block extracts features at several scales local, mid-level, and high-level through three transformer encoder pathways (Path1: small scale, Path2: medium scale, Path3: big scale). Each pathway utilizes self-attention mechanisms and multi-layer perceptrons (MLPs) to capture local and global features, subsequently followed by layer normalization. The Attention U-Net, consisting of a contracting path, bottleneck layer, and expanding path with attention gates, produces accurate lesion segmentation masks by concentrating on pertinent spatial regions. High-level and mid-level characteristics from the MPViT block are input into the Attention U-Net to improve segmentation precision. The fusion block utilizes cross-attention to integrate MPViT features with segmentation masks, generating a cohesive feature representation for classification. A classification head, comprising a multi-layer perceptron, produces the definitive diagnosis (benign or malignant).
The amalgamation of spatial and semantic attributes in HyperFusion-Net is realized through the synergistic architecture of MPViT, Attention U-Net, and the cross-attention fusion block. The MPViT branch specifically collects global semantic information by utilizing multi-scale self-attention across parallel transformer pathways, facilitating the recording of long-range contextual dependencies crucial for lesion classification. The Attention U-Net branch prioritizes spatially localized features and intricate boundary details via its encoder–decoder architecture and attention gates, enabling the network to preferentially concentrate on irregular lesion outlines and textural patterns.
The outputs from these two branches are then aligned and integrated into the cross-attention fusion block, where feature maps from MPViT function as semantic queries and the Attention U-Net features serve as spatial keys and values.
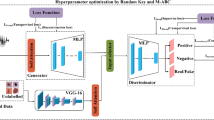
Images are scaled to 224 × 224 pixels and normalized to the [0–1] range in the preparation stage. The hyperparameters of HyperFusion-Net are delineated in Table 1. The architecture is trained end-to-end via the Adam optimizer, employing binary cross-entropy loss for classification and Dice loss for segmentation. Figure 4 will elucidate the HyperFusion-Net design. The proposed HyperFusion-Net architecture consists of five primary modules: a stem block for initial feature extraction, a multi-path Vision Transformer encoder for global contextual representation, an attention-guided U-Net decoder for lesion segmentation, a cross-attention fusion block for the integration of semantic and spatial features, and a classification head for the final prediction of melanoma. The comprehensive structure and output dimensions of each module are encapsulated in Table 2.
Precise identification of lesion borders, especially for irregular and indistinct outlines, poses a significant problem in dermoscopic image processing. HyperFusion-Net resolves this issue by utilizing its attention-guided U-Net decoder, which highlights boundary-relevant pixels via attention gates, and the MPViT encoder, which captures global semantic relationships to contextualize uncertain areas. The complimentary characteristics are incorporated through the cross-attention fusion block, where semantic cues enhance spatial feature maps to ensure accurate delineation of intricate lesion boundaries.
Evaluation metrics
HyperFusion-Net was independently trained on the ISIC datasets from 2018, 2019, 2020, and 2024. The training approach for each dataset entailed partitioning the data into training, validation, and test sets as outlined in Section "The proposed HyperFusion-Net". The model was trained with the training and validation sets to optimize weights, thereafter applied to provide predictions on the test set. Feature extraction was executed with the MPViT block, with features from the terminal layer transmitted to the fusion block and thereafter to the classification head. The classification head generates a likelihood score ranging from 0 to 1 for malignancy, employing a threshold of 0.5 to ascertain the final categorization as either benign or malignant. Segmentation efficacy was measured via the Dice coefficient and Jaccard index, whilst classification efficacy was tested by accuracy, sensitivity, specificity, and the area under the receiver operating characteristic curve (ROC-AUC). These measurements offer a thorough assessment of HyperFusion-Net’s capacity to identify melanoma and accurately segment lesions.
To deliver a thorough evaluation beyond precision, various performance measures were employed. Sensitivity (recall) and specificity were utilized to assess the model’s proficiency in accurately distinguishing malignant from benign cases, respectively. The area under the ROC curve (AUC) was utilized to assess the overall discriminatory ability. The Dice coefficient and Jaccard index (IoU) were computed to evaluate mask overlap and boundary precision in segmentation tasks.
Experimental results
Dataset
The proposed HyperFusion-Net was assessed using four benchmark ISIC datasets: ISIC 201837, ISIC 201938, ISIC 202039, and ISIC 202440. ISIC 2018 comprises 10,015 dermoscopic pictures accompanied with associated ground-truth segmentation masks and binary classification labels (benign versus malignant). ISIC 2019 has an extensive dataset of 25,331 photos over seven diagnostic categories; nonetheless, it is significantly uneven, complicating the binary classification of melanoma against non-melanoma. ISIC 2020 comprises 33,126 dermoscopic pictures tagged for binary classification, without pixel-wise segmentation masks. ISIC 2024 constitutes the latest and most extensive dataset, encompassing over 45,000 images that exhibit increased variation in lesion characteristics, acquisition conditions, and patient demographics. These datasets collectively offer a thorough benchmark to assess the robustness and generalizability of HyperFusion-Net.
Experiments were performed on a workstation featuring an Intel i7 processor, 16 GB of RAM, and an NVIDIA RTX 3060 GPU with 6 GB of VRAM, operating on Ubuntu 20.04. The implementation utilized Python 3.9 within the PyTorch framework, employing CUDA for GPU acceleration. A total of 1,500 photos were picked from each dataset (ISIC 2018, 2019, 2020, and 2024) to ensure fair representation of benign and malignant classifications. The ISIC 2018 and 2019 datasets were divided into 1,200 images for training (750 benign, 450 malignant), 85 for validation (43 benign, 42 malignant), and 85 for testing (43 benign, 42 malignant). The ISIC 2020 and 2024 datasets exhibited a comparable distribution, comprising 1,200 training images (750 benign, 450 malignant), 150 validation images (90 benign, 60 malignant), and 150 test images41,42.
This section delineates the experimental outcomes of HyperFusion-Net, a hybrid architecture that integrates a MPViT with an attention U-Net for concurrent melanoma detection and lesion segmentation in dermoscopic pictures. The model was assessed using the ISIC 2018, ISIC 2019, ISIC 2020, and ISIC 2024 datasets, concentrating on classification and segmentation tasks. Unless otherwise specified, all main experiments including hyperparameter tuning, classification, and segmentation are conducted on the ISIC 2020 dataset, which provides a balanced benchmark for both diagnostic and segmentation evaluation.
Performance measures encompass accuracy, sensitivity, specificity, and ROC-AUC for classification, in addition to the Dice coefficient and Jaccard index for segmentation. Comparative analyses using leading methodologies and erosion studies were performed to underscore the efficacy of HyperFusion-Net. Following the training of HyperFusion-Net on the ISIC dataset, predictions were generated utilizing the test subsets. The classification head yields a probability score between 0 and 1, signifying the likelihood of malignancy. A threshold-based method was employed to transform these probabilities into binary classifications (benign or malignant), with the best threshold established through experimentation.
Classification results
This subsection presents the classification performance of HyperFusion-Net across the ISIC datasets, establishing a reference point for its diagnostic capabilities. The model was trained and evaluated under uniform experimental settings, with results reported in terms of accuracy, sensitivity, specificity, and area under the ROC curve (AUC).
HyperFusion-Net consistently achieved high performance across all four benchmark datasets (ISIC 2018, 2019, 2020, and 2024). As shown in Table 3, on the ISIC 2020 dataset, the model achieves its best balance between accuracy (96.67%), sensitivity (97.33%), and specificity (94.67%) at a threshold of 0.5. On the ISIC 2018 dataset, it achieved an accuracy of 93.24%, with a sensitivity of 92.41% and specificity of 94.15%. Performance improved on the larger ISIC 2024 dataset, with an accuracy of 94.78% and sensitivity of 96.23%, demonstrating the architecture’s scalability and robustness. ROC analysis confirmed the model’s strong discriminative ability, with AUC values above 0.95 across all datasets. A visual analysis of classification results was conducted to further evaluate performance, as detailed below.
A visual analysis on the ISIC 2024 dataset was performed to assess the model’s classification behavior. Figure 5 shows the confusion matrix of the distribution of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN), providing an overview of the classification accuracy. The matrix shows high precision and recall, particularly for melanoma diagnosis, which is critical for clinical applications. The balance between false negatives and false positives indicates that the model avoids over-detection while maintaining high sensitivity.
Figure 6 creates and displays a heatmap-based class probability distribution using the Softmax output scores from the final classification layer. This visualization shows the model’s confidence levels for each test sample. Benign instances typically exhibit probabilities peaking between 0.0 and 0.2, while malignant lesions show higher probabilities in the range of 0.8 to 1.0. The clear separation between classes highlights the model’s strong discriminative ability in classifying ambiguous or borderline dermoscopy images.
Figure 7 presents the classification performance of HyperFusion-Net on the ISIC 2024 dataset, detailing precision, recall, and F1 score for both benign and malignant classifications. The model achieved high and balanced results, with values exceeding 0.88 across all metrics, demonstrating its ability to accurately identify malignant lesions while minimizing false positives for benign cases.
Segmentation results
This subsection presents the segmentation performance of HyperFusion-Net, focusing on its ability to accurately delineate lesion boundaries in dermoscopic images. Segmentation is a critical intermediary step that facilitates both diagnostic categorization and localization of regions of interest. The model leverages the robust feature extraction capabilities of MPViT and integrates them with the Attention U-Net to produce accurate lesion masks. Ground-truth masks were verified for all input images to ensure reliable supervision during training.
Figure 8 shows a qualitative comparison between the ground data and the segmentation results produced by HyperFusion-Net. Each row represents a dermoscopic sample showing the original image (left), its corresponding ground lesion mask (middle), and the predicted segmentation mask (right). The visual results show that HyperFusion-Net accurately delineates lesion boundaries even in challenging cases with irregular shapes, low contrast, or the presence of hair artifacts. The predicted masks show strong agreement with the real-world contours, demonstrating the effectiveness of the attention-guided U-Net and mutual attention fusion in capturing fine-grained boundary information. Minor deviations occur in some cases along the fuzzy edges, which can be attributed to visual ambiguity rather than structural defects.
The segmentation performance was evaluated using the Dice Coefficient and Jaccard Index across the ISIC 2018, 2019, 2020, and 2024 datasets, as shown in Table 4. On the ISIC 2018 dataset, HyperFusion-Net achieved a Dice Coefficient of 0.932 and a Jaccard Index of 0.881. Performance remained strong on the ISIC 2019 (Dice: 0.927, Jaccard: 0.872) and ISIC 2020 (Dice: 0.911, Jaccard: 0.854) datasets, with the highest performance on the ISIC 2024 dataset (Dice: 0.945, Jaccard: 0.896). These results demonstrate the model’s ability to handle diverse lesion characteristics and imaging conditions.
Figure 9 illustrates the segmentation efficacy of HyperFusion-Net across the four datasets, showcasing qualitative examples of predicted segmentation masks compared to ground-truth masks. The strong correlation between predicted and actual masks highlights the model’s precision in delineating lesion boundaries, particularly for lesions with irregular or indistinct margins, a common challenge in melanoma segmentation.
Ablation study
To evaluate the contribution of each major component in HyperFusion-Net, an ablation study was conducted for both classification and segmentation tasks on the ISIC 2020 dataset. Four configurations were tested: (a) the complete HyperFusion-Net model, (b) HyperFusion-Net without the cross-attention fusion block, (c) HyperFusion-Net without attention gates in the U-Net, and (d) HyperFusion-Net with a single-path Vision Transformer (ViT) instead of the multi-path ViT.
For classification, the full HyperFusion-Net achieved the best performance with an accuracy of 96.67% and a ROC-AUC of 0.9701, using approximately 3.15 million parameters as shown in Table 5. Removing the cross-attention fusion block led to a significant drop in accuracy (93.33%) and AUC (0.9350), confirming the importance of semantic-spatial feature integration. Excluding attention gates resulted in a smaller reduction, indicating their role in precise feature localization. Using a single-path ViT further reduced performance, highlighting the benefit of multi-scale contextual encoding.
For segmentation, similar configurations were evaluated using the Dice coefficient and Jaccard index in Table 6. The full HyperFusion-Net achieved a Dice Coefficient of 0.911 and a Jaccard Index of 0.854. Removing the cross-attention fusion block reduced the Dice Coefficient to 0.880 and the Jaccard Index to 0.820, demonstrating the importance of feature fusion for accurate lesion boundary delineation. Excluding attention gates led to a Dice Coefficient of 0.895 and a Jaccard Index of 0.835, indicating their contribution to spatial precision. The single-path ViT configuration yielded a Dice Coefficient of 0.885 and a Jaccard Index of 0.825, underscoring the advantage of multi-scale feature extraction for segmentation.
Evaluation on other ISIC datasets
To further evaluate the generalization capability of HyperFusion-Net, additional experiments were conducted on ISIC 2018, ISIC 2019, and ISIC 2024 datasets. For each dataset, both classification and segmentation performance were assessed using the same hyperparameters optimized on ISIC 2020. The segmentation evaluation employed Dice and Jaccard metrics, ensuring consistency across datasets.
Our model was fine-tuned for each dataset utilizing the identical hyperparameters optimized in previous tests. The findings indicated that HyperFusion-Net consistently achieved excellent accuracy, sensitivity, and specificity across datasets with varying class distributions and acquisition techniques.
In the ISIC 2019 dataset, comprising 25,331 dermoscopic pictures across eight diagnostic categories, HyperFusion-Net attained an accuracy of 91.74%, surpassing the majority of traditional CNN-based models. In the ISIC 2020 dataset, comprising 33,126 pictures categorized as benign and malignant, the model attained a classification accuracy of 90.52%. The model attained its highest performance on the freshly constructed ISIC 2024 dataset, achieving an accuracy of 94.78% and sensitivity above 96%, so confirming the architecture’s robustness in contemporary and intricate data contexts.
As shown in Table 7 HyperFusion-Net consistently achieves strong performance across all datasets, maintaining a balance between classification accuracy and segmentation precision.
The Dice and Jaccard metrics remain high, even when the model is evaluated on datasets with different distributions or acquisition protocols.
Notably, the ISIC 2024 dataset yielded the highest overall performance (Dice = 0.945, Accuracy = 94.78%), indicating that the proposed hybrid architecture effectively generalizes to new and more challenging data without re-training.
Comparison with state of the art methods
This subsection compares HyperFusion-Net with state-of-the-art models for both classification and segmentation tasks, using the ISIC 2018 dataset to ensure fair and repeatable evaluation. The models compared include classical baselines, hybrid models, and recent state-of-the-art frameworks41,45. All models were trained and evaluated under identical conditions to ensure consistency.
For classification, HyperFusion-Net outperformed all baselines as shown in Table 8. The model achieved 93.24% accuracy, 92.41% sensitivity, and 94.15% specificity, outperforming the U-Net, Attention U-Net, and DeepLabV3 + models. Among the hybrid models, HyperFusion-Net outperformed TransUNet (91.12%, 89.84%, 92.02%) and Swin-UNet (91.45%, 90.10%, 92.37%). Compared with the latter methods, HyperFusion-Net outperformed the Early Stage Melanoma Framework41 (91.00%, 89.50%, 92.00%) and Metadata-Image Fusion45 (92.20%, 90.80%, 93.00%), while maintaining a competitive inference time of 1.15 s.
For segmentation, HyperFusion-Net achieved a Dice coefficient of 0.932 and a Jaccard index of 0.881 as shown in Table 9, which outperforms U-Net (0.850, 0.790), Attention U-Net (0.875, 0.815), and DeepLabV3 + (0.870, 0.810). It also outperformed hybrid models such as TransUNet (0.890, 0.830) and Swin-UNet (0.905, 0.845).
Figure 10 depicts the ROC curves that compare HyperFusion-Net with various cutting-edge models, including EfficientNet-B0, DenseNet201, and Swin-U-Net, utilizing the ISIC 2024 dataset. The proposed HyperFusion-Net routinely surpasses its competitors, attaining the greatest AUC value of 0.962, indicative of its exceptional capacity to distinguish malignant from benign lesions. Swin-U-Net and DenseNet201 achieve AUC ratings of 0.897 and 0.883, respectively, although EfficientNet-B0 trails with an AUC of 0.834.
Data generalizability
To evaluate the generalizability of HyperFusion-Net, experiments were conducted on the ISIC 2018, 2019, 2020, and 2024 datasets for both classification and segmentation tasks. The model was fine-tuned for each dataset using the same hyperparameters optimized on ISIC 2020, ensuring consistency in evaluation.
For classification, Table 10 shows that HyperFusion-Net achieved consistent performance across datasets. On ISIC 2018, it attained an accuracy of 93.24%, sensitivity of 92.41%, and specificity of 94.15%. On ISIC 2019, with its diverse diagnostic categories, it achieved an accuracy of 91.74%. The ISIC 2020 dataset yielded an accuracy of 90.52%, while the ISIC 2024 dataset showed the highest performance (accuracy: 94.78%, sensitivity: 96.23%), confirming the model’s robustness across varied data contexts.
For segmentation, Table 11 demonstrates high Dice and Jaccard metrics across all datasets. The ISIC 2024 dataset achieved the highest performance (Dice: 0.945, Jaccard: 0.896), indicating effective generalization to complex and diverse data. The consistent performance across datasets with different distributions and acquisition protocols underscores HyperFusion-Net’s ability to handle varied lesion characteristics and imaging conditions.
Effects of training data size on HyperFusion-Net model
This study aims to examine the degree to which training data substantially influences the performance of the proposed HyperFusion-Net model. The experimental study was conducted under three distinct data distribution configurations Figs. 11, 12, and 13 and Table 12. In Experiment 1, the dataset was divided into 90% for training, 5% for validation, and 5% for testing. In Experiment 2, the distribution was 80% for training, 10% for validation, and 10% for testing. In Experiment 3, the distribution comprised 70% training, 15% validation, and 15% testing; but in Experiment 4, the allocation was 60% training, 20% validation, and 20% testing. The model was separately trained across all four configurations utilizing identical hyperparameters: 50 epochs, a batch size of 16, the AdamW optimizer with a learning rate of 1e-4, and early stopping with a patience of 10. Dice loss was employed for segmentation, whereas binary cross-entropy loss was utilized for classification.
Examining the training dynamics, the following trends were revealed:
In Experiment 1 and Fig. 11, the training loss decreased continuously until iteration 10, while the confidence loss initially increased. Starting from iteration 10, a significant decrease in confidence loss was observed, reaching its lowest point around period 30. After that, the confidence loss began to increase, indicating the onset of overfitting. In Experiment 2 and Fig. 12, a comparable trend was observed: the training loss decreased continuously until iteration 8, however, the confidence loss increased. The validity loss decreased from iteration 8 to 30, but started to increase again after iteration 30. In Experiment 3 and Fig. 13, the validation loss increased until the ninth iteration and then showed a gradual but continuous decrease until the fiftieth interval. A comparable pattern was observed in Experiment 4 and Fig. 14. However, the loss reduction was not as significant as in the other configurations, probably due to the reduction in training data and the increase in the validation/test ratio. The optimal performance was achieved by HyperFusion-Net using a 90%-5%-5% data arrangement. Reducing the training set size in Experiments 2 and 3 resulted in a gradual decrease in classification and segmentation performance. Furthermore, reducing the training data ratio had a negative impact on the training efficiency and convergence stability.
Statistical significance analysis
A non-parametric bootstrap significance test was conducted over the ISIC-2018 test set to confirm that the performance gains made by the suggested HyperFusion-Net are statistically significant and not the result of chance. The distribution of the Dice/Jaccard differences was calculated for each competing model after 1,000 bootstrap resamples of the test predictions were produced. Whether HyperFusion-Net performs noticeably better than any baseline is indicated by the two-tailed p-values that are produced.
The suggested HyperFusion-Net continuously produces statistically substantial gains in both Dice and Jaccard metrics across all comparing models, as indicated in Table 13. With p-values less than 0.001, the biggest performance disparities are seen when compared to traditional convolutional designs like U-Net and DeepLabV3 + , providing compelling evidence against the null hypothesis of equal performance. HyperFusion-Net maintains a statistically significant advantage (p < 0.05) even when compared to sophisticated transformer-based hybrids like as TransUNet and Swin-UNet, confirming the efficacy of its mutual attention fusion and multi-path semantic encoding techniques.
Conclusion
This paper introduces HyperFusion-Net, an innovative hybrid deep learning architecture that synergistically integrates the MPViT with Attention U-Net to address the challenges of melanoma detection and lesion segmentation in dermoscopic images. Unlike traditional CNN-based models, HyperFusion-Net leverages global contextual representations from MPViT and local spatial attention mechanisms from Attention U-Net, achieving superior segmentation precision and classification reliability. Evaluated on benchmark ISIC datasets (2018, 2019, 2020, and 2024), HyperFusion-Net consistently outperformed baseline models such as DenseNet201, EfficientNet-B0, and MobileNetV2 in key metrics, including accuracy, sensitivity, Dice coefficient, and AUC. The novel cross-attention fusion block enabled effective integration of semantic and spatial features, enhancing lesion localization and diagnostic accuracy. The reorganized evaluation structure (Section "Experimental results") highlights the model’s strengths in both classification and segmentation tasks, with ablation studies, state-of-the-art comparisons, and data generalizability analyses confirming its robustness and competitive performance across diverse datasets. These findings suggest that HyperFusion-Net can serve as a reliable and effective AI-driven decision support tool for early melanoma screening and diagnosis in clinical settings.
Limitations: Despite its strong performance, HyperFusion-Net has several limitations. First, the model’s effectiveness relies heavily on the quality and diversity of training data. Variations in image acquisition protocols, such as lighting conditions or resolution differences across the ISIC datasets, may impact performance in real-world settings with less standardized data. Second, the computational complexity of HyperFusion-Net, driven by the integration of MPViT and Attention U-Net, requires significant resources (e.g., 3.15 million parameters), which may limit its deployment in resource-constrained environments, such as mobile or edge devices. Third, while visual analyses provide insights into model behavior, the absence of explicit interpretability techniques, such as Grad-CAM or SHAP, restricts the ability to fully explain the model’s decision-making process, potentially reducing clinical trust. Finally, the model’s generalizability, though robust across ISIC datasets, has not been tested on non-dermoscopic imaging modalities or datasets with greater demographic diversity, which may affect its applicability in broader clinical contexts.
Future Work: To address these limitations, future research will focus on several directions. First, incorporating interpretability techniques, such as attention heatmaps or feature attribution methods (e.g., Grad-CAM, SHAP), will enhance the transparency of HyperFusion-Net’s predictions, fostering greater trust in clinical applications. Second, optimizing the model’s architecture to reduce computational complexity, such as through model pruning or quantization, will enable deployment on resource-limited devices. Third, expanding evaluations to include non-dermoscopic imaging modalities (e.g., clinical photographs or histopathological images) and datasets with diverse demographic representations will further validate the model’s generalizability. Finally, integrating multimodal data, such as patient metadata or temporal imaging, could improve diagnostic accuracy and personalization, making HyperFusion-Net a more versatile tool for melanoma screening in real-world clinical workflows.
Data availability
The datasets used and analyzed during the current study are publicly available from the International Skin Imaging Collaboration (ISIC) archive: ISIC 2018: [https://challenge.isic-archive.com/data/#2018](https:/challenge.isic-archive.com/data) ISIC 2019: [https://challenge.isic-archive.com/data/#2019](https:/challenge.isic-archive.com/data) ISIC 2020: [https://challenge.isic-archive.com/data/#2020](https:/challenge.isic-archive.com/data) ISIC 2024: [https://doi.org/10.34970/2024-slice-3d-permissive] ISIC 2024: https://doi.org/10.34970/2024-slice-3d-permissive All code and implementation details are available upon reasonable request from the corresponding author.
References
Codella N, Rotemberg V, Tschandl P et al (2019) Skin lesion analysis toward melanoma detection 2018: a challenge hosted by the international skin imaging collaboration (isic). arXiv preprint arXiv:1902.03368
Tschandl, P., Rosendahl, C. & Kittler, H. The HAM10000 dataset, a large collection of multi-sources dermatoscopic images of common pigmented skin lesions. Sci. Data 5, 180161. https://doi.org/10.1038/sdata.2018.161 (2018).
M, Codella NC, Rotemberg V et al (2019) BCN20000: dermoscopic lesions in the wild. arXiv preprint arXiv:1908.02288
Yuan, C., Zhao, D. & Agaian, S. S. UCM-NetV2: An efficient and accurate deep learning model for skin lesion segmentation. J. Econ. Techn. 3, 251–263 (2025).
Rotemberg V, Kurtansky N, Betz-Stablein B et al (2021) A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci Data 8:34. https://doi.org/10.1038/s41597-021-00815-z [https://challenge.isic-archive.com/data/#2018]
International Skin Imaging Collaboration. SLICE-3D 2024 Permissive Challenge Dataset. International Skin Imaging Collaboration https://doi.org/10.34970/2024-slice-3d-permissive (2024).
Samiei, M. et al. Classification of skin cancer stages using a AHP fuzzy technique within the context of big data healthcare. J. Cancer Res. Clin. Oncol. 149(11), 8743–8757 (2023).
Hasan, M. K., Dahal, L., Samarakoon, P. N., Tushar, F. I. & Martí, R. DSNet: Automatic dermoscopic skin lesion segmentation. Comput. Biol. Med. 120, 103738 (2020).
Al-Masni, M. A., Al-Antari, M. A., Choi, M. T., Han, S. M. & Kim, T. S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 162, 221–231 (2018).
Song, Z., & Yang, B. Ant colony based fish crowding degree optimization algorithm for magnetic resonance imaging segmentation in sports knee joint injury assessment. Expert Systems, 40(4), e12849. https://doi.org/10.1111/exsy.12849. (2023).
Shu, M., Fan, Z., Huang, B. & Wang, C. Retrospective analysis of clinical features of pembrolizumab induced psoriasis. Invest. New Drugs https://doi.org/10.1007/s10637-025-01536-5 (2025).
Luan, S. et al. Deep learning for fast super-resolution ultrasound microvessel imaging. Phys. Med. Biol. 68(24), 245023. https://doi.org/10.1088/1361-6560/ad0a5a (2023).
Yu, X. et al. Deep learning for fast denoising filtering in ultrasound localization microscopy. Phys. Med. Biol. 68(20), 205002. https://doi.org/10.1088/1361-6560/acf98f (2023).
Wu, Z., Li, X., Huang, R., He, B. & Wang, C. Clinical features, treatment, and prognosis of pembrolizumab -induced Stevens-Johnson syndrome / toxic epidermal necrolysis. Invest. New Drugs 43(1), 74–80. https://doi.org/10.1007/s10637-024-01499-z (2025).
Xu, X. et al. Large-field objective lens for multi-wavelength microscopy at mesoscale and submicron resolution. Opto-Electronic Advances. 7(6), 230212. https://doi.org/10.29026/oea.2024.230212 (2024).
Lama, N., Hagerty, J., Nambisan, A., Stanley, R. J. & Van Stoecker, W. Skin lesion segmentation in dermoscopic images with noisy data. J. Digit. Imaging 36(4), 1712–1722 (2023).
Bi, L. et al. Dermoscopic image segmentation via multistage fully convolutional networks. IEEE Trans. Biomed. Eng. 64(9), 2065–2074 (2017).
Nawaz, M. et al. Skin cancer detection from dermoscopic images using deep learning and fuzzy k-means clustering. Microsc. Res. Tech. 85(1), 339–351 (2022).
Wang, J., Chen, Y. & Zou, Q. Inferring gene regulatory network from single-cell transcriptomes with graph autoencoder model. PLoS Genet. 19(9), e1010942 (2023).
Nawaz, K. et al. Skin cancer detection using dermoscopic images with convolutional neural network. Sci. Rep. 15(1), 7252 (2025).
Wang, S., Dong, B., Xiong, J., Liu, L., Shan, M., Koch, A. W.,... Gao, S. Phase manipulating Fresnel lenses for wide-field quantitative phase imaging. Optics Letters, 50(8), 2683-2686. https://doi.org/10.1364/OL.555558. (2025).
Bakkouri, I. & Afdel, K. MLCA2F: Multi-level context attentional feature fusion for COVID-19 lesion segmentation from CT scans. SIViP 17(4), 1181–1188 (2023).
Bakkouri, I., Afdel, K., Benois-Pineau, J. & Initiative, G. C. F. T. A. S. D. N. BG-3DM2F: bidirectional gated 3D multi-scale feature fusion for Alzheimer’s disease diagnosis. Multimed. Tools Appl. 81(8), 10743–10776 (2022).
Chen, Y., Li, M., Li, J. "Fault-tolerant control design for nonlinear multilateral teleoperation system with unreliable communication channels and actuator constraints," International Journal of Machine Learning and Cybernetics, 16: 1991-2007, https://doi.org/10.1007/s13042-024-02373-3. (2025).
Khan, S., Ali, H. & Shah, Z. Identifying the role of vision transformer for skin cancer—A scoping review. Front. Artif. Intell. 6, 1202990 (2023).
Yizheng Wang, Xin Zhang, Ying Ju, Qing Liu, Quan Zou, Yazhou Zhang, Yijie Ding, Ying Zhang. Identification of human microRNA-disease association via low-rank approximation-based link propagation and multiple kernel learning. Frontiers of Computer Science. 2024, 18(2): 182903
Dakhli, R., & Barhoumi, W. (2024, October). Toward a quantitative trustworthy evaluation of post-hoc XAI feature importance maps using saliency-based occlusion. In 2024 IEEE/ACS 21st International Conference on Computer Systems and Applications (AICCSA) (pp. 1–8). IEEE.
Dakhli, R., & Barhoumi, W. (2024, April). Exploring XAI Attention Maps to Investigate the Effect of Distance Metric and Lesion-Shaped Border Expansion Size for Effective Content-Based Dermatological Lesion Retrieval. In Asian Conference on Intelligent Information and Database Systems (pp. 126–138). Singapore: Springer Nature Singapore.
Dakhli, R. & Barhoumi, W. Improving skin lesion classification through saliency-guided loss functions. Comput. Biol. Med. 192, 110299 (2025).
Dakhli, R. & Barhoumi, W. A skin lesion classification method based on expanding the surrounding lesion-shaped border for an end-to-end inception-ResNet-V2 classifier. SIViP 17(7), 3525–3533 (2023).
Wang, J., Wei, L., Wang, L., Zhou, Q., Zhu, L., & Qin, J. (2021, September). Boundary-aware transformers for skin lesion segmentation. In International conference on medical image computing and computer-assisted intervention (pp. 206–216). Cham: Springer International Publishing.
Ayas, S. Multiclass skin lesion classification in dermoscopic images using swin transformer model. Neural Comput. Appl. 35(9), 6713–6722 (2023).
Jiang, R. et al. A transformer-based weakly supervised computational pathology method for clinical-grade diagnosis and molecular marker discovery of gliomas. Nat. Mach. Intell. 6(8), 876–891. https://doi.org/10.1038/s42256-024-00868-w (2024).
Yang, J., Wang, G., Xiao, X., Bao, M. & Tian, G. Explainable ensemble learning method for OCT detection with transfer learning. PLoS ONE 19(3), e0296175. https://doi.org/10.1371/journal.pone.0296175 (2024).
Zhao, S., Sun, W., Sun, J., Peng, L. & Wang, C. Clinical features, treatment, and outcomes of nivolumab induced psoriasis. Invest. New Drugs 43(1), 42–49. https://doi.org/10.1007/s10637-024-01494-4 (2025).
Nie, Y., Sommella, P., Carratù, M., O’Nils, M. & Lundgren, J. A deep CNN transformer hybrid model for skin lesion classification of dermoscopic images using focal loss. Diagnostics 13(1), 72 (2022).
Ali, R., Hardie, R. C., De Silva, M. S., & Kebede, T. M. (2019). Skin lesion segmentation and classification for ISIC 2018 by combining deep CNN and handcrafted features. arXiv preprint arXiv:1908.05730.
Kassem, M. A., Hosny, K. M. & Fouad, M. M. Skin lesions classification into eight classes for ISIC 2019 using deep convolutional neural network and transfer learning. IEEE Access 8, 114822–114832 (2020).
Cassidy, B., Kendrick, C., Brodzicki, A., Jaworek-Korjakowska, J. & Yap, M. H. Analysis of the ISIC image datasets: Usage, benchmarks and recommendations. Med. Image Anal. 75, 102305 (2022).
Rashid, J., Boulaaras, S. M., Saleem, M. S., Faheem, M. & Shahzad, M. U. A novel transfer learning approach for skin cancer classification on ISIC 2024 3D total body photographs. Int. J. Imaging Syst. Technol. 35(2), e70065 (2025).
Khan, A. R., Mujahid, M., Alamri, F. S., Saba, T. & Ayesha, N. Early-stage melanoma cancer diagnosis framework for imbalanced data from dermoscopic images. Microsc. Res. Tech. 88(3), 797–809 (2025).
Amin, J., Azhar, M., Arshad, H., Zafar, A. & Kim, S. H. Skin-lesion segmentation using boundary-aware segmentation network and classification based on a mixture of convolutional and transformer neural networks. Front. Med. 12, 1524146 (2025).
Rasul, M. F., Dey, N. K., & Hashem, M. M. A. (2020, June). A comparative study of neural network architectures for lesion segmentation and melanoma detection. In 2020 IEEE Region 10 Symposium (TENSYMP) (pp. 1572–1575). IEEE.
Thapar, P. et al. A hybrid grasshopper optimization algorithm for skin lesion segmentation and melanoma classification using deep learning. Healthcare Analytics 5, 100326 (2024).
Ahmad, M., Ahmed, I., Chehri, A., & Jeon, G. (2025). Fusion of metadata and dermoscopic images for melanoma detection: Deep learning and feature importance analysis. Information Fusion, 103304.
Funding
The authors did not receive any financial support for this study.
Author information
Authors and Affiliations
Contributions
All authors conceived and designed the study. **Min Li, Yinping Jiang, Ge Cao, Tao Xu** and **Ruiqiang Guo** collected, simulated, and analyzed data. " **Ruiqiang Guo** " wrote the first draft, and all authors reviewed it.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Ethics approval
This study does not involve any human participants, personal data, or clinical interventions requiring ethical approval. The research is based entirely on publicly available dermoscopic image datasets (ISIC 2018, 2019, 2020, and 2024), which are anonymized and open-access, and thus exempt from institutional review board (IRB) approval.
Consent to publish
Not applicable. This study does not involve any individual person’s data in any form, including images, videos, or personal identifiers, that would require consent for publication.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, M., Jiang, Y., Cao, G. et al. HyperFusionNet combines vision transformer for early melanoma detection and precise lesion segmentation. Sci Rep 16, 505 (2026). https://doi.org/10.1038/s41598-025-30184-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-30184-1