Abstract
Lumpy skin disease (LSD) is a highly contagious viral disease of cattle and causes significant economic losses in the livestock industry, around the globe. Early and accurate detection is critical for effective disease management and control in a timely manner. Early-warning digital detection approach such as Convolutional Neural Networks (CNNs) have shown promising results in medical and veterinary diagnostics, Vision Transformers (ViT) remain relatively unexplored in this field. In the proposed research, we used a total of 8000 images of cattle (retrieved from Kaggle) and trained the model to achieve the optimal detection of infections. We used data pre-processing techniques of resizing, normalisation, and augmentation followed by the ViT architecture as a classifier. The model provided excellent performance that showed a 98.12% accuracy, 98.5% precision, 98.5% recall, and a 98.5 F1 score. We have shown that ViT achieves better results in LSD classification compared to multiple approaches that are traditionally used, providing greater accuracy and precision. To encourage adaptation and apply model easier in the field conditions, a mobile application was created and validated on PyTorch Lite and Flutter. Collectively, this powerful approach would change the health management of livestock and allow swifter, and more accurate diagnosis not only to contain the infection but also to counteract its transmission in susceptible livestock.
Similar content being viewed by others
Introduction
Lumpy skin disease (LSD) is a highly contagious disease in cattle due to lumpy skin disease virus (LSDV) which is a member of the genus Capripoxvirus. In disease-endemic countries, it causes extreme economic losses in livestock populations and manifestations of fever and nodules on the skin1. The disease is mostly transmitted by blood sucking insects thus posing a big problem in both tropical and temperate areas. Vulnerable calves include those with unvaccinated calves who tend to be complicated even though the mortality rates of LSD are still low2,3. The economic impact is substantial and is attributed to reduced milk yield, and damaged hides, and resulting trade restrictions exacerbate the impact of LSD on economies4.
Early and precise diagnosis is crucial for managing outbreaks effectively. Traditional laboratory methods, though reliable, are slow, expensive, and often inaccessible in resource-limited areas5. Recent advances in artificial intelligence (AI), particularly Convolutional Neural Networks (CNNs), have shown promise in veterinary diagnostics. Pre-trained models including Xception, VGG16, and MobileNetV2 have achieved up to 96.39% accuracy in detecting LSD, with strong sensitivity and specificity6. A smart intelligence system of breed and disease identification of hen was suggested by Himel et al.7 by using an ensemble method of Extra Tree classifier that provided high accuracy in determining poultry diseases. However, CNNs failed to capture long-range spatial patterns in images and demanded heavy computational power8.
Vision Transformers (ViTs) offer a compelling alternative to CNNs for on-site, economical and timely digital diagnosis of infectious diseases. Using self-attention mechanisms, global relationships in images can effectively be mapped by ViTs. In medical imaging, ViTs have outperformed CNNs in tasks like glioma classification and renal pathology analysis, reaching accuracies of 81.8% and 96–99%, respectively9,10. Their ability to interpret complex visual patterns makes them ideal for veterinary diagnostics. Despite their success in plant disease detection and other related fields such as image classification, object detection, and segmentation, ViTs applications to LSD classification is underexplored11. Several recent molecular investigations highlight emerging trends in disease dynamics and thus necessitating the application of advanced and highly accurately trained digital diagnostic platforms. For example, Abdallah et al.12 have found genetic differences in LSDV strains isolated from vaccinated Egyptian cattle and linked them to African and European variants of the LSD. Recently, Bich et al.13 have identified a unique 12-nucleotide insertion in Vietnamese LSDV strains, stressing the importance of region-specific monitoring and vaccine evaluation14. Himel et al.15 have come up with a classification model of skin cancer based on the architectures of Vision Transformer and Segment Anything Model (SAM) to segment the lesion. With the HAM10000 dataset, their methodology attained a Dice coefficient of 98.14 and an accuracy of 96.15 using Google ViT patch-32 model and compared to other superior conventional deep learning models. Such findings outline the potential of ViTs in learning multidimensional spatial relationships and enhancing diagnostic performance in very difficult to classify tasks.
This study explores ViT for classifying LSD in cattle, aiming to improve early detection and outbreak management. We propose a ViT-based model and compare its performance with existing CNN architectures. Our results show that ViTs achieve 98.12% accuracy, surpassing traditional CNNs by better identifying intricate LSD patterns in images. We have developed a mobile application that enables farmers and veterinarians to capture images and perform real-time, on-site diagnosis and monitoring of livestock diseases. This breakthrough could transform livestock health management, enabling faster, more accurate diagnoses and scalable prevention strategies during outbreaks.
Methods
A range of tools, manual and automated curations and models were sequentially and retrospectively applied to optimize and validate a model that aim to achieve early and reliable diagnostic system. A step-by-step application of these approaches is outlined in Fig. 1.
Data collection
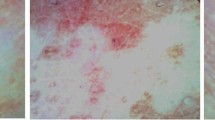
The dataset was sourced from Kaggle16, that was comprised of labelled images of either of the two types of cattle: LSD-affected cattle (Fig. 2) and healthy cattle (Fig. 3). In total, the dataset contained 8,000 images, with an equal number of 4,000 LSD-positive images and 4,000 healthy cattle images, ensuring a perfectly balanced class distribution. Data is easily loaded and pre-processed in a form of ImageFolder module available through directories PyTorch-compatible. The different categories were well distinguished to facilitate the process of training a model. Prior to training, we thoroughly prepared the dataset, so that we could be sure about the integrity of images and exclude corrupted ones or those which were missing. Such measure was needed to make the preprocessing and model evaluation consistent and reliable.
Data preprocessing
To improve performance of the model, we applied a few pre-processing techniques on the stored dataset. To begin with, all the images were transformed to a universal size of 224 by 224, i.e., the input dimensions needed by the Vision Transformer (ViT)17. The standardised sizing made sure that it was consistent among differently sized original size images. There was also data augmentation with random horizontal flipping and rotation (up to 10 degrees) to enable the model to handle more diverse representations of data. Data transformation pipeline was implemented by utilizing the PyTorch Tensor function that converts images to tensors and normalizes pixel values to [-1, 1] range by using a mean and standard deviation of 0.5. This normalization became useful towards stabilizing the training process. Lastly, to attain a balanced distribution of data, the data was partitioned into a training set and testing set.
Model selection
Based on good results in image classification, particularly with intricate patterns in data fetching, we selected the Vision Transformer (ViT) of this research. Among models available in the Timm library (Torch Image Models) we have chosen the variant ViT-Base (vit_base_patch16_224) due to its pre-trained transformer architecture. The way that this model works is that it decomposes images into smaller patches thereby allowing its self-attention mechanism to comprehend spatial positions within the image. The difference between ViT and normal Convolutional Neural Networks (CNNs) is that it does not use local feature extraction but rather apply self-attention mechanism. ViT differs in the common treatment of image patches as sequences, so its approach differs fundamentally to CNNs and their localized receptive fields. This algorithm has proved to be outstanding in many of image classification tasks, with larger image data sets. An overview of classification of healthy and affected cattle using a vision transformer is shown in Fig. 4.
Image patch embedding
The ViT processes images by dividing them into fixed size, non-overlapping patches that cover the entire image. For an input image of spatial resolution \(\:\text{H}\:\times\:\text{W}\:\), the image is partitioned into patches of size \(\:\text{P}\times\:\text{P}\:\). The total number of patches is given by:
Where N is the total number of patches. The image is resized or padded such that both H and W are multiples of P, ensuring N is a positive integer.
Each patch is flattened into a 1-D vector and then mapped to a D-dimensional embedding space through a learnable linear transformation, shown in 1:
where \(\:{x}_{i}\in\:{\mathbb{R}}^{{P}^{2}\cdot\:C}\) denotes the flattened vector of the \(\:i\)-th image patch (with \(\:C\) color channels), \(\:{W}_{p}\in\:{\mathbb{R}}^{D\times\:\left({P}^{2}C\right)}\) is a trainable weight matrix, and \(\:{b}_{p}\in\:{\mathbb{R}}^{D}\) is a learnable bias.
A learnable class token, \(\:{x}_{\text{cls\:}}\), is prepended to the sequence of patch embeddings. This token serves as a global representation that aggregates information from all image patches through the self-attention mechanism. After the transformer encoder, the final hidden state corresponding to this class token is used for image-level prediction, similar to the [CLS] token in BERT (Large Language Models).
Positional encoding
Since transformer architectures lack an inherent notion of spatial order, positional embeddings are added to each patch embedding to retain spatial information. The resulting sequence is expressed as
.
where \(\:{E}_{i}\in\:{\mathbb{R}}^{D}\) represents the positional encoding corresponding to the \(\:i\)-th patch. The operator " + " denotes elementwise addition between the patch embedding and its positional encoding. This enables the model to learn spatial relationships between patches and capture both local and global dependencies.
Multi-head self-attention (MSA)
The multi-head self-attention (MSA) mechanism enables global interactions among all patch embeddings, allowing the model to integrate contextual information across the entire image.
For a given input sequence \(\:Z\in\:{\mathbb{R}}^{(N+1)\times\:D}\), the Query \(\:\left(Q\right)\), Key \(\:\left(K\right)\), and Value \(\:\left(V\right)\) matrices are computed using separate linear projections:
where \(\:{W}_{Q},{W}_{K},{W}_{V}\in\:{\mathbb{R}}^{D\times\:{D}_{k}}\) are trainable projection matrices and \(\:{D}_{k}\) is the dimension of the key vectors.
The self-attention operation is defined as
.
To enhance the model’s representational power, multiple attention heads are applied in parallel:
where each attention head is computed as
.
and \(\:{W}_{O}\) denotes a learnable projection matrix applied to the concatenated heads. The matrices \(\:{W}_{{Q}_{j}},{W}_{{K}_{j}},{W}_{{V}_{j}}\) are head-specific projections.
Feedforward network (FFN)
After the attention operation, each transformer block processes its output through a feed-forward network (FFN) comprising two linear layers separated by a Gaussian Error Linear Unit (GELU) activation as shown in 6:
where W₁ and W₂ denote the learnable weight matrices, with b₁ and b₂ representing their corresponding bias terms. This FFN architecture enables non-linear feature transformations, refining the representations before propagation to subsequent layers.
Layer normalization and residual connections
Each transformer block employs layer normalization (LN) and residual connections to maintain stable gradient flow and mitigate degradation effects. This was formally expressed as shown in 7 and 8:
Where \(\:{\text{z}}_{\text{l}}\) is the final output of the transformer block at layer \(\:\text{l}\).
Classification head
Once the class token has been processed through multiple transformer layers, its final representation was retrieved and input into a softmax-activated fully connected layer for classification (9):
Where Wc and bc are trainable parameters, and \(\:{\text{z}}_{\text{L}}^{0}\) is the final class token embedding. The output \(\:\text{y}\) represents the probability distribution over the target classes.
Model training
For training and testing, 6,000 images were used as the training set, while 12,00 and 800 images were reserved for validation and testing respectively. The training process for ViT involves carefully designed steps, such as setting up loss functions, selecting optimization strategies, and implementing performance-enhancing methods. By training on various image categories, the model learns discriminative features to achieve better classification results. Figure 5 shows the flowchart of the proposed algorithm, starting from input data and data augmentation to model training and testing.
Loss function
This classification approach employs Cross-Entropy as its core loss function, defined by comparing the model’s predictions yi against the actual labels ti through the following formulation (10):
Where, N is a number of classes, \(\:{\text{y}}_{\text{i}}\)is the predicted probability output of class i, and \(\:{t}_{i}\) is a one-hot encoded true label. The loss function becomes effective by reducing the discrepancy between the actual probability of classes and the predictions made by the model.
Optimizer and learning rate strategy
Due to the maximized behavior with sparse gradients and the built-in weight decay regularization, which is paramount in reducing the overfitting, AdamW optimizer was chosen to be used in training the model. The parameter updates follow this mathematical formulation:
In which, θ itself is the parameter of the model, \(\:{\upeta\:}\) is the learning rate, \(\:{\text{m}}_{\text{t}}\) and \(\:{\text{v}}_{\text{t}}\) are the first and second estimates of the moment respectively, and \(\:\epsilon\) is a small constant that is used numerically maintained.
Training was performed with a batch size of 32, an initial learning rate of 0.0003, and a weight decay of 0.0001. To further enhance convergence and generalization, a cosine annealing learning rate scheduler was applied, gradually reducing the learning rate to near zero over the training period. This combination of optimizer choice, batch configuration, and learning rate scheduling ensured smooth convergence, efficient GPU utilization, and robust performance on the validation and test sets.
Data augmentation and batch processing
We use data augmentation which entails random horizontal flipping as well as random rotation to help in generalization and reduce overfitting. The data is split into 32 mini-batches which makes it easy to train on a GPU. This divide into batch-wise processing can enable updated parameters more frequently, a factor that enhances the ability to converge within a short period and without compromising on the computational effort.
Training procedure
The training of the model occurs in iterations and lasts 50 epochs, one of them containing the steps below:
-
1.
Forward Pass: Input images were processed by the Vision Transformer (ViT) to generate class probability distributions.
-
2.
Loss Computation: The Cross-Entropy Loss quantifies the discrepancy between predictions and ground-truth labels.
-
3.
Backward Pass: Gradients of the loss with respect to model parameters were computed via backpropagation.
-
4.
Weight Update: The optimizer (AdamW) adjusts the model weights based on the derived gradients.
This iterative design alters the parameters of the model in small steps, and their performance improves over time across different epochs.
Model evaluation
An evaluation assessment was used to analyze the success of the model to fit images of cattle into the Lumpy Skin Disease Dataset. The assessment includes interpreting the key performance measure, comparing the model predictions with the actual labels to find out the ability of the method to generalize new and unseen data.
Evaluation metrics
-
(a)
Accuracy: This metric is computed by dividing the number of accurate predictions with the total number of predictions by the percentage. It gives a rough estimate of the skill of the model in classify the images correctly as evidenced in formula 12.
$$\:\text{A}\text{c}\text{c}\text{u}\text{r}\text{a}\text{c}\text{y}=\frac{\text{T}\text{P}+\text{T}\text{N}}{\text{T}\text{P}+\text{T}\text{N}+\text{F}\text{P}+\text{F}\text{N}}$$(12)Where TP = True positives (correctly predicted positive cases). TN = True negatives (correctly predicted negative cases). FP = False positives (incorrectly predicted positive cases). FN = False negatives (incorrectly predicted negative cases).
-
(b)
Precision: This metric takes into account the quotient of precise, accurate prediction (true positives) related to all positive predictions (true positives + false positives). It is especially relevant in cases when the consequences of the false positive occurrence are high.
$$\:\text{P}\text{r}\text{e}\text{c}\text{i}\text{s}\text{i}\text{o}\text{n}=\:\frac{\text{T}\text{P}}{\text{T}\text{P}+\text{F}\text{P}}$$(13) -
(c)
Recall (Sensitivity): As the number of correctly predicted positives divided by the total amount of real positives, recall measures the extent to which the model identifies the actual instances exhaustively. It is crucial to perform under such applications that the cost of a failed positive (FN) is greater than the cost of false alarms.
$$\:\text{R}\text{e}\text{c}\text{a}\text{l}\text{l}=\:\frac{\text{T}\text{P}}{\text{T}\text{P}+\text{F}\text{N}}$$(14) -
(d)
F1-Score: It is the harmonic mean of precision and recall, balancing these two measures, which is why this metric is useful in particular when considering a model on data imbalanced in its classes.
$$\:\text{F}1\:\text{S}\text{c}\text{o}\text{r}\text{e}=2\:\text{X}\frac{\text{P}\text{r}\text{e}\text{c}\text{e}\text{s}\text{i}\text{o}\text{n}\:\text{x}\:\text{R}\text{e}\text{c}\text{a}\text{l}\text{l}}{\text{P}\text{r}\text{e}\text{c}\text{e}\text{s}\text{i}\text{o}\text{n}+\:\text{R}\text{e}\text{c}\text{a}\text{l}\text{l}}$$(15) -
(e)
Confusion Matrix: The Confusion matrix represents an appearance of a tabular display of model performance, exhibiting the allocation of accurate predictions and mistake in each class. Such decomposition allows pinpointing the patterns of misclassification as well as problematic categories with full accuracy.
Model deployment
This study contributes to the development of a mobile application for automated detection of Lumpy Skin Disease (LSD) in cattle. By integrating deep learning-based image classification with an intuitive mobile interface, the solution aims to support real-time LSD identification for livestock handlers, veterinary professionals, and agricultural field staff.
Mobile application development
Flutter was used to develop a mobile application as it provides better OS to be compatible across different platforms, making it usable on Android and iOS devices. The deep learning model trained using pytorch was converted to pytorch lite for easy mobile deployment. The app utilizes the device’s camera for image capture, pre-processes input and does a real-time inference for the embedded model used. Workflow of the application is demonstrated in Fig. 6. The application provides two major ways of uploading images into the mobile application:
The mobile application was developed using Flutter framework to ensure cross-platform compatibility across both Android and iOS devices. The deep learning model, originally trained using PyTorch, was converted to PyTorch Lite format for optimized mobile deployment. The application leverages the device’s built-in camera for direct image capture, incorporating automated input preprocessing and real-time inference capabilities through its embedded model. As shown in Fig. 6, the system architecture supports two primary image upload methods:
-
i.
Take Picture: direct capture using the device’s camera for real-time cattle assessment.
-
ii.
Selecting Image from Gallery: gallery selection for analyzing pre-existing images.
After the image acquisition, using either of the methods, the backend classification model integrated in the application executed. This dedicated deep learning model, trained on large datasets, conducts a thorough comparative analysis by detecting typical LSD lesions and differentiating them from normal cattle skin patterns. The entire workflow from taking the image to generating a diagnostic recommendation runs smoothly within the mobile platform, delivering quick, practical insights to users.
Image classification outcomes
The image was analysed and classified into one of the following categories:
-
1.
LSD-Positive: Indicates the presence of LSD symptoms, such as nodules, lesions, or abnormal skin texture, as identified by the model.
-
2.
LSD-Negative: Confirms that the cattle image shows no signs of LSD, indicating a healthy condition.
-
3.
Non-Cattle Image: The model determines that the uploaded image does not contain cattle or is unsuitable for classification (e.g., other animals or objects). This ensures diagnostic accuracy by filtering out irrelevant inputs.
Results are delivered in real time, facilitating early clinical decisions or additional investigations.
Application interface
The application has a simple interface which is user friendly, and there are clear visual indicators. The main screen of the application has two primary buttons to import, an image, and a check result with buttons of the camera, a gallery, and an about us section. The information section about us will summarize the details about the app and those behind the app and the intent. The use of icons is being utilized to improve navigation enabling the user to more easily use the system who does not qualify as a technical expert.
Practical utility and impact
Here the system provides a mobile solution to monitoring and surveillance of LSD outbreaks through introduction of AI-driven diagnostics to a smartphone application. It is faster in detecting the presence of an animal because it minimizes the need to use manual checks and laboratory tests, which makes it easier to provide veterinary aid.
Results and discussion
ViT model was evaluated by four main indicators: accuracy, precision, recall, and F1-score. The evaluation findings show that the model is effective at classifying images in their proper categories. Further validation using the confusion matrices and class-specific measures proves the resilience of the model in class-separation. Review of the classified and not-classified correctly generates concerns that could be improved. The excellent performances on classification in the ViT model is due to its structure, self-attention in which essential features of visual nature are successfully realized.
Qualitative results
In the qualitative assessment of the model, both correct and incorrect classifications are discussed. Analysis of prediction samples has revealed a high capability of the model to create class distinctions. The predictions performed by the ViT model for the LAD-infected cattle is shown in Fig. 7 while Fig. 8 provide an overview of the classification of normal cattle using ViT model. Self-attention maps produced by the self-attention layers suggested which regions of the photograph were the most essential in the prediction of ViT model showing its ability to grasp the important spatial relations and patterns of lesions. A close examination of such cases of misclassification, some patterns were observed. False negative responses were common where early-stage LSD lesions were very small, partly covered by hair, and/or in image regions with poor illumination, and therefore visually resembled normal skin. Other factors that influenced the visibility of subtle lesions include motion blur, or low-resolution images in some instances. However, false positives were often related to the fact that cattle with some other normal skin condition with the appearance of LSD nodules like healed scar or insect-derived inflammation and little rupture superficially resembled LSD nodules. Moreover, there are environmental conditions including spotting of dirt or splashing of mud on the skin that sometimes confused the model. Such observations indicate that increasing the variety and difficulty of the images in the dataset, especially by adding early infections, differing light settings, and representations of typical non-LSD skin anomalies could increase robustness. Further options to minimize such misclassification cases are advanced data augmentation techniques, refinement of the attention mechanisms to better selectively focus on lesion morphology, and inclusion of multi-modal data (e.g., thermal imaging) as possible future directions.
Quantitative results
The performance of the trained ViT model’ was assessed through standard classification measures such as accuracy, precision, recall, F1-score, and confusion matrix analysis. Each metric is explained in detail in the following section.
Performance matrices
As shown in Table 1, classification accuracy by the trained model was vast ranging to 98.12%, meaning that the model could identify a large portion of the images in the dataset. The model produces low false positive values (98.5%) and hence reliable predictions. Similarly, its recall score of 98.5% shows a good presence of relevant instances by few false negatives. 98.5% is defined as an incredible F1 score that characterizes the performance of the model and guarantees fantastic overall performance and no overfitting. The combination of these metrics confirms the high performance of the model to be used in the intended classification task, making it one of the candidates to be used as a viable approach in real-life cases. Analysis of confusion matrix can be further done to give insights into relationships between classes and their performance.
Confusion matrix
The confusion matrix shows the classification output of the model (Fig. 9) with four important parameters namely, True Positive (TP), True Negative (TN), False Positive (FP) and False Negative (FN). The model is strongly sensitive and specific in that it accurately determines 388 cases that were infected and 401 were normal. But it gave error in the sense that it incorrectly identified 10 infected samples as normal (False Negative) and one as infected (False Positive) where there was high overall precision less errors. There was also a significant finding of a small bias to the misclassification of normal cases as infected which led to a large number of false negatives than false positives. This is more so especially in medical diagnostics with the attendant implications of false negatives being worse than false positives. In order to counter this, we could consider improvement ideas to update thresholds of classification or rebalance the class weights. On the whole, the results of the confusion matrix prove once again that the model can offer great and reliable classification capabilities, and only slight aspects can be optimized in it.
Comparative analysis
The comparative analysis with other models clearly demonstrated that the ViT outperforms the others, achieving an accuracy of 98%, precision of 98%, recall of 98%, and a perfect F1-score of 98% (Table 2). MobileNetV2 ranks second, achieving 96% accuracy, 99% precision, but a somewhat lower recall (88%), yielding an F1-score of 94%. DenseNet201 attains 94% accuracy, with balanced precision (93%) and recall (95%), also resulting in a 94% F1-score. Xception, despite its lower accuracy (93%), excels in recall (96%) and achieves a 93% F1-score. ResNetV2 performs the weakest, with 92% accuracy, 88% precision, and 95% recall, culminating in a 91% F1-score. In summary, ViT emerges as the top-performing model in this evaluation.
Comparative performance of deep learning models across four key metrics was assessed (Fig. 10): Accuracy, Precision, Recall, and F1-score. The graphical analysis clearly highlights ViT as the top-performing model, achieving the highest scores in Accuracy, Precision, and Recall. Although its Recall is marginally lower than MobileNetV2’s, ViT maintains robust overall performance. Meanwhile, DenseNet201 showed a balanced performance across all metrics, albeit slightly below MobileNetV2 and ViT. In contrast, Xception and ResNetV2 exhibit relatively weaker performance, particularly in Accuracy and Precision. However, Xception compensates with a higher Recall score. Overall, the visual comparison underscores ViT’s dominance, demonstrating its consistent superiority across all evaluated metrics.
Performance evaluation of the mobile application
The performance of mobile application was evaluated based on inference speed, accuracy, and usability. Real-world testing across multiple devices showed an average prediction time of under one second, with accuracy comparable to desktop-based systems. Field trials participants reported positive experiences, particularly regarding the app’s ease of use and dependable disease detection, underscoring its practical benefits for livestock management. To illustrate user interface (UI) of the mobile application and its real-world functionality, several screenshots are presented in Fig. 11. These results validate an end-to-end workflow of the application, showcasing both its intuitive design and the effective integration of AI-driven classification.
From the application’s central hub, users can access the main features of the application including images importing, real time capture, results checking and application information in a fast manner (Fig. 11a). Figure 11b shows the camera interface that allows users to take live pictures of cattle, an important aspect for field based and on-site disease diagnosis. Alternatively, users can also choose any previously stored pictures from their device’s gallery to further ease the image selection process (Fig. 11c). After submitting an image, the application processes the input from the embedded classification model and returns one of three possible outputs. It can output where the model identifies correctly the symptoms of the Lumpy Skin Disease (LSD) (Fig. 11d) or it can illustrate a situation in which the model classifies the image of the cattle as healthy (No-LSD) (Fig. 11e). In the cases of non-cattle input or incorrectly framed photos of cattle, in cases where the input image is irrelevant, the application treats the situation gracefully presenting informative error message as shown in Fig. 11(f).
Illustration of the key screens of the mobile application, showcasing its user interface and functionality. The main page (a) features an image display area alongside navigation and function icons. Users can either capture real-time images through the camera interface (b) or select existing photos from the gallery page (c). After processing, the app displays one of three possible outputs: a detection of Lumpy Skin Disease (LSD) in cattle (d), a confirmation of a healthy (No-LSD) image (e), or an error message for irrelevant/non-cattle inputs (f). This structured workflow ensures intuitive navigation, from image acquisition to accurate disease assessment, enhancing user experience and diagnostic efficiency.
Conclusion
This paper presents an automated methodology of classifying LSD through a ViT model. Our pre-processing model included simple pre-processing in order to optimize the dataset of around 8,000 images available on Kaggle by resizing, normalizing and performing a data augmentation. The ViT architecture was utilized as a kind of a classifier that can classify healthy and diseased cattle skin images. The dataset was divided into 6000, 1200 and 800 training, validation and testing sets respectively. It was trained with more than 50 epochs and was tested with the classical performance measures (accuracy, precision, recall, F1-score), as well as a confusion matrix. The findings were very good and gave a ratio ranging between 98 and 98, which equalled the rate of accuracy 98.12, precision 98.5, recall 98.5, and F1-score 98.5. When compared to existing research, the proposed model outperformed previous methods across all evaluation metrics. To bridge the gap between AI-assisted cattle disease detection and real-world application, a supporting mobile application was developed, offering an accessible and time-efficient solution for livestock disease screening. However, this study has certain limitations. The dataset size (8,000 images) remains relatively small and expanding it could further enhance model performance. Additionally, computational resource constraints during training suggest that more advanced hardware could yield even better results. While a mobile app was developed, it has not yet been tested in real-world agricultural environments on a scale, and early user feedback comes from a small group. The model also risks false reassurance if used without veterinary confirmation, especially in the case of false negatives. To build on this work, we plan to collect more diverse, real-life images across multiple regions and conditions, ideally with lab-confirmed cases. We also intend to expand the model to detect a wider range of cattle diseases, support low-end devices more efficiently, and integrate geotagging for outbreak monitoring. Field testing in collaboration with veterinary teams and refining the app based on real user input are also priorities.
Data availability
The dataset used in this study is publicly available on Kaggle at https://www.kaggle.com/datasets/warcoder/lumpy-skin-images-dataset, and all experiments were performed using this dataset.
References
Adamu, K. et al. Lumpy skin disease virus isolation, experimental infection, and evaluation of disease development in a calf. Sci. Rep. 14, 20460. https://doi.org/10.1038/s41598-024-60994-8 (2024).
Haider, A. et al. Lumpy skin disease: insights into molecular pathogenesis and control strategies. Veterinary Sci. https://doi.org/10.3390/vetsci11110561 (2024).
Ahmed, M. S. Lumpy skin disease: A review of epidemiological study and preventive measures. Bangladesh J. Veterinary Med. 22 (1), 7–16. https://doi.org/10.33109/bjvmjj2024fam2 (2024).
Di Giuseppe, A., Zenobio, V., Dall’Acqua, F., Di Sabatino, D. & Calistri, P. Lumpy skin disease. Veterinary Clin. North. America-Food Anim. Pract. 40 (2), 261–276. https://doi.org/10.1016/j.cvfa.2024.01.002 (2024).
Bora, M., Mathew, D. D., Das, H., Bora, D. P. & Barman, N. N. Point of care diagnostics and non-invasive sampling strategy: A review on major advances in veterinary diagnostics. Acta Vet. Brno. 91 (1), 17–34. https://doi.org/10.2754/avb202291010017 (2022).
Senthilkumar, C., Vadivu, C. S. & Neethirajan, S. G. Early detection of lumpy skin disease in cattle using deep Learning—A comparative analysis of pretrained models. Vet. Sci. https://doi.org/10.3390/vetsci11100510 (2024).
Himel, G. M. S. & Islam, M. M. A smart intelligence system for Hen breed and disease classification using extra tree classifier-based ensemble technique. J. Electr. Syst. Inform. Technol. 12 (2), 2. https://doi.org/10.1186/s43067-025-00191-3 (2025).
Chibuike, O. & Yang, X. Convolutional neural Network–Vision transformer architecture with gated control mechanism and Multi-Scale fusion for enhanced pulmonary disease classification. Diagnostics 14 (24), 2790. https://doi.org/10.3390/diagnostics14242790 (2024).
Wickramasinghe, W. M. S. P. B. & Dissanayake, M. B. Vision Transformers for glioma classification using T1 magnetic resonance imaging. Deleted J.. https://doi.org/10.36922/aih.4155 (2024).
Zhang, J. et al. Vision transformer introduces a new vitality to the classification of renal pathology. BMC Nephrol. 25 (1). https://doi.org/10.1186/s12882-024-03800-x (2024).
Fu, X. et al. Crop pest image recognition based on the improved ViT method. Inform. Process. Agric. 11 (2), 249–259. https://doi.org/10.1016/j.inpa.2023.02.007 (2024).
Abdallah, F. M., Damaty, E., Kotb, G. F. & H. M., & Sporadic cases of lumpy skin disease among cattle in Sharkia province, egypt: genetic characterization of lumpy skin disease virus isolates and pathological findings. Veterinary World. 11 (8), 1150–1158. https://doi.org/10.14202/vetworld.2018.1150-1158 (2018).
Bich, T. N. et al. Characterization and molecular identification of the lumpy skin disease virus in cattle in the Mekong delta of Vietnam. Open. Veterinary J. 14 (8), 1877–1895. https://doi.org/10.5455/OVJ.2024.v14.i8.16 (2024).
Sharma, S., Joshi, K., Singh, R., Chauhan, P. & Garg, P. In depth analysis of recent models for lumpy skin disease virus and stage detection: A comparative study. In Emerging Trends in Expert Applications and Security. ICE-TEAS 2024. Lecture Notes in Networks and Systems (eds Rathore, V. S. et al.), vol 1030. https://doi.org/10.1007/978-981-97-3745-1_29 (Springer, 2024).
Himel, G. M. S., Islam, M. M., Al-Aff, K. A., Karim, S. I. & Sikder, M. K. U. Skin cancer segmentation and classification using vision transformer for automatic analysis in dermatoscopy-based noninvasive digital system. Int. J. Biomed. Imaging 2024 (1), 3022192. https://doi.org/10.1155/2024/3022192 (2024).
Kaggle. Lumpy Skin Disease in Cattle (LSD). https://www.kaggle.com/datasets/andrewmvd/lumpy-skin-disease-in-cattle (n.d.).
Chen, T. et al. A vision transformer machine learning model for COVID-19 diagnosis using chest X-ray images. Healthc. Analytics. 5, 100332. https://doi.org/10.1016/j.health.2024.100332 (2024).
Author information
Authors and Affiliations
Contributions
Asad Ullah and Fakhre Alam: Conceptualization, Methodology, Data Curation, Writing Original Draft.Numan Iqbal and Yoga Rahulamathavan: Formal Analysis, Software, Validation, Visualization.Mohammed A Rohaim: Project AdministrationReview & Editing.Muhammad Munir: Supervision, Conceptualization, Data Collection, Project Administration, Funding Acquisition, Review & Editing.Ali Haider, Muhammad Waqas Khokhar: Investigation, Resources, Data Collection.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ullah, A., Haider, A., Rahulamathavan, Y. et al. A vision transformer model-integrated mobile application for early and accurate detection of lumpy skin disease in cattle. Sci Rep 16, 683 (2026). https://doi.org/10.1038/s41598-025-30259-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-30259-z