Abstract
Antimicrobial resistance (AMR) in Enterobacterales poses serious public health, agricultural, and environmental threats. In Southeast Asia, a coordinated “One Health” approach is lacking, and fragmented evidence hampers targeted interventions. This study systematically quantify and analyse AMR prevalence across human, animal, and environmental sectors in Southeast Asia by conducting a meta-analysis of 137 observational studies from 2013 to 2023. We found that Ceftriaxone resistance in E. coli was highest in human samples (49.3%, 95% CI: 37.3–61.3; N = 2,640), followed by environmental (37.1%, 95% CI: 8.4–72.2; N = 288) and animal sources (11.2%, 95% CI: 1.6–27.9; N = 923). In humans, meropenem resistance was 13.0% in K. pneumoniae (95% CI: 2.0–31.3; N = 7,803) and 1.4% in E. coli (95% CI: 0.1–4.4; N = 13,696). Resistance increased over time in human (p = 0.009) and animal sectors (p = 0.004). blaCTX-M and blaTEM were reported across all sectors. This synthesis also highlights a critical evidence gap: most studies focused on Thailand (67) and Vietnam (42). Samples came mostly from animals (62) and humans (59), with limited multi-sector studies. Only one study assessed all four sectors (human, animal, environment, food). Our study reveals an escalating AMR crisis alongside critical research gaps across Southeast Asia. Future efforts must therefore strengthen both integrated surveillance to understand transmission and regional health systems to implement effective One Health action.
Similar content being viewed by others
Introduction
Antimicrobial resistance (AMR) poses a severe global health threat. Enterobacterales frequently cause serious infections, including urinary tract infections, sepsis, and pneumonia, amounting to high clinical and economic burdens1,2. Beyond humans, these pathogens can facilitate the transfer of resistance genes across human, animal, and environmental boundaries, and negatively impact animal and environmental health3,4. This potential interspecies and environmental mobility of AMR necessitates a comprehensive approach that extends beyond human medicine to include veterinary and environmental sciences5. In line with the Sustainable Development Goals ‘Good Health and Well-being’, ‘Clean Water and Sanitation’, and ‘Responsible Consumption and Production’6, addressing AMR holistically with a One Health approach is crucial for promoting public health and sustainable living practices.
The Southeast Asia region (SEAR), a centre for global biodiversity characterized by varied socioeconomic landscapes, faces unique challenges that exacerbate the spread of AMR7. The region’s high population density and significant role in global trade and travel can facilitate international dissemination of antibiotic resistance, posing public health threats beyond geographical borders. Most countries within SEAR are of low- and middle-income with limited antibiotic resistance surveillance and regulation7,8. The recent surge in antibiotic resistance in SEAR, has been described as ‘burgeoning and often neglected’ 9. As a result, SEAR bears high AMR burdens with multidrug-resistant Enterobacterales as a leading cause of infections7,8,9,10.
To effectively tackle AMR in SEAR, a critical first step is to understand the epidemiology and resistance transfer amongst various reservoirs of AMR collectively as a region5,11. This includes the integration and synthesis of diverse and fragmented datasets, accounting for inherent biases, and addressing gaps in AMR surveillance11. Current studies often restrict to specific sectors, such as human health, agriculture, or the environment10,12, and localized areas within the region13,14. This piecemeal approach prevents the quantification of the overall burden from a One Health perspective, hampering targeted control strategies in this region.
We conducted a systematic review and meta-analysis to describe the epidemiology of antibiotic resistance in Enterobacterales from a One Health perspective in SEAR. We aimed to provide comprehensive estimates for the prevalence of antibiotic resistance in Enterobacterales in SEAR across human, animal, and environmental sectors, identify data gaps, and inform the development of targeted control strategies for the region.
Results
Study characteristics
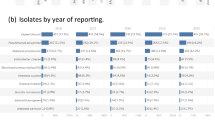
The initial search identified a total of 844 citations. Of these, 137 studies were included in the systematic review and analysis (Fig. 1). The number of studies increased yearly from two in 2015 to 47 in 2022 (Fig. 2A). The most common countries where these studies were conducted were Thailand with 67 studies, followed by Vietnam with 42 studies, and Indonesia with 19 studies. In contrast, Laos (n = 7 studies) and Myanmar (n = 4 studies) published few studies. No studies were reported from Brunei (Fig. 2B).
In terms of pathogens, Escherichia coli (E. coli) was the most frequently reported (n = 123 studies), followed by Salmonella spp. (n = 44 studies) and Klebsiella pneumoniae (K. pneumoniae) (n = 27 studies). Of the 137 studies included, 59 reported data from the human sector and 62 from the animal sector, with some studies covering multiple sectors. Specifically, 29 studies (21% of included studies) investigated antibiotic resistant Enterobacterales across more than one sector (Fig. 1; Table 1). Among these multi-sector studies, only one study examined all four sectors (human, animal, environment, and food). Additionally, five examined AMR prevalence in animals and their human contactors (table S2).
Antibiotic resistance in the human sector
Human studies were primarily conducted in hospitals (n = 39), using clinical samples (n = 36) and faecal samples (n = 21) (Table 1). Carbapenems were the most frequently tested class of antibiotics (number of isolates (N) = 62,060), followed by third generation cephalosporins (3GC, N = 48,547), and fluoroquinolones (N = 25,489) (Fig. 3, table S3). We found an increasing trend in carbapenem resistance over the past ten years (p = 0·009) (Fig. 4, table S4). No significant temporal linear changes were observed for 3GC and fluoroquinolone resistance (p = 0.304, p = 0.975, respectively).
Reported prevalence of antibiotic resistance in Enterobacterales by Sector and Antibiotic Class in Southeast Asia, 2013–2023. This boxplot illustrates the distribution of antibiotic resistance across various sectors for different antibiotic classes in the SEAR over the last decade. Each bubble represents the reported proportion of antibiotic resistance in individual samples, while transparency indicates the total number of isolates for each sample, with higher transparency corresponding to smaller sample sizes. The boundaries of the boxes denote the interquartile range, i.e., the 25th and 75th percentiles, while the median resistance is indicated by a horizontal line within each box. Whisker lengths extend to 1.5 times the interquartile range, with outliers depicted as separate points beyond these boundaries. 1GC = first generation cephalosporins. 2GC = second generation cephalosporins. 3GC = third generation cephalosporins. 4GC = fourth generation cephalosporins. BL-BLI = β-lactam or β-lactamase inhibitors. ESBLs = extended-spectrum beta-lactamases.
Temporal trend of antibiotic resistance in Enterobacterales in the Southeast Asia region from 2013–2023. This figure illustrates the temporal trends in antibiotic resistance among Enterobacterales in the Southeast Asia region from 2013 to 2023. LOESS smoothing (blue line) and weighted linear regression (red dashed line) are used to depict resistance trends, with the corresponding p-value for the linear trend presented. 3GC = third generation cephalosporins.
Pooled estimation found that cephalosporin resistance was higher than carbapenem across the region (Fig. 5, table S5). Cephalosporin resistance levels for E. coli were 45.9% (95% CI: 25.9–66.7%, N = 1,039, I2 = 97.0%) resistance for cefepime (4GC) and 49.3% (95% CI: 37.3–61.3, N = 2,640, I2 = 97.0%) for ceftriaxone (3GC). K. pneumoniae showed 44.3% (95% CI: 14.1–77.0, N = 592, I2 = 98.3%) resistance to cefepime and 51.1% (95% CI: 31.5–70.5, N = 1,896, I2 = 97.2%) to ceftriaxone. For resistance to carbapenem, meropenem resistance was 7.2% (95% CI: 1.8–15.9%, N = 7,803, I2 = 98.4%) for K. pneumoniae, while for E. coli, meropenem resistance was 1.4% (95% CI: 0.1–4.4%, N = 13,696, I2 = 96.7%) (Fig. 5, table S5).
The pooled prevalence of bacteria-specific antibiotic resistance in Enterobacterales across sectors in the Southeast Asia region from 2013 to 2023. This heatmap describes the pooled prevalence of bacteria-specific antibiotic resistance in Enterobacterales in humans, animals, environments, and food. Colors represent the proportion of antibiotic resistance. The size of the circle represents the sample size of each estimation.
Country-specific data revealed varying resistance levels (Fig. 6, table S6). For cephalosporin resistance, Myanmar, Vietnam, and Indonesia, showed high resistance to ceftriaxone in E. coli at 82.2% (75.8–87.7%, N = 157, I2 = 0.0%), 70.9% (95% CI: 51.4–87.0%, N = 550, I2 = 85.1%), and 62.4% (95% CI: 26.4–91.9%, N = 525, I2 = 97.6%), respectively. Resistance toward carbapenem was relatively low across different countries.
The country-specific pooled prevalence of bacteria-specific antibiotic resistance in Enterobacterales across sectors in the Southeast Asia region from 2013 to 2023. Country-level pooled analysis of antibiotic resistance within Enterobacterales, differentiated across the human, animal, environment, and food sectors. Colours represent the sample size of each estimation. The proportion of antibiotic resistance is displayed as a percentage in the box. Only sample size larger than 100 are included in the plot. Estimations for Klebsiella spp. and Nontyphoidal Salmonella can be found in table S5. SMZ-TMP: Sulfamethoxazole-Trimethoprim. TZP: Piperacillin-tazobactam. AMC: Amoxicillin-clavulanic acid.
Antibiotic resistance in the animal sector
Animal studies focused on livestock (n = 21) and poultry farms (n = 17), using faecal samples (n = 27) and rectal swabs (n = 14) (Table 1). 3GC was the most frequently tested antibiotic (N = 15,308). Other commonly tested antibiotics include tetracyclines (N = 10,903) and sulfonamides (N = 10,715) (Fig. 3, table S3). 3GC resistance increased over time (p = 0·004). Carbapenem and fluoroquinolone resistance remained consistently low without significant variation (p = 0·922, p = 0·230, respectively) (Fig. 4, table S4).
High resistance to sulfonamides and tetracyclines was observed across the region (Fig. 5, table S5). E. coli demonstrated resistance rates of 73.7% to sulfamethoxazole (95% CI: 65.4–81.3%, N = 4,198, I2 = 97.1%), while Salmonella spp. showed 62.0% resistance to sulfamethoxazole (95% CI: 24.1–92.9%, N = 1700, I2 = 99.4%). Tetracycline resistance was also prevalent, with E. coli and Salmonella spp. showing resistance rates of 60.8% (95% CI: 49.2–71.9, N = 6,125, I2 = 98.4%)) and 53.1% (95% CI: 37.0-68.9, N = 2,675, I2 = 98.0%), respectively. In contrast, resistance to 3GC in E. coli was relatively low throughout the region, with 11.2% resistance to ceftriaxone (95% CI: 1.6–27.9%, N = 923, I2 = 98.1%).
Several country-specific hotspots were identified (Fig. 6, table S6). Sulfonamide resistance was high among E. coli in Vietnam, Thailand, and Lao PDR. Resistance to sulfamethoxazole was 87.0% in Vietnam (95% CI: 80.9–92.2, N = 1,092, I² = 86.3%), 75.6% in Thailand (95% CI: 65.5–84.8, N = 523, I² = 80.7%), and 75.8% in Lao PDR (95% CI: 58.3–89.7, N = 1,979, I² = 98.8%). Lao PDR showed high tetracycline resistance for both E. coli (85.2%, 95% CI: 74.0-93.7, N = 1,979, I2 = 98.0%) and Salmonella spp. (74.8%, 95% CI: 45.4–95.2, N = 1,430, I2 = 99.2%).
Antibiotic resistance in the food sector
Food sector research centred on markets (n = 18) and mostly food samples (n = 25) (Table 1). 3GC was the most frequently reported class of antibiotics (N = 4,746) (Fig. 3). Other commonly tested antibiotics included tetracycline (N = 2,373), fluoroquinolones (N = 2,133), and carbapenems (N = 1,668) (Fig. 3, table S3). No significant temporal changes were observed in 3GC (p = 0·076), carbapenem (p = 0·754), or fluoroquinolone (p = 0·094) resistance over time (Fig. 4, table S4).
High resistance to various antibiotics were observed in both Salmonella spp. and E. coli from food sources (Fig. 5, table S5). Sulfamethoxazole resistance was 84.1% in Salmonella spp. (95% CI: 48.9–99.9%, N = 422, I2 = 99.0%) and 56.8% in E. coli (95% CI: 51.5–62.0%, N = 459, I2 = 24.1%). Tetracycline resistance was 66.0% in nontyphoidal Salmonella (95% CI: 37.2–89.4%, N = 203, I2 = 92.8%) and 61.9% in Salmonella spp. (95% CI: 45.1–77.4%, N = 824, I2 = 96.2%). 3GC and carbapenem resistance were relatively low, with Salmonella spp. showing 4.1% resistance to ceftazidime (95% CI: 0.2–12.2%, N = 536, I2 = 86.4%)) and 4.5% to imipenem in E. coli (95% CI: 0.3–13.4%, N = 694, I2 = 95.1%). Country-specific analysis revealed that sulfonamide and tetracycline resistance were high among Salmonella spp. in Lao PDR (Fig. 6, table S6). Resistance to sulfamethoxazole was 98.9% (95% CI: 91.0–99.1%, N = 178, I² = 86.6%), while resistance to tetracyclines was 95.8% (95% CI: 91.2–98.7%, N = 178, I² = 38.5%).
Antibiotic resistance in the environmental sector
Samples from the environment were mostly derived from aquatic ecosystems and wastewater
3GC (N = 2,552), fluoroquinolones (N = 2,899), and carbapenems (N = 1,450) were frequently tested among selected studies (Fig. 3, table S3). 3GC and fluoroquinolone resistance showed a decreasing trend over the decade (p < 0·001, p = 0.002, respectively). Carbapenem resistance followed a fluctuating pattern over time without significant linear temporal changes (p = 0·592) (Fig. 4, table S4).
In E. coli across the region, there was prevalent resistance to 3GC (ceftazidime 21.3%, 95% CI: 1.3–56.0%, N = 516, I2 = 97.7%; ceftriaxone 37.1%, 95% CI: 8.4–72.2%, N = 288, I2 = 92.2%), tetracycline (40.8%, 95% CI: 31.0-51.1%, N = 1,050, I2 = 86.1%), and fluoroquinolone (ciprofloxacin 19.6%, 95% CI: 10.5–30.6%, N = 1,888, I2 = 92.6%) (Fig. 5, table S5). Country-specific data (Fig. 6, table S6) for E. coli showed high resistance toward tetracycline in Cambodia (54.6%, 95% CI: 44.1–64.9%, N = 99, I2 = 21.3%) and Vietnam (56.1%, 95% CI: 35.8–75.3%, N = 233, I2 = 90.1%).
Characteristics of resistance genes
Among studies that reported resistance genes, 18 included animal isolates, 12 human isolates, 14 environmental isolates, and 4 food sector isolates (table S7). The blaCTX-M gene family is common across various sectors. In hospitals, blaCTX-M-15 showed the highest prevalence (48.3%, 95% CI: 36.4–60.2%, N = 433, I² = 73.7%), followed by blaCTX-M-9 (20.0%, 95% CI: 0.0–62.7%, N = 628, I² = 98.9%) and blaCTX-M-55 (13.7%, 95% CI: 6.5–23.0%, N = 1,306, I² = 93.9%). Other variants including blaCTX-M-27, blaCTX-M-14, blaCTX-M-3, blaCTX-M-24, and blaCTX-M-65 were detected at lower frequencies. In poultry farms, blaCTX-M-15 and blaCTX-M-1 were both detected at 15.5% (95% CI: 5.2–29.9%, N = 312, I² = 89.9%), followed by blaCTX-M-2 (14.0%, 95% CI: 5.1–26.2%, N = 312, I² = 87.1%), blaCTX-M-9 (11.4%, 95% CI: 6.9–16.9%, N = 312, I² = 50.8%), and blaCTX-M-8 (4.7%, 95% CI: 2.0–8.3%, N = 312, I² = 44.5%) (Fig. 7, Table S8).
The pooled prevalence of resistance genes and their origins across study settings in the SEAR (2013–2023). This figure illustrates two key aspects of resistance gene data. Due to sample limitations, we reported both gene families and their specific subtypes. In cases where the raw data only identified the gene families without specifying subtypes, the data was reported at the family level. (A) Shows the pooled prevalence of resistance genes across different study settings. Colours represent the sample size of each estimation. The proportion of resistance genes is displayed as a percentage in the box. (B) Shows a chord plot depicting the origins of resistance genes from various study settings. The width of the links represents the number of isolates with resistant genes.
The blaTEM gene showed the highest prevalence in fish farms (59.7%, 95% CI: 43.9–74.5%, N = 188, I² = 73.1%), followed by hospitals (50.9%, 95% CI: 28.4–73.2%, N = 860, I² = 96.4%) and poultry farms (40.2%, 95% CI: 5.8–81.7%, N = 467, I² = 97.2%). Lower prevalence was observed in lakes (38.3%, 95% CI: 28.5–48.5%, N = 947, I² = 76.2%), livestock farms (21.5%, 95% CI: 0.8–58.9%, N = 529, I² = 99.0%), and marine environments (7.6%, 95% CI: 5.3–50.5%, N = 96, I² = 91.3%). Similarly, the tetA gene was common in fish farms (61.9%, 95% CI: 47.2–75.6%, N = 188, I² = 75.3%), with lower levels in livestock farms (31.3%, 95% CI: 14.6–50.9%, N = 517, I² = 96.5%), and rivers (18.0%, 95% CI: 6.3–34.0%, N = 535, I² = 92.7 (Fig. 7, Table S8).
Discussion
In this systematic review and meta-analysis of antibiotic resistant Enterobacterales in SEAR, we found a high prevalence of resistance in key antibiotic classes across the region. Resistance to some 3GCs was over 50% in Enterobacterales isolated from human sectors, while carbapenem resistance was relatively lower in each sector. Substantial sulfonamides resistance was found mainly in animal and food sectors, while tetracyclines resistance was found in animal, food, and environmental sectors. Resistance genes such as blaCTX-M and blaTEM were reported from various source settings, indicating shared ecological niches across hosts and sectors. In addition, we identified data gaps across pathogens, countries, and sectors, with a notable scarcity of studies exploring cross-sectoral transmission especially in low-income settings.
Our finding of the high prevalence of antibiotic resistance in SEAR amongst the human sector superseded that in North America, Western Europe. Specifically, 4GCs and 3GCs showed a higher resistance levels near 50% compared to the 10–30% resistance reported in North America, Western Europe15,16, and aligned more closely with the patterns observed in sub-Saharan Africa (40.6–84.9%)17. In terms of carbapenem resistance, the prevalence in the SEAR was lower than that reported in Africa, which showed a pooled prevalence of 30.34%18. The overall antibiotic resistance pattern in the SEAR likely stemmed from unregulated antibiotic distribution and insufficient hospital infection control19,20. Our study’s findings, which mirror the rise in antibiotic use in SEAR21, underscores the need for targeted interventions to mitigate the region’s high resistance levels.
The high sulfonamide and tetracycline resistance across animal and food sector were concerning. These resistance likely stemmed from the extensive usage of these antimicrobials in livestock operations, attributed to their economic accessibility, disease prevention, and growth-enhancement properties22,23. Recent systematic analyses of regional AMR action plans identified critical gaps in governance frameworks and policy implementation across Southeast Asian nations, which could harbour systemic deficiencies in regulatory oversight and surveillance infrastructure24. While the ASEAN Working Group on Livestock has established a regional framework, prioritizing standardized surveillance systems, and harmonized regulatory controls are critical to mitigate escalating antimicrobial resistance patterns in Southeast Asian livestock production25.
The widespread distribution of resistance genes such as blaCTX-M and blaTEM across various environments, including hospitals, farming, and food markets, highlighted the role of human activities and food supply chains in disseminating these mobile genetic elements [5, 29–31]. Extensive use of antibiotics, agricultural runoff, untreated wastewater, and improper disposal of pharmaceutical waste, might facilitate the persistence and spread of resistance genes in microbial communities [26, 31–33]. These findings highlighted the importance of genomic surveillance, which will deepen our understanding of how antimicrobial resistance crosses the environment, human activities, and agricultural boundaries to cause human disease27.
This study has several implications for public health. First, our study provided a region-specific estimation from a One Health perspective, forming a baseline with which future surveillance research can be compared. This approach complements the WHO Global Antimicrobial Resistance and Use Surveillance System (GLASS), which primarily records data from clinical isolates31. This baseline is essential for tracking temporal changes, evaluating interventions, and guiding future research priorities in AMR. Second, our study revealed significant gaps in current AMR surveillance, particularly in tracking transmission pathways across different sectors. To address these gaps, future studies should aim to elucidate these relationships through integrated investigations focusing on the characterization of plasmids and hyperendemic clones from environmental, agricultural, and clinical data27,32. Lastly, the disparities in AMR research across SEAR underscore the need for concerted efforts to strengthen health systems and policies, especially in low-income countries. These countries require may extra support to implement robust national action plans and establish comprehensive AMR surveillance systems24,33. Drawing from models like the European Antimicrobial Resistance Surveillance Network (EARS-Net)34, we propose to establish a regional surveillance network that implements standardized protocols and structured capacity-building programs across different sectors35. This evidence-based framework can guide targeted investments in health infrastructure and foster regional collaboration essential for improving AMR management capacity across the region24,36,37.
Our study has several limitations. First, estimates with small sample sizes were prone to sampling bias, and should be interpreted with caution. Second, our prevalence estimates showed high heterogeneity due to variations in study populations, periods, and settings. To address this, we conducted several subgroup analyses including sector, specific bacteria-antibiotic combinations, and country. However, residual heterogeneity may persist due to limited information on other factors such as study designs, prior antibiotic exposure38, local antimicrobial stewardship practices39, and socioeconomic factors40 that may affect AMR prevalence. These factors were inconsistently reported in our included studies. Third, though similar resistance genes were reported in multiple settings (Fig. 7), without high-resolution phylogenetic analysis, transmissions could not be identified between these sectors.
In conclusion, AMR is highly prevalent in Enterobacterales across SEAR, highlighting the need for coordinated, multisectoral efforts to combat AMR using a One Health approach. Our study provided valuable baseline data for developing surveillance frameworks and identified critical gaps in the current research landscape. The disparities in AMR research across SEAR highlighted the need to strengthen health systems and policies, especially in low-income countries. Additionally, more evidence on inter-sectoral resistance transfer is essential to understand One Health’s role in AMR dynamics.
Methods
Search strategy and selection criteria
This study was performed following the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. The protocol was registered in the PROSPERO International Prospective Register of Systematic Reviews (number CRD42024499521). Searches were conducted in PubMed and Web of Science using free text and controlled vocabulary terms (MeSH) between January 1, 2013, to October 31, 2023. No language restrictions were applied. The search strategy used the following terms: ‘Enterobacterales bacteria’, ‘antimicrobial resistance’ descriptions, countries within the ‘Southeast Asia’ region as defined by the Association of Southeast Asian Nations (ASEAN), ‘animal’, ‘human’, and ‘environment’ sectors41. The main search strategy, initially conducted in PubMed, was adapted for other databases (appendix pp 8). Additionally, we manually searched the reference lists of shortlisted articles to identify additional relevant studies.
We included peer-reviewed articles and published abstracts that met the following inclusion criteria: observational studies that reported the prevalence of antibiotic resistant Enterobacterales, provided data from either human, animal, or environmental sectors, and were conducted in the SEAR. Exclusion criteria included dissertation theses, systematic reviews, and meta-analyses, studies that did not report data required for prevalence estimation, duplicate publications of the same data, randomised controlled trials, and studies with unknown sample collection periods.
Title and abstract screening, as well as full-text evaluation of retained studies, were independently conducted by two authors (YX, CM, YP, JP, XL) using Covidence. Disagreements were resolved through discussion or involvement of a third reviewer (CM, XL, YX). Included abstracts were searched to determine whether the same data had been subsequently published as a full-text article in a peer-reviewed journal. In such cases, the abstract was excluded, and the full-text article was included in the review.
Data analysis
Data extraction was carried out by independent reviewers (YX, XW, YC, FJ, YD, IS, CM, AG) using a standardised data extraction form. This form included the article’s year of publication, location/country, study settings, type of study samples, pathogens, number of isolates tested for antibiotic susceptibility, number of antibiotic resistant isolates, and resistance genes, when available.
We classified AMR using antimicrobials identified as priority agents by the 2023 WHO Access, Watch, Research (AWaRe) classification42. Multidrug resistance was defined as resistance to three or more classes of antibiotics43. The classification of each sector was based on the origin of the samples.
The pooled prevalence of AMR for each organism-antibiotic combination was reported separately for humans, animals, food, and the environment. The prevalence of resistance genes was also pooled across different study sectors. We conducted an inverse variance weighted random-effects model to address the expected heterogeneity stemming from variations in study populations and settings. Due to the highly skewed sample sizes across studies and frequent reports of single proportions in each study, arcsine transformation was used to stabilise prevalence variances. The arcsine transformation provided a more accurate representation of the data, particularly when dealing with weighted studies18. Heterogeneity among studies was quantified using the I² statistic. Subgroup analyses were conducted by stratifying studies by bacteria type, country, and sector. We conducted a temporal trend analysis by stratifying studies according to calendar year, determined using the midpoint of their respective sampling periods. To characterize temporal patterns, we implemented two complementary analytical approaches: (1) a locally weighted scatterplot smoothing (LOESS) regression to present potential non-linear temporal associations, and (2) a linear regression model, weighted by the sample size of each estimate and fitted using ordinary least squares, was used to quantify the overall linear trend. Statistical analyses were conducted using the R software, version 4.3.344.
Role of funding sources
There was no role of funding source for this study. All authors confirm that they had full access to all the data in the study and accept responsibility for the decision to submit it for publication.
Data availability
Requests for data by researchers with proposed use of the data can be made to the corresponding author with specific data needs, analysis plans, and dissemination plans.
References
Iredell, J., Brown, J. & Tagg, K. Antibiotic resistance in enterobacteriaceae: mechanisms and clinical implications. BMJ 352, (2016).
Jonas, O., Irwin, A., Berthe, F., Le Gall, F. & Marquez, P. Drug-Resistant Infections: A Threat To our Economic Future (World Bank Group, 2017).
Müller, A., Stephan, R. & Nüesch-Inderbinen, M. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci. Total Environ. 541, 667–672 (2016).
Dorado-García, A., Smid, J. & Van Pelt, W. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J. Antimicrob. Chemother. 73, 339–347 (2018).
Arnold, K. E. et al. The need for one health systems-thinking approaches to understand multiscale dissemination of antimicrobial resistance. Lancet Planet. Health. 8, e124–e133 (2024).
Assembly, U. Transforming our world: the 2030 agenda for sustainable development. (2015).
Zellweger, R. M. et al. A current perspective on antimicrobial resistance in Southeast Asia. J. Antimicrob. Chemother. 72, 2963–2972 (2017).
Gandra, S. et al. Antimicrobial resistance surveillance in Low- and Middle-Income countries: progress and challenges in eight South Asian and Southeast Asian countries. Clin. Microbiol. Rev. 33, e00048–e00019 (2020).
Antimicrobial resistance global report on surveillance. 2014 summary. https://www.who.int/publications/i/item/WHO-HSE-PED-AIP-2014.2
Hsu, L. Y. et al. Carbapenem-Resistant acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin. Microbiol. Rev. 30, 1–22 (2017).
Birkegård, A. C., Halasa, T., Toft, N., Folkesson, A. & Græsbøll, K. Send more data: a systematic review of mathematical models of antimicrobial resistance. Antimicrob. Resist. Infect. Control. 7, 117 (2018).
Schar, D. et al. Twenty-year trends in antimicrobial resistance from aquaculture and fisheries in Asia. Nat. Commun. 12, 5384 (2021).
One Health Antimicrobial Resistance Workgroup, S. One Health Report on Antimicrobial Utilisation and Resistance. (2019).
Phu, D. H. et al. A systematic review and meta-analysis of integrated studies on antimicrobial resistance in Vietnam, with a focus on Enterobacteriaceae, from a one health perspective. One Health. 15, 100465 (2022).
OneHealthTrust & ResistanceMap Antibiotic resistance.https://resistancemap.onehealthtrust.org/CountryPageSub.php?country=United%20States (2024).
Antimicrobial resistance surveillance in. Europe 2023 – 2021 data. https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2023-2021-data
Kowalski, M. et al. Antimicrobial resistance in enterobacterales infections among children in sub-Saharan africa: a systematic review and meta-analysis. EClinicalMedicine 70, 102512 (2024).
Sisay, A. et al. Prevalence of carbapenem-resistant gram-negative bacteria among neonates suspected for sepsis in africa: a systematic review and meta-analysis. BMC Infect. Dis. 24, 838 (2024).
Vilar-Compte, D., Camacho-Ortiz, A. & Ponce-de-León, S. Infection control in limited resources countries: challenges and priorities. Curr. Infect. Dis. Rep. 19, 1–7 (2017).
Haenssgen, M. J., Charoenboon, N. & Zanello, G. Antibiotic knowledge, attitudes and practices: new insights from cross-sectional rural health behaviour surveys in low-income and middle-income South-East Asia. BMJ Open. 9, e028224 (2019).
Browne, A. J. et al. Global antibiotic consumption and usage in humans, 2000–18: a Spatial modelling study. Lancet Planet. Health. 5, e893–e904 (2021).
Schar, D., Sommanustweechai, A., Laxminarayan, R. & Tangcharoensathien, V. Surveillance of antimicrobial consumption in animal production sectors of low- and middle-income countries: optimizing use and addressing antimicrobial resistance. PLOS Med. 15, e1002521 (2018).
Kasimanickam, V., Kasimanickam, M. & Kasimanickam, R. Antibiotics use in food animal production: escalation of antimicrobial resistance: where are we now in combating AMR? Med. Sci. 9, 14 (2021).
Chua, A. Q., Verma, M., Hsu, L. Y. & Legido-Quigley, H. An analysis of National action plans on antimicrobial resistance in Southeast Asia using a governance framework approach. Lancet Reg. Health - West. Pac. 7, 100084 (2021).
Strategic Plan of Action for ASEAN Cooperation in Crops. 2016–2020. | FAOLEX. https://www.fao.org/faolex/results/details/en/c/LEX-FAOC197568/
Cox, G. & Wright, G. D. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 303, 287–292 (2013).
Djordjevic, S. P. et al. Genomic surveillance for antimicrobial resistance — a one health perspective. Nat. Rev. Genet. 25, 142–157 (2024).
Nnadozie, C. F. & Odume, O. N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut.. 254, 113067 (2019).
Nuangmek, A., Rojanasthien, S. & Yamsakul, P. Perspectives on antimicrobial use in pig and layer farms in thailand: legislation, policy, regulations and potential. Vet. Integr. Sci. 19, 1–21 (2021).
Gwenzi, W., Musiyiwa, K. & Mangori, L. Sources, behaviour and health risks of antimicrobial resistance genes in wastewaters: a hotspot reservoir. J. Environ. Chem. Eng. 8, 102220 (2020).
Organization, W. Global antimicrobial resistance surveillance system (GLASS) report: early implementation. (2020).
Castañeda-Barba, S., Top, E. M. & Stalder, T. Plasmids, a molecular cornerstone of antimicrobial resistance in the one health era. Nat. Rev. Microbiol. 22, 18–32 (2024).
Seale, A. C., Hutchison, C. & Fernandes, S. & et al. Supporting surveillance capacity for antimicrobial resistance: laboratory capacity strengthening for drug resistant infections in low and middle income countries. Wellcome Open. Res 2, (2017).
Mader, R. et al. Building the European antimicrobial resistance surveillance network in veterinary medicine (EARS-Vet). Eurosurveillance 26, 2001359 (2021).
Delpy, L. et al. Integrated surveillance systems for antibiotic resistance in a one health context: a scoping review. BMC Public. Health. 24, 1717 (2024).
Shrestha, P., He, S. & Legido-Quigley, H. Antimicrobial resistance research collaborations in asia: challenges and opportunities to equitable partnerships. Antibiotics 11, 755 (2022).
Kesselheim, A. S. & Outterson, K. Fighting antibiotic resistance: marrying new financial incentives to meeting public health goals. Health Aff (Millwood). 29, 1689–1696 (2010).
Costelloe, C., Metcalfe, C., Lovering, A., Mant, D. & Hay, A. D. Effect of antibiotic prescribing in primary care on antimicrobial resistance in individual patients: systematic review and meta-analysis. BMJ 340, (2010).
Baur, D., Gladstone, B. P. & Burkert, F. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect. Dis. 17, 990–1001 (2017).
Collignon, P., Beggs, J. J., Walsh, T. R., Gandra, S. & Laxminarayan, R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet. Health. 2, e398–e405 (2018).
About, A. S. E. A. N. ASEAN Main Portal https://asean.org/about-asean/
AWaRe classification of antibiotics for evaluation and monitoring of use. https://www.who.int/publications/i/item/WHO-MHP-HPS-EML-2023.04 (2023).
Magiorakos, A. P., Srinivasan, A. & Carey, R. B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281 (2012).
Schwarzer, G. Meta: an R package for meta-analysis. R News. 7, 40–45 (2007).
Acknowledgements
National Centre for Infectious Diseases, Singapore (OHARP-2022-003) and SingHealth Duke-NUS Global Health Institute (SDGHI_PHDRG_FY2024_0004 − 01) funded this study. The authors are entirely responsible for the content of this manuscript, and the views and opinions expressed in the publication are solely those of the authors and not of the funders.
Author information
Authors and Affiliations
Contributions
MY, NG, and YX designed the study and developed the protocol and the data extraction form. NG, MY, and YX acquired funding for this study. YX created the search strategy and did the literature search in electronic databases. YX, CM, YP, JP, and XL screened all studies for inclusion in the systematic review. CM, XL, and YX provided input on studies where consensus could not be reached. YX, FJ, XW, YC, YD, IS, CM, and AG extracted data. NG, MY, and YX contributed to data interpretation and data analysis. YX drafted the manuscript, and all authors revised it critically for content. All authors had full access to all data and could take responsibility for the integrity of the data and the accuracy of the data analysis. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xie, Y., Srivastava, I.M., Jing, F. et al. One health perspective of antibiotic resistance in enterobacterales from Southeast Asia: a systematic review and meta-analysis. Sci Rep 16, 1694 (2026). https://doi.org/10.1038/s41598-025-31195-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-31195-8