Abstract
Agroforestry systems are multifunctional land-use systems that promote soil life. Despite their large potential spatio-temporal complexity, the majority of studies that investigated soil organisms in temperate cropland agroforestry systems focused on rather non-complex systems. Here, we investigated the topsoil and subsoil microbiome of two complex and innovative alley cropping systems: an agrosilvopastoral system combining poplar trees, crops, and livestock and a syntropic agroforestry system combining 35 tree and shrub species with forage crops. Increasing soil depth resulted in a decline of bacterial and fungal richness and a community shift towards oligotrophic taxa in both agroforestry systems, which we attribute to resource-deprived conditions in subsoil. At each soil depth, the microbiome of the tree rows was compositionally distinct from the crop rows. We detected a shift towards beneficial microorganisms as well as a decline in putative phytopathogens under the trees as compared to the crop rows. Finally, based on our results on community dissimilarity, we found that compared to an open cropland without trees, spatial heterogeneity introduced by the tree rows in the agrosilvopastoral system translated into a compositionally less homogeneous soil microbiome, highlighting the potential of agroforestry to counteract the homogenization of the soil microbiome through agriculture.
Similar content being viewed by others
Introduction
Agroforestry is an umbrella term that describes the fusion of agriculture and forestry on the same land that is practiced by millions of farmers around the globe and includes a large variety of transformative land-use systems. In the temperate zone, silvoarable systems that simultaneously grow woody perennials and crops are gaining increasing popularity1,2. Although the intentional spatial integration of tree rows into arable land (i.e. alley cropping) through cropland agroforestry can be associated with specific challenges (e.g. resource competition between trees and crops, labor-intensive management of trees, and lack of financial incentives), it offers numerous environmental benefits as compared to open croplands without trees3. For example, well-designed tree rows in alley cropping systems can effectively reduce wind speeds and the associated risk for soil erosion4 and improve microclimatic conditions5. Further environmental benefits include sequestration of C in the biomass of the trees as well as in soil6, promotion of biodiversity and related ecosystem services7,8,9, and improvement of soil health10. Another key benefit of trees in alley cropping systems is that they can reduce nutrient leaching as their deep rooting system can act as a “safety-net” for leached nutrients that are inaccessible to crops and would otherwise be lost from the system11,12. Although the “safety-net”-role of trees was first mentioned in the scientific literature decades ago13, this mechanism still requires further investigation14,15. Overall, integrating trees into annual cropping systems through alley cropping increases multifunctionality as compared to open croplands3.
Multiple ecosystem functions are mediated by above- and belowground biota including microorganisms in soil16,17. Soil microorganisms constitute a substantial part of global biodiversity18 and contribute to key soil functions such as nutrient cycling19. Therefore, their abundance, diversity, and functions are an integral20 but underrepresented21 part of soil health assessments. Temperate alley cropping agroforestry systems have consistently shown to promote the abundance, diversity, and functions of soil microorganisms as reviewed by Beule et al.22. As the integration of tree rows into arable land through agroforestry increases the complexity of the systems and therefore introduces spatial heterogeneity, sampling efforts in agroforestry systems are greater than in open croplands in order to capture spatial heterogeneity. This was already recognized over a quarter of a century ago23 and found implementation in the sampling designs of numerous recent studies on soil microorganisms in alley cropping agroforestry systems as reviewed recently22. Commonly, spatial gradients from the trees into the crops as well as different soil depths are investigated. Realizing such a study design, several studies reported that soil microbial population size and activity were not just promoted under the trees but that the beneficial effects of the trees gradually extend into the crop rows24,25. In addition, Beule et al.14 found that microbial communities in subsoil were stronger promoted by trees in agroforestry systems than those in topsoil. The extent to which beneficial effects of tree rows on soil microbial communities expand into the crop rows of agroforestry systems is hypothesized to be mainly depending on the distribution of below- (rhizodeposits and root litter) and aboveground tree litter inputs (leaf litter)22.
Despite an ever-growing body of literature on soil microorganisms in temperate cropland agroforestry systems, most research efforts focused on conventionally farmed alley cropping systems that combine a single tree species (mostly poplar or walnut) with a rather narrow crop rotation of annual crops (mostly small-grain cereals or maize). This ignores the large diversity in spatial and temporal configuration and management of agroforestry systems in the temperate zone. For instance, agrosilvopastoral systems, which integrate trees with crops and livestock, are highly understudied although such integrated crop-livestock systems offer several benefits26. A prominent example for such benefits is the grazing of cover crops grown between cash crops, which offers economic returns without compromising soil health27,28,29. Grazing is also known to influence microbial communities30. For instance, grazing of cover crops has recently been shown to increase microbial biomass31. Furthermore, carefully selected tree species can benefit animal welfare through, inter alia, provision of shelter, improved digestion, and enhanced parasite control32. Similar to agrosilvopastoral systems, syntropic agroforestry systems, in which complex tree components are designed to mimic natural succession and stratification in order to maximize synergies among plants, are not yet in the focus of research. This is surprising given that syntropic agriculture, also referred to as dynamic or successional agroforestry, is gaining increasing popularity and is perceived as an innovative approach to agroecology33. Syntropic agroforestry involves a complex and diverse planting scheme that aims to mimic a forest ecosystem to create a resilient, self-sufficient, and constantly evolving system that is supposed to produce long-term yields without external inputs (e.g. fertilizer and irrigation). Due to greater plant diversity and density compared to open croplands, syntropic agroforestry systems can be expected to have a strong impact on soil microbial communities, especially through increased quality and quantity of rhizodeposition and leaf litter inputs34,35,36. Finally, organic agriculture, which –unlike in conventional agriculture– abandons synthetic fertilizers and pesticides, has shown a significant increase during the last 25 years37. However, studies investigating soil microbial communities in organically farmed temperate cropland agroforestry systems are scarce. Recently, Rosati et al.38 collected evidence that despite challenges, the adoption of agroforestry in organic agriculture can result in multiple environmental benefits. The authors further argued that organic farmers are expected to be more open to the adoption of agroforestry as compared to their conventional counterparts38. As highlighted above, despite their large potential spatial and temporal complexity, the majority of studies that investigated soil life in temperate agroforestry systems is at the lower end of the spectrum in terms of system complexity.
Our study aims to investigate the soil microbiome in topsoil and subsoil of two temperate alley cropping agroforestry systems (an agrosilvopastoral and a syntropic system) under organic farming in Germany. We determined the population size of soil archaea, bacteria, and fungi using real-time PCR and assess the diversity and community composition of soil bacteria and fungi by amplicon sequencing. We hypothesized that (i) microbial population size decreases with soil depth due to resource limitations in deeper soil layers. Furthermore, we hypothesized that (ii) microbial populations size increases with decreasing distance to the trees due to increased tree litter inputs (rhizodeposition and leaf litter) in close proximity to the trees. We further expected that (iii) microbial communities within topsoil have greater community similarity than those within subsoil as a result of homogenization of topsoil communities due to soil management and that (iv) tree rows promote beneficial microorganisms rather than putative plant pathogens.
Materials and methods
Study sites
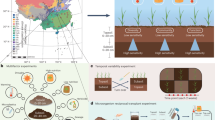
Our study was conducted at (1) a paired agrosilvopastoral alley cropping system (28.4 ha, 52°22’37.70"N, 14°17’25.29"E, m.a.m.s.l.: 65 m) and open cropland system and (2) a syntropic alley cropping system (3.2 ha, 52°22’58.24"N, 14°17’12.52"E, m.a.m.s.l.: 66 m). Both, the agrosilvopastoral and syntropic alley cropping system, were under agricultural use for at least 40 years prior to conversion of open cropland to agroforestry in 2019. Both study sites were under organic farming since 2004 and located on Eutric Retisol and Geoabruptic Luvisol soils near Briesen (Mark), in the federal state of Brandenburg, Germany (mean annual air temperature 1991–2020: 9.4 °C; mean annual precipitation 1991–2020: 545.6 mm). Brandenburg is characterised by its sandy and sandy loamy soils that have low water-holding capacity39. In combination with low annual precipitation mostly below 600 mm and a decreasing trend in available soil water40, Brandenburg is among the most vulnerable regions to climate change in Germany41.
Agrosilvopastoral alley cropping system
The agrosilvopastoral alley cropping system comprised six 4-m wide rows of trees that were planted in North-South orientation and alternated with 36-m wide rows of arable crops. The tree rows of the agrosilvopastoral system covered 8.7% of the total area of the system and consisted of 30-m long segments of different tree species for biomass production. The majority of trees were poplar (Populus maximowiczii × P. trichocarpa (clone Androscoggin), P. trichocarpa (clone Columbia River and Muhle Larsen), and P. x euramericana (clone Jacometti 78 B)), willow (Salix purpurea and S. caprea), and alder trees (Alnus incana). Each tree row comprised two tree lines that were planted by hand using a dibble bar. Prior to planting, a planting furrow was created for the trees utilizing a forest plough and trees were planted at 1 m distance within each tree line. Soil sampling was conducted in segments of poplar clones Columbia River, Jacometti 78 B, and Muhle Larsen. Within these segments, the poplar clones were alternated with an alder tree every four trees. At the time of sampling, the aboveground biomass of the four-year-old trees has not been harvested yet. The agrosilvopastoral system was spatially paired with an adjacent open cropland system that served as a reference land use without trees. The crop rotation of the crop rows of the agrosilvopastoral system was identical to that of the adjacent open cropland since 2022. In 2022, winter rye (Secale cereale) with an undersown grass-clover mixture was cultivated. From October 2022 until March 2023, the undersown grass-clover mixture at both the agrosilvopastoral system and the open cropland were grazed by a beef cattle (Bos taurus) herd of 140 individuals of Angus and Salers breed. The herd was managed under year-round grazing with grazing on cover crops and undersown crops and additional hay and silage feeding during winter. After the herd was moved from the study site, the crop row of the agrosilvopastoral system and the open cropland did not receive any fertilizer prior to drilling of summer oat (Avena sativa) in spring 2023. The crop rows of the agrosilvopastoral system as well as the open cropland were managed under reduced tillage since the establishment of the agroforestry system.
Syntropic alley cropping system
The syntropic alley cropping system comprised 19 hand-planted 1 m-wide rows of trees in North-South orientation that were alternated with 10 m-wide rows of forage crops (grass-alfalfa mixture). The tree rows covered 9.2% of the total area of the system and comprised a single line of trees bordered by a layer of mulch comprising wood chips and cut biomass of the forage crops (Fig. 1). The system is specialized on fruit and nut production and applies the principles of syntropic agroforestry. The system can be classified as a successional agroforestry system due to the integration of more than two stratification levels (‘strata’) of trees and shrubs, the plant selection which is based on ecological succession, and the dynamic management approach42.
Location of the study sites near Alt Madlitz in the federal state of Brandenburg (highlighted in grey), Germany (A, B). Study design and photos of the syntropic alley cropping system (C), the open cropland (D), and the agrosilvopastoral and alley cropping system (E). Photo credit: Sam Waltl (Finck Foundation) and Anna Vaupel (JKI). Maps were created using Natural Earth (naturalearthdata.com) and QGIS version 3.4.4-Madeira.
Within the tree rows of the syntropic system, fruit and nut trees were the main tree crops, which were densely grown along with timber trees and ‘support species’ that are supposed to promote ecosystem functions. The selection of plant species as well as the spatial arrangement of trees and shrubs followed their position in stratification and ecological succession. As support species, the system included undemanding plants such as herbs, sea buckthorn, and blackberries as well as fast-growing pioneer tree species such as poplar, birch, and wild cherry. As succession proceeds, pioneer trees will be superseded by fruit, nut, and timber trees, and finally by oak and maple trees, which were grown from seeds and reach maturity after several decades. Plants are further cultivated at four strata according to their individual light requirements. For example, ground-covering herbaceous plants such as mugwort, mallow, tarragon, and comfrey grow at the lowest strata, followed by sea buckthorn or berries at the second lowest strata, followed by fruit and nut trees, and finally pioneer trees at the highest strata. The management of the tree rows included manual weeding, dynamic pruning of timber and support species to maintain stratification as well as fruit and nut trees to ensure yield production, and adding the pruned biomass to the layer of mulch bordering the tree rows. The forage crop rows were cut up to four times a year for hay production. Forage crop cuttings within 2 m distance to the tree rows were not used for hay production and swathed onto the layer of mulch. Overall, the syntropic system comprised 35 tree and shrub species of different varieties as well as 20 herbaceous plant species.
Soil sampling was conducted in the fourth tree row from the eastern end of the system (Fig. 1) with plums and pears as main fruit crops. Within the tree row, 18 plum varieties were spaced at a distance of 2 m and six pear varieties were spaced at a distance of 10 m. Between plum and pear trees, birch, poplar, and wild cherry trees were planted that were in turn alternated with blackberries and diverse herbaceous plants as ground cover.
Soil sampling and general soil properties
At each sampling location, soil samples were collected in topsoil (0–30 cm) and subsoil (30–60 cm) on 20.09.2023 using stainless steel augers. Furthermore, three soil cores per soil depth were collected at each sampling location (technical replicates), transferred into a sterile polyethylene bag, and thoroughly homogenized in the field to obtain a composite sample per sampling location (biological replicate). To account for spatial heterogeneity introduced by the trees24,25, soil samples within the agroforestry system were collected at different sampling locations along six transects spanning from the centre of the tree row into the centre of the crop row. The transects were delineated orthogonal to the north-south orientation of the tree rows and covered the tree row as well as 1, 4, 8, and 18 m distance (centre of the crop row) from the trees into the crop row. Furthermore, an adjacent open cropland cultivated with identical crops and under identical management practices as the crop row of the agroforestry system served as a reference land use. In the open cropland, soil samples were collected at six sampling locations. Thus, we collected 72 soil samples in the agrosilvopastoral alley cropping system (6 sampling locations (tree row, 1, 4, 8, and 18 m crop row and open cropland) × 6 replicates × 2 soil depths = 72 samples). In contrast to the agrosilvopastoral alley cropping system, crop rows in the syntropic alley cropping system were only 10 m wide. Therefore, soil samples in the syntropic alley cropping system were only collected in the tree row as well as in the centre of the crop rows at 5 m distance from the trees using six replicates. For this system, an open cropland serving as a reference land use was unavailable. Thus, we collected 24 soil samples in the syntropic alley cropping system (2 sampling locations (tree row, 5 m crop row) × 6 replicates × 2 soil depths = 24 samples). From each of the 96 soil samples (72 from the agrosilvopastoral and 24 from the syntropic system), an aliquot of approx. 50 g fresh soil was transferred into a sterile 50-ml Falcon tube and frozen at −20 °C in the field for molecular analyses. The remaining soil was used for the analyses of general soil properties.
Soil pH, SOC, and total N were determined from air-dried, sieved (< 2 mm) soil. Soil pH was determined in 0.01 M CaCl2 at a ratio of 1:2.5 (soil: CaCl2(w/v)). For SOC and total N, air-dried and sieved soil was finely ground and subjected to acid fumigation43 to remove inorganic C. Following acid fumigation, SOC and total N were determined utilizing a CNS elemental analyzer (Vario EL Cube, Elementar, Germany).
Soil DNA extraction
Upon arrival at the laboratory, samples were stored at −20°C until freeze-drying for 72 h. Freeze-dried soil was finely ground using a vortexer as described earlier44. Soil DNA was extracted using an extraction protocol tailored to subsoil samples as per Guerra et al.45. Briefly, 200 mg freeze-dried and finely ground soil were suspended in 250 µL phosphate lysis buffer (1 M phosphate buffer with 0.5% SDS (w/v)) and incubated at 65 °C for 10 min. The suspension was centrifuged, the supernatant was transferred into a new reaction tube, diluted 1:10 (v/v) in double-distilled H2O (ddH2O) and extracted twice with chloroform-isoamyl alcohol (24:1 (v/v)). Following this, DNA was allowed to precipitate for 15 min at room temperature under the presence of 30% (w/v) polyethylene glycol (PEG 6000) and 5 M NaCl (2:1 (v/v)). Precipitated DNA was pelleted using centrifugation and obtained pellets were washed twice with 80% (v/v) EtOH prior to drying the pellets in a vacuum centrifuge. Dried pellets were resuspended in 50 µL of 1× TE buffer (10 mM Tris, 1 mM EDTA, adjusted to pH 8.0 with HCl) and incubated for 42 °C for 2 h to facilitate resuspension of DNA. The quantity and quality of the extracted DNA was visually checked on 1% (w/v) agarose gels stained with SYBR Green I solution (Thermo Fisher Scientific GmbH, Dreieich, Germany).
Real-time PCR assays for soil archaea, bacteria, and fungi
Soil archaea, bacteria, and fungi were quantified using real-time PCR using the primer sets ARC787F/ARC1059R46, Bac349F/Bac806R47 and ITS3/ITS448,49, respectively. To overcome PCR inhibition, DNA extracts were diluted 1:50 (v/v) in ddH2O prior to PCR45. PCR reactions were performed in a 384-Well real-time PCR cycler (qTOWER3 84 G, Analytik Jena GmbH + Co. KG, Jena, Germany) in 5-µL reaction volumes. The reaction volumes were comprised of 4 µL master mix (ddH2O, primer (0.3, 0.5, and 0.3 µM of each primer for archaea, bacteria, and fungi, respectively), and 2.5 µL 1× Luna® Universal qPCR Master Mix (New England Biolabs, Beverly, Massachusetts, USA)) and 1 µL template DNA or ddH2O for a negative control. The thermocycling conditions were as follows: initial denaturation at 95 °C for 5 min followed by 40 cycles of denaturation at 95 °C for 40 s, annealing at 60, 53, and 58 °C for 30 s for archaea, bacteria, and fungi, respectively, and elongation at 68 °C for 30 (archaea) or 60 s (bacteria and fungi). Final elongation was carried out at 68 °C for 5 min. Following amplification, melting curves were generated by step-wise increasing the temperature from 65 °C to 95° at a rate of 1 °C per step under continuous fluorescence measurement.
Amplicon sequencing of soil bacteria and fungi
Sequencing libraries for soil bacteria and fungi were constructed using primer pairs 341F/785R50and ITS1-F_KYO2/ITS86R51,52, respectively. PCR was performed using an Eppendorf Mastercycler EP Gradient S thermocycler (Eppendorf, Hamburg, Germany) in 25-µL reaction volumes containing 18.75 µL master mix and 6.25 µL template DNA or ddH2O for a negative control. The master mix contained ddH2O, buffer (10 mM Tris–HCl, 50 mM KCl, 2.0 mM MgCl2, pH 8.3 at 25°C), 100 µM of each deoxynucleoside triphosphate (New England Biolabs, Beverly, Massachusetts, USA), 0.5 µM of each primer, 1 mg mL−1 bovine serum albumin, and 0.03 u µL−1 Hot Start Taq DNA Polymerase (New England Biolabs, Beverly, Massachusetts, USA). As per Bednar et al.53, each primer was a mixture of the primer with (50%) and without (50%) Illumina TruSeq 5’-end adapters. We used a touch-up thermocycling protocol54 to amplify bacteria and fungi. The thermocycling conditions were as follows: initial denaturation at 95 °C for 2 min, 3 touch-up cycles (denaturation: 95 °C for 20 s, annealing: 50 °C for 30 s, and elongation: 68 °C for 60 s), 25 and 27 cycles (denaturation: 95 °C for 20 s, annealing: 58 °C for 30 s, and elongation: 68 °C for 60 s) for bacteria and fungi, respectively. Final elongation was carried out at 68 °C for 5 min. Amplicons were screened on 1.7% (w/v) agarose gels as described above. Sequencing libraries were subjected to a second amplification with standard i7- and i5- sequencing adapters, multiplexed, and sequenced on an Illumina MiSeq (V3 chemistry, 2 × 300 bp) (Illumina, Inc., San Diego, CA, USA) at the facilities of LGC Genomics (Berlin, Germany). Amplicon sequencing data have been deposited at NCBI’s Sequence Read Archive (BioProject numbers PRJNA1142703 for bacteria and PRJNA1142770 for fungi).
Processing of amplicon sequencing data and statistical analysis
Obtained sequence reads were processed as detailed in Bednar et al.53. First, data were demultiplexed utilizing Illumina’s bcl2fast version 2.20 (Illumina, San Diego, CA, USA). Demultiplexed data were cleaned from one-sided and conflicting barcodes as well as barcodes containing more than two mismatches. Furthermore, sequencing adapter and primer sequences as well as sequence reads with < 100 bp were removed. The cleaned sequence reads were processed in QIIME 2 (version 2022.255). We used DADA256 to quality filter, merge, and remove chimeras and singletons from the data. Amplicon sequencing variants (ASVs) were taxonomically classified against the SILVA ribosomal RNA gene database release 13857 for bacteria and UNITE database version 9.058 for fungi. We utilized a scikit-learn Naive Bayes machine-learning classifier (‘q2-fit-classifier-naive-bayes’ and ‘q2- classify-sklearn’ plugin) in the ‘balanced’ configuration ([7,7]; 0.7 for bacteria and [6,6]; 0.96 for fungi as suggested by Bokulich et al.59) for classification. Non-bacterial and non-fungal sequence reads were removed from the bacterial and fungal data set, respectively.
Downstream analyses of the obtained ASV tables were performed in R version 4.4.160 using the ‘microeco’ R package version 1.7.161. ASV tables were normalized to account for uneven sequencing depth using scaling with ranked subsampling (SRS)62 using the ‘SRS’ R package version 0.2.363 as implemented in ‘microeco’. The bacterial ASV table was normalized to 30,098 sequence reads. The fungal ASV table was normalized to 55,287 sequence reads. Alpha diversity indices (Shannon diversity, Chao1 index, and Pielou’s evenness) and Bray-Curtis dissimilarity were calculated in ‘microeco’. Differences in microbial community composition among different soil depths and sampling locations within each agroforestry system were identified with Permutational Multivariate Analysis of Variance (PERMANOVA) based on the ‘adonis 2’-function of the R package ‘vegan’. Prior to using PERMANOVA, permutation tests for homogeneity of multivariate dispersions were performed using the ‘permdisp’ function of the ‘vegan’ package. Differences in Bray-Curtis dissimilarities were identified using Wilcoxon Rank Sum test for pairwise comparisons (i.e. soil depth or sampling locations within the syntropic system) and Kruskal-Wallis Rank Sum test followed by Dunn’s multiple comparisons test for more than two groups (i.e. the six sampling locations within the paired agrosilvopastoral and open cropland system). Differences in alpha diversity indices, microbial population sizes, and general soil properties between different soil depths and sampling locations within each agroforestry system were identified using one-way ANOVA followed by Tukey HSD test or Student’s t-test for pairwise comparisons (i.e. soil depth or sampling locations within the syntropic system). For the analysis of different soil depths, linear mixed-effect models were used with soil depth as a fixed effect and sampling location as random effect. Before performing ANOVA, all data were tested and visually checked to meet the criteria of normality of residuals (Shapiro-Wilk test) and homogeneity of variance (Levene’s test). Data not meeting the criteria were either log- (soil microbial population sizes) or square root-transformed (alpha diversity indices). Differential abundance analysis was performed at all taxonomic levels utilizing linear models for differential abundance analysis (LinDA)64. For differential abundance analysis of more than two groups (i.e. among the six sampling locations within the paired agrosilvopastoral system and open cropland), tree row was set as reference group. For differential abundance analysis of different soil depths, linear mixed-effect models were used with soil depth as a fixed effect and sampling location as random effect. For all tests, statistical significance was considered at p < 0.05.
Results
General soil properties
In both agroforestry systems (agrosilvopastoral and syntropic alley cropping system), SOC and total N were greater in topsoil than subsoil (p < 0.001) (Figure S1 A, B, C, D). In topsoil of the paired agrosilvopastoral and open cropland system, few sporadic differences in SOC and total N among sampling locations were found with slightly lower SOC (p < 0.0021) and total N (p < 0.0027) in the open cropland as compared to the crop row of the agrosilvopastoral system (Figure S1 A, C). In contrast, no statistically significant differences in SOC and total N among sampling locations were detected in subsoil. Soil pH remained unaffected by the agrosilvopastoral system at both soil depths. In the syntropic agroforestry systems, SOC (p < 0.001) and total N (p < 0.016) were greater under the trees as compared to the crop row at both soil depths (Figure S1 A, B, C, D). Furthermore, the tree row of the syntropic system had a higher soil pH than its corresponding crop row in topsoil (p < 0.001) (Figure S1 E).
Population size of soil bacteria, fungi, and archaea
Soil microbial population size was overall greater in topsoil than subsoil for bacteria, fungi, and archaea (p < 0.001). Differences between the tree row and the crop row of the syntropic system were more pronounced in subsoil than in topsoil, where soil microbial population sizes of bacteria, fungi and archaea were greater under the trees than in the centre of the crop row (p < 0.001, p = 0.011, and p = 0.048, respectively) (Fig. 2B, D, F). In the agrosilvopastoral system, soil microbial population size did not differ between different sampling locations (tree row, 1, 4, 8, and 18 m crop row and open cropland) regardless of soil depth, except for archaea in topsoil, which showed slightly lower gene copy numbers under the trees than at 4 m distance in the crop row (p = 0.015) and the open cropland (p = 0.008) (Fig. 2E).
Soil microbial population size of total bacteria (A, B), fungi (C, D) and archaea (E, F) in topsoil and subsoil of an agrosilvopastoral and syntropic alley cropping system near Alt Madlitz, Germany (n = 6). Dots indicate individual data points. Different letters indicate statistically significant differences among the six sampling locations within the agrosilvopastoral alley cropping system (ANOVA followed by Tukey’s HSD test; p ≤ 0.05). Asterisks indicate statistically significant differences between the two sampling locations within the syntropic alley cropping system (Student’s t-test; p ≤ 0.05). Absence of letters or asterisks indicates no statistically significant differences among sampling locations (p > 0.05). Icons were created with BioRender.com.
Alpha diversity of soil bacteria and fungi
Alpha diversity indices (Shannon diversity, Chao1 index, and Pielou’s evenness) of soil bacterial ASVs were greater in topsoil than subsoil in the agrosilvopastoral system (p ≤ 0.001) but did not differ between soil depths in the syntropic system (Fig. 3A, G, M). In the agrosilvopastoral system, bacterial alpha diversity indices showed no statistically significant differences among sampling location in both topsoil and subsoil (Fig. 3B, C, H, I, N, O). In subsoil of the syntropic system, bacterial Shannon diversity was greater under the trees than in the centre of the crop row (p = 0.034) (Fig. 3C). Shannon diversity of soil fungi remained unaffected by soil depth and sampling location in both agroforestry systems (Fig. 3D, E, F). However, fungal ASV richness (Chao1 index) was greater in topsoil then subsoil in both systems (p < 0.001) (Fig. 3J) as well as in the tree row as compared to the centre of the crop row within the syntropic system at both soil depths (p = 0.046 and p < 0.001 for topsoil and subsoil, respectively) (Fig. 3K, L). Evenness of fungal ASVs (Pielou’s evenness) was greater in subsoil than topsoil of the agrosilvopastoral system (p < 0.001) (Fig. 3P) as well as in the centre of the crop row than under the trees in subsoil of the syntropic system (p = 0.004) (Fig. 3R).
Alpha diversity indices (Shannon diversity (A, B, C, D, E, F), Chao1 index (G, H, I, J, K, L), and Pielou’s evenness (M, N, O, P, Q, R)) of soil bacterial and fungal amplicon sequencing variants (ASVs) in topsoil and subsoil of an agrosilvopastoral and syntropic alley cropping system near Alt Madlitz, Germany (n = 6). Dots indicate individual data points. Different letters indicate statistically significant differences among the six sampling locations within the agrosilvopastoral alley cropping system (ANOVA followed by Tukey’s HSD test; p ≤ 0.05). Asterisks indicate statistically significant differences between the two soil depths across sampling locations (linear mixed effect models; p ≤ 0.05) as well as the two sampling locations within the syntropic alley cropping system (Student’s t-test; p ≤ 0.05). Absence of letters or asterisks indicates no statistically significant differences between soil depths or sampling locations (p > 0.05). Icons were created with BioRender.com.
Community composition of soil bacteria and fungi
Across all samples, Acidobacteria (20.68 ± 2.91%), Proteobacteria (15.77 ± 1.87%), and Actinobacteria (14.84 ± 3.51%) were the most abundant bacterial phyla, whereas fungal communities were dominated by the phyla Ascomycota (40.52 ± 13.10%), Basidiomycota (20.44 ± 17.79%), and Mortierellomycota (9.59 ± 6.01%). In both agroforestry systems, soil bacterial community composition differed between topsoil and subsoil (p < 0.001) (supplementary Table S1). The compositional differences in soil bacterial communities between soil depths were also reflected by the clustering of the data in the NMDS plot (Fig. 4, A, B) as well as by the number of unique and shared ASVs between topsoil and subsoil (Figure S2, A, B). Within each agroforestry system, topsoil and subsoil each had at least twice as much unique bacterial ASVs than shared ones across depths; however, the shared ASVs across depths accounted for approximately three quarters of total ASV counts (Figure S2 A, B). The composition of the soil bacterial community in topsoil of the agrosilvopastoral systems was strongly affected by the sampling location as the tree row, the different distances from the trees within the crop row, and the open cropland all harboured compositionally distinct communities (supplementary Table S2, Fig. 4E). Similarly, community composition of bacteria in the syntropic system differed among the tree and crop row at both soil depths (supplementary Table S3, Fig. 4F, J). Community dissimilarity of soil bacteria was greater in subsoil than topsoil across sampling locations in both agroforestry systems (p < 0.001) (Fig. 5A, B). Within each soil depth of the agrosilvopastoral system, community dissimilarity of soil bacteria gradually decreased with increasing distance from the trees (Fig. 5E, I). In subsoil of the syntropic system, bacterial community dissimilarity was greater in the crop row than in the tree row (p = 0.016) (Fig. 5J).
Non-metric multidimensional scaling (NMDS) of Bray–Curtis dissimilarities of bacterial and fungal communities among sampling depths (A, B, C, D) of an agrosilvopastoral (n = 36) and syntropic alley cropping system (n = 12) as well as among sampling locations in topsoil (E, F, G, H) and subsoil (I, J, K, L) of both systems (n = 6) near Alt Madlitz, Germany. Dots indicate individual data points.
Community dissimilarity of soil bacteria and fungi within each soil depth (A, B, C, D) and at each sampling location in topsoil (E, F, G, H) and subsoil (I, J, K, L) of an agrosilvopastoral and syntropic alley cropping system near Alt Madlitz, Germany. Community dissimilarity was assessed using pairwise Bray-Curtis distances between individual samples. Dots indicate pairwise Bray-Curtis distances between samples. Different letters indicate statistically significant differences among the six sampling locations within the agrosilvopastoral alley cropping system (Kruskal-Wallis test followed by Dunn’s test; p ≤ 0.05). Asterisks indicate statistically significant differences between the two soil depths across sampling locations (linear mixed effect models; p ≤ 0.05) as well as the two sampling locations within the syntropic alley cropping system (Wilcoxon Rank Sum test; p ≤ 0.05). Absence of letters or asterisks indicates no statistically significant differences between soil depths or sampling locations (p > 0.05). Icons were created with BioRender.com.
Soil depth was a strong driver of the fungal community composition in the agrosilvopastoral but not in the syntropic system (supplementary Table S5, Fig. 4C, D). In both agroforestry systems, topsoil harboured more unique fungal ASVs than subsoil whereas the number of ASVs shared between both soil depths was greater than in subsoil but lower than in topsoil and accounted for more than 90% of total ASV counts (Figure S2, C, D). At both soil depths, community composition of soil fungi in the agrosilvopastoral was distinct among the tree row, the crop row, and the open cropland (supplementary Table S6). Likewise, in the syntropic system, the composition of the fungal community was different between the tree and crop row at both topsoil and subsoil (supplementary Table S7, Fig. 4H, L). Soil fungi showed greater community dissimilarity in subsoil than in topsoil across all sampling locations of the agrosilvopastoral system (p < 0.001) (Fig. 5C), whereas no differences were found between soil depths in the syntropic system (p = 0.191) (Fig. 5D). Fungal community dissimilarity was greater under the trees than in the crop rows of the agrosilvopastoral system and the adjacent open cropland in topsoil (Fig. 5G). While no decline in community dissimilarity with increasing distance from the trees was observed in topsoil, a similar decline as for bacteria was found for soil fungi in subsoil of the agrosilvopastoral system (Fig. 5K). At both soil depths, community dissimilarity of soil fungi was greater in the crop row than in the tree row of the syntropic system (p < 0.001) (Fig. 5, H, L).
Differential abundance analysis
Soil depth strongly influenced the relative abundance of multiple bacterial groups in both agroforestry systems. In the agrosilvopastoral system, relative abundance of members of the bacterial phyla Acidobacteriota, Bacteroidota, Patescibacteria, and Planctomycetota were greater in topsoil than in subsoil (Fig. 6A). Likewise, members of the genera Bryobacter, Chthoniobacter, Microbacterium, Microlunatus, Pirellula, and RB41 (Acidobacteriota) showed lower relative abundance in subsoil than topsoil of the same agroforestry system (Fig. 6D). Conversely, members of the phyla Actinobacteriota, Chloroflexi, Gemmatimonadota, Latescibacterota, Methylomirabilota, Nitrospirota, and Verrucomicrobiota and of the genera Candidatus Udaeobacter (Verrucomicrobiota), Gaiella, Massilia, Nitrospira, and Nocardioides showed greater relative abundance in subsoil of the agrosilvopastoral system than in topsoil (Fig. 6A, D). In the syntropic system, only the phylum Planctomyctota showed greater relative abundance in topsoil than subsoil, while the phyla Chloroflexi, Firmicutes, Gemmatimonadota, Latescibacterota, Methylomirabilota, Myxococcota, Nitrospira, and Verrucomicrobiota were promoted in subsoil (Fig. 6B). In contrast, on genus level, relative abundance of Microlunatus, Microbacterium, MND1 (Proteobacteria), Pirellula, and Sphingomonas were greater in topsoil as compared to subsoil (Fig. 6E).
Differentially abundant soil bacterial (A, B, D, E) and fungal (C, F, G) phyla and genera in topsoil and subsoil of an agrosilvopastoral (n = 36) and syntropic alley cropping system (n = 12) near Alt Madlitz, Germany. Bars left from the dashed line show greater relative abundance in topsoil than subsoil, whereas bars right from the dashed line show greater relative abundance in subsoil than topsoil. The p-values between bars indicate statistically significant difference between topsoil and subsoil for the relative abundance of the respective phyla or genera. Statistically significant differences were determined using linear models for differential abundance analysis (LinDA) utilizing linear mixed-effect models with soil depth as a fixed effect and sampling location as random effect (p < 0.05). Icons were created with BioRender.com.
Within the fungal community of the agrosilvopastoral system, only the genera Lectera showed greater relative abundance in topsoil than subsoil, whereas members of Filobasidium and Mortierella were greater in subsoil than topsoil (Fig. 6F). Similarly, the phylum Mortierellomycota showed greater relative abundance in subsoil of the syntropic system than in topsoil (Fig. 6C), whereas the genus Cercophora showed the opposite pattern (Fig. 6G).
In topsoil of the paired agrosilvopastoral and open cropland system, sampling location influenced the relative abundance of several bacterial phyla and genera (Fig. 7A, I). The relative abundance of the phyla Methylomirabilota and Verrucomicrobiota as well as the genera Chthoniobacter and Mycobacterium showed a pattern of greater relative abundance in the tree row as compared to the crop row and open cropland. Conversely, the relative abundance of the phyla Chloroflexi and Myxococcota were generally lower in the tree row than at sampling locations in the crop row and open cropland. Similarly, the genus Sphingomonas showed lower relative abundance under the trees of the agrosilvopastoral system as compared to all other sampling locations except for the distance closest to the trees in the crop row. In the syntropic system, however, only the phyla Bacteroidota and Proteobacteria were influenced by the sampling location, showing greater relative abundance in topsoil of the tree row as compared to the crop row (Fig. 7B). Similar results were obtained for the genus Microbacterium, whereas the genera Candidatus Solibacter (Acidobacteriota), Nitrospira, and RB41 showed the opposite pattern of lower relative abundance under the trees than in the crop row (Fig. 7J).
Relative abundance of bacterial (A, B, C, D, I, J, K, L) and fungal (E, F, G, H, M, N, O, P) phyla and genera in topsoil and subsoil of an agrosilvopastoral and syntropic alley cropping system near Alt Madlitz, Germany (n = 6). The p-values above bars indicate statistically significant difference between the tree row and the other sampling locations within the respective alley cropping system. Statistically significant differences were analysed using linear models for differential abundance analysis (LinDA) with tree row set as reference group for differential abundance analysis (p < 0.05).
In subsoil of the paired agrosilvopastoral and open cropland system, relative abundance of the phylum Acidobacteriota and the genus Streptomyces were affected by the trees (Fig. 7C, K). In the syntropic system, relative abundance of members of the phyla Actinobacteriota, Bacteroidota, Firmicutes, Planctomycetota, and Proteobacteria were greater in the tree row as compared to the crop row while Chloroflexi and Nitrospira showed the opposite trend (Fig. 7D). Similarly, members of the genera Bacillus, Microbacterium, Microlunatus, MND1 (Proteobacteria), Pirellula, Sphingomonas, and Streptomyces were promoted under the trees, whereas, Nitrospira, Massilia, and RB41 showed greater relative abundance in the crop row than in the tree row (Fig. 7L).
Sampling locations in both alley cropping systems contributed to differentially abundant fungal groups in topsoil and subsoil. In the paired agrosilvopastoral and open cropland system, relative abundance of the phylum Basidiomycota was greater under the trees than in the arable land, whereas the opposite pattern emerged for Ascomycota (Fig. 7E). In subsoil, relative abundance of Basidiomycota and Olpidiomycota gradually declined with increasing distance from the trees (Fig. 7G). Ascomycota and Basidiomycota showed opposing patterns in the syntropic system with greater abundance of Ascomycota and lower abundance of Basidiomycota in the tree row than in the crop row in topsoil and subsoil, respectively (Fig. 7F, H). In both topsoil and subsoil of the agrosilvopastoral system, relative abundance of Hebeloma and Tuber spp. were strongly increased under the trees as compared to the arable land, whereas relative abundance of Lectera spp. was greater in the open cropland than in the tree row (Fig. 7M, O). Sampling location further affected the relative abundance of Fusarium spp. but only in topsoil (Fig. 7M). In the syntropic system, relative abundance of Agrostalagmus spp. increased under the trees as compared to the crop row in both topsoil and subsoil, while Agremonium and Cercophoria spp. showed only effects in topsoil (Fig. 7N, P). In subsoil of the syntropic system, relative abundance of Mortierella and its phylum Mortierellomycota as well as Filobasidium were lower in the tree row than in the crop row (Fig. 7H, P).
Discussion
Despite the large diversity of temperate alley cropping systems, so far most research focused on rather non-complex systems in terms of management and tree species composition. In this work, we investigated the topsoil and subsoil microbiome of two complex alley cropping systems, namely, an agrosilvopastoral and a syntropic system. Overall, we found that the richness of bacterial and fungal taxa declined from topsoil to subsoil in both agroforestry systems, which we attribute to resource-deprived conditions in subsoil. Accompanied with the decline in taxa richness, we detected a community shift for both bacterial and fungal communities from topsoil to subsoil. In both agroforestry systems, the tree rows harboured bacterial and fungal communities that were compositionally distinct from those in the crop rows. Furthermore, in the agrosilvopastoral system, the soil microbiome became compositionally less homogeneous with decreasing distance to the trees.
Patterns of SOC in two young and differently managed alley cropping systems
SOC in topsoil and subsoil of the agrosilvopastoral system remained largely unaffected by the integration of trees (Fig S1A, B), which is likely due to the young age of the system (4 years). Indeed, recent literature suggests that increases in SOC stocks in alley cropping systems are usually detectable after one decade post their establishment65. In contrast, SOC under the trees in the syntropic system was greater than in the crop row although both systems are of the same age. The intensive mulching of tree rows most likely caused the rapid increase in SOC under the trees. Although we did not determine different SOC fractions, we expect that mainly labile SOC fractions increased through mulching as these fractions can respond rapidly to environmental changes. Furthermore, labile SOC fractions are considered indicators of long-term SOC trends66, however, their fast turnover and sensitivity towards environmental changes also makes them vulnerable to loss67. In a case study from 2015, Cardinael and others68 suggested that the additional SOC storage under the trees of an 18-year-old walnut alley cropping system in France was mostly due to an increase in labile SOC fractions, raising questions regarding its stability. We therefore suggest that future research on SOC stocks in alley cropping systems should consider differentiating between labile and stable SOC fractions in order to infer information on SOC stabilization dynamics in alley cropping systems.
Soil depth as a key driver of the abundance and community composition of bacteria and fungi
Soil depth is a key driver of the assembly of the soil microbiome since multiple influencing factors such as the availability of oxygen and other resources generally decrease as soil depth increases. The scarcity of resources in subsoil serves as a reasonable explanation for the observed decline in population size of bacteria, fungi, and archaea with increasing soil depth (Fig. 2) that confirms our first hypothesis and is congruent with previous studies69,70,71,72. Furthermore, specialization to resource-deprived conditions in subsoil also explains the decline in bacterial species richness from topsoil to subsoil of the agrosilvopastoral system (Fig. 3) as only a limited number of taxa can thrive in subsoil72.
In addition to changes in alpha diversity, soil depth led to compositionally distinct bacterial communities in both agroforestry systems. For instance, multiple phyla associated with an oligotroph lifestyle such as Chloroflexi, Verrucomicrobiota, Gemmatimonadota, Nitrospirota, and Latescibacterota showed greater relative abundance in subsoil than topsoil, highlighting their ability to adapt to resource-deprived soil environments. Similarly, in the agrosilvopastoral system, Gaiella and Nocardioides spp. as well as their affiliated phylum Actinobacteria showed a similar pattern of promotion in subsoil, which is in line with their presumed oligotrophic lifestyle. However, it should be noted that the generalized assignment of a single lifestyle to a broad taxonomic group may not match the lifestyles of all its members73,74. Although our results for certain phyla mentioned above (Actinoabacteria, Chloroflexi, Nitrospirota, and Latescibacterota) are in line with previous studies that investigated bacterial communities at different soil depths69,75, Lopes et al.76 reported opposing results for Verrucomicrobiota and Gemmatimonadota (i.e. greater relative abundance in topsoil than subsoil). For instance, the authors found the genus Candidatus Udaeobacter (Verrucomicrobiota) to be depleted in deeper soil layers and attributed this finding to its aerobic lifestyle76,77. However, in the agrosilvopastoral system, relative abundance of Candidatus Udaeobacter was greater in subsoil than topsoil (Fig. 6D). Such a discrepancy may partly be due to differences in sampling depth: while Lopes and co-authors76 sampled up to 240 cm deep, subsoil samples in our study were collected at 30–60 cm depth, which likely did not cover anaerobic conditions. Likewise, in 2015, Navarette et al.78 reported decreasing relative abundance of Verrucomicrobiota with increasing soil fertility, which coincides with the observed increased proportion of this phylum in resource-deprived subsoil (Fig. 6A, B).
In contrast to bacteria, only very few fungal phyla and genera were differentially abundant between topsoil and subsoil; however, in the agrosilvopastoral and syntropic system, members of Mortierellomycota and Mortierella showed greater relative abundance in subsoil than topsoil, respectively (Fig. 6C, F). Although information on the vertical distribution of Mortierellomycota and Mortierella spp. in agricultural soils is scarce, Mortierella spp. are widely recognized as important decomposers as well as for their benefits as plant-growth-promoting fungi79,80. Another fungal genus that was dominant in subsoil of the agrosilvopastoral system was Filobasidium (Fig. 6F). The genus Filobasidiumharbours members with the ability to produce extracellular polymeric substances, which enable them to form biofilms81, a survival mechanism by which these organisms may cope with the resource-deprived conditions in subsoil.
In addition to a distinct community composition between soil depths, community similarity of bacteria and fungi was greater within topsoil than subsoil (Fig. 5), revealing greater compositional homogenization of the topsoil than the subsoil microbiome and thereby confirming our third hypothesis. As our study sites are located in a glacial landscape that is characterized by small-scale spatial variability of soil properties, we expected a comparatively large dissimilarity in microbiome composition among samples. Although our study sites were managed under reduced tillage for five years, the previous decade-long tillage regime likely reduced the degree of spatial variability of soil properties in topsoil through mechanical homogenization while subsoil remained undisturbed. We therefore propose that spatial homogenization of topsoil through tillage translated into compositional homogenization of topsoil biota. Likewise, we argue that greater spatial variability of soil properties in deeper soil layers contributed to greater community dissimilarity in subsoil as compared to topsoil.
Distance to trees shapes the soil microbiome in agroforestry systems
The distance to the tree rows influenced bacterial and fungal community composition in both systems. For bacterial communities, most striking effects of the trees were observed in subsoil of the syntropic system: Actinobacteria and its affiliated genera Microbacterium, Microlunatus, and Streptomyces showed greater relative abundance under the trees than in the crop row. Members of these groups are often found in rhizosphere and include species that are recognized for their antimicrobial properties as well as their plant growth promoting effects82,83. We found similar effects for Proteobacteria and its affiliated genera Sphingomonas and MND1 that are also frequently recovered from rhizosphere with some strains showing beneficial effects on plant growth84,85,86. The syntropic system is characterized by high diversity of plants, which may explain the promotion of putative beneficial bacteria that are known for root interactions. We expect the aboveground plant diversity and density to be mirrored belowground through the formation of a complex rooting system along the soil profile that creates numerous habitats for soil organisms. In line with this assumption, we observed greater mean population size of bacteria, fungi, and archaea in the tree row than the crop row of the syntropic system, partly confirming our second hypothesis (Fig. 2). The promotion of microbial population size was likely due to increased root biomass and root exudates as plant diversity increased as shown previously for bacteria and fungi in a microcosm experiment35.
In topsoil of the agrosilvopastoral system, trees specifically promoted ectomycorrhizal fungi (EMF) of the genera Hebeloma, Laccaria, and Tuber (Fig. 7M). This finding is not surprising considering the symbiotic association that EMF form with poplar trees and agrees with our previous findings87. Furthermore, for Hebeloma and Tuber spp., the promotion through the trees was also detected in subsoil, indicating that the formation of beneficial tree–microbe relationships extended into deeper soil layers. In contrast, the genus Fusarium, which harbours numerous economically relevant plant pathogens, showed a pattern of lower relative abundance under the trees than in the crop row and open cropland in topsoil. While this pattern could simply emerge due to the absence of their host plants in the tree rows, increased microbial antagonism as well as enhanced biological control through improved soil faunal activity under the trees as proposed by Vaupel et al.87 may have contributed to the observed pattern. Together with our results on potentially beneficial bacteria and EMF, lower abundance of Fusarium spp. under the trees than the crops confirms our fourth hypothesis that trees promote beneficial and symbiotic microorganisms rather than putative phytopathogens.
The introduction of tree rows in arable land through agroforestry is frequently described as a diversification measure that increases spatial heterogeneity by enabling biological interactions between trees and crops88. Consequently, biological interactions within alley cropping systems can be expected to be spatially depended on the distance from the trees into the crop rows. In order to capture such spatial heterogeneity, sampling soil along linear transects spanning from the tree rows into the crop rows is advisable. Using transect sampling within the agrosilvopastoral systems, we here for the first time report that (i) tree rows in a temperate agroforestry system not only promote microbial community dissimilarity in both topsoil and subsoil but that (ii) community dissimilarity decreased with increasing distance from the trees into the crop rows and the open cropland (Fig. 5). In other words, compared to the open cropland system, spatial heterogeneity introduced by the tree rows in the agrosilvopastoral system translated into a compositionally less homogeneous soil microbiome that became increasingly homogenous as the distance from the trees increased. However, we did not observe this pattern in the syntropic system, which we expect to be due to the intensive mulching of the tree rows that has a strong impact on soil microbial communities89,90. Nevertheless, our finding on community dissimilarity in the agrosilvopastoral system is of particular interest given that land-use change from natural ecosystems to agriculture has recently been shown to result in taxonomic homogenization of soil microorganisms91. Agroforestry practice may counteract the homogenization of the soil microbiome through agriculture.
Conclusion
Soil depth strongly affected bacterial and fungal richness and community composition in both the agrosilvopastoral and syntropic agroforestry system. In addition to a decline in microbial richness from topsoil to subsoil, we observed a community shift towards oligotrophic microorganisms with increasing soil depth, as only a limited number of taxa can thrive in the resource-deprived conditions in subsoil. At both soil depths, tree rows of both systems harboured compositionally distinct microbiomes from those of the crop rows. The soil microbiome modulated by the trees showed a shift towards beneficial microorganisms (potential plant growth-promoting bacteria and EMF) while putative phytopathogens were found in lower relative abundance as compared to the crop rows. Finally, we show that compared to an open cropland system without trees, spatial heterogeneity introduced by the tree rows in the agrosilvopastoral system translated into a compositionally less homogeneous soil microbiome that became increasingly homogenous as the distance from the trees increased. Therefore, we suggest that agroforestry can counteract the homogenization of the soil microbiome through agriculture.
Data availability
Amplicon sequencing data have been deposited at NCBI’s Short Read Archive (BioProject https://dataview.ncbi.nlm.nih.gov/object/PRJNA1142703?reviewer=8tmcmrc85peublr8q712s4immh for bacteria and https://dataview.ncbi.nlm.nih.gov/object/PRJNA1142770?reviewer=ss0detkc3eqfdj833q4rs2nbs8 for fungi).
References
Tsonkova, P., Böhm, C., Quinkenstein, A. & Freese, D. Ecological benefits provided by alley cropping systems for production of woody biomass in the temperate region: a review. Agroforest Syst. 85, 133–152 (2012).
Reisner, Y., de Filippi, R., Herzog, F. & Palma, J. Target regions for silvoarable agroforestry in Europe. Ecol. Eng. 29, 401–418 (2007).
Veldkamp, E. et al. Multifunctionality of temperate alley cropping agroforestry outperforms open cropland and grassland. Commun. Earth Environ. 4, 20 (2023).
van Ramshorst, J. G. V. et al. Reducing wind Erosion through Agroforestry: a Case Study using large Eddy Simulations. Sustainability 14, 13372 (2022).
Kanzler, M., Böhm, C., Mirck, J., Schmitt, D. & Veste, M. Microclimate effects on evaporation and winter wheat (Triticum aestivum L.) yield within a temperate agroforestry system. Agroforest Syst. 93, 1821–1841 (2019).
Mayer, S. et al. Soil organic carbon sequestration in temperate agroforestry systems – A meta-analysis. Agric. Ecosyst. Environ. 323, 107689 (2022).
Kletty, F., Rozan, A. & Habold, C. Biodiversity in temperate silvoarable systems: a systematic review. Agric. Ecosyst. Environ. 351, 108480 (2023).
Varah, A., Jones, H., Smith, J. & Potts, S. G. Temperate agroforestry systems provide greater pollination service than monoculture. Agric. Ecosyst. Environ. 301, 107031 (2020).
Staton, T., Walters, R. J., Smith, J. & Girling, R. D. Evaluating the effects of integrating trees into temperate arable systems on pest control and pollination. Agric. Syst. 176, 102676 (2019).
Dollinger, J. & Jose, S. Agroforestry for soil health. Agroforest Syst. 92, 213–219 (2018).
Allen, S. C. et al. Safety-net role of tree roots: evidence from a pecan (Carya Illinoensis K. Koch)–cotton (Gossypium hirsutum L.) alley cropping system in the southern United States. For. Ecol. Manag. 192, 395–407 (2004).
Wang, Y., Zhang, B., Lin, L. & Zepp, H. Agroforestry system reduces subsurface lateral flow and nitrate loss in Jiangxi Province, China. Agric. Ecosyst. Environ. 140, 441–453 (2011).
Young, A. Agroforestry for soil Conservation (CAB international, 1989).
Beule, L., Guerra, V., Lehtsaar, E. & Vaupel, A. Digging deeper: microbial communities in subsoil are strongly promoted by trees in temperate agroforestry systems. Plant. Soil. 480, 423–437 (2022).
Rivest, D. & Martin-Guay, M-O. Nitrogen leaching and soil nutrient supply vary spatially within a temperate tree-based intercropping system. Nutr. Cycl. Agrosyst. 128, 217–231 (2024).
Mace, G. M., Norris, K. & Fitter, A. H. Biodiversity and ecosystem services: a multilayered relationship. Trends Ecol. Evol. 27, 19–26 (2012).
Nielsen, U. N., Wall, D. H. & Six, J. Soil Biodiversity and the Environment. Annu. Rev. Environ. Resour. 40, 63–90 (2015).
Anthony, M. A., Bender, S. F. & van der Heijden, M. G. A. Enumerating soil biodiversity. Proceedings of the National Academy of Sciences 120, e2304663120 (2023).
Van Der Heijden, M. G. A., Bardgett, R. D. & Van Straalen, N. M. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310 (2008).
Pankhurst, C. E. Biodiversity of soil organisms as an indicator of soil health. In: Biological Indicators of soil Health (eds (eds Pankhurst, C., Doube, B. & Gupta, V.) CAB International, Wallingford (1997).
Lehmann, J., Bossio, D. A., Kögel-Knabner, I. & Rillig, M. C. The concept and future prospects of soil health. Nat. Reviews Earth Environ. 1, 544–553 (2020).
Beule, L., Vaupel, A., Moran-Rodas, V. E. & Abundance Diversity, and function of Soil microorganisms in Temperate Alley cropping Agroforestry systems: a review. Microorganisms 10, 616 (2022).
Stamps, W. T. & Linit, M. J. The problem of experimental design in temperate agroforestry. Agroforest Syst. 44, 187–196 (1998).
Beule, L. et al. Poplar rows in Temperate Agroforestry croplands promote Bacteria, Fungi, and denitrification genes in soils. Front. Microbiol. 10, 3108 (2020).
Guillot, E. et al. Spatial heterogeneity of soil quality within a Mediterranean alley cropping agroforestry system: comparison with a monocropping system. Eur. J. Soil Biol. 105, 103330 (2021).
Lemaire, G., Franzluebbers, A., Carvalho, P. C. F. & Dedieu, B. Integrated crop–livestock systems: strategies to achieve synergy between agricultural production and environmental quality. Agric. Ecosyst. Environ. 190, 4–8 (2014).
Franzluebbers, A. J. & Stuedemann, J. A. Soil physical responses to cattle grazing cover crops under conventional and no tillage in the Southern Piedmont USA. Soil Tillage. Res. 100, 141–153 (2008).
Franzluebbers, A. J. & Stuedemann, J. A. Does grazing of cover crops impact biologically active soil carbon and nitrogen fractions under inversion or no tillage management? J. Soil Water Conserv. 70, 365 (2015).
Tobin, C., Singh, S., Kumar, S., Wang, T. & Sexton, P. Demonstrating short-term impacts of grazing and cover crops on soil health and economic benefits in an integrated crop-livestock system in South Dakota. Open. J. Soil. Sci. 10, 109 (2020).
Yang, Y. et al. Responses of the functional structure of soil microbial community to livestock grazing in the tibetan alpine grassland. Glob. Change Biol. 19, 637–648 (2013).
Mubvumba, P., DeLaune, P. B. & Gentry, T. J. Grazing cover crops increases soil microbial biomass in Texas semiarid ecoregion. Agrosystems Geosci. Environ. 7, e20538 (2024).
Vandermeulen, S., Ramírez-Restrepo, C. A., Beckers, Y., Claessens, H. & Bindelle, J. Agroforestry for ruminants: a review of trees and shrubs as fodder in silvopastoral temperate and tropical production systems. Anim. Prod. Sci. 58, 767–777 (2018).
Andrade, D., Pasini, F. & Scarano, F. R. Syntropy and innovation in agriculture. Curr. Opin. Environ. Sustain. 45, 20–24 (2020).
Steinauer, K., Chatzinotas, A. & Eisenhauer, N. Root exudate cocktails: the link between plant diversity and soil microorganisms? Ecol. Evol. 6, 7387–7396 (2016).
Eisenhauer, N. et al. Root biomass and exudates link plant diversity with soil bacterial and fungal biomass. Sci. Rep. 7, 44641 (2017).
Santonja, M. et al. Plant litter diversity increases microbial abundance, fungal diversity, and carbon and nitrogen cycling in a Mediterranean shrubland. Soil Biol. Biochem. 111, 124–134 (2017).
Willer, H., Trávníček, J. & Schlatter, B. The World of Organic Agriculture. Statistics and Emerging Trends 2024 (Research Institute of Organic Agriculture FiBL and IFOAM-Organics International, 2024).
Rosati, A., Borek, R. & Canali, S. Agroforestry and organic agriculture. Agroforest Syst. 95, 805–821 (2021).
Gutzler, C. et al. Agricultural land use changes – a scenario-based sustainability impact assessment for Brandenburg, Germany. Ecol. Ind. 48, 505–517 (2015).
Holsten, A., Vetter, T., Vohland, K. & Krysanova, V. Impact of climate change on soil moisture dynamics in Brandenburg with a focus on nature conservation areas. Ecol. Model. 220, 2076–2087 (2009).
Reyer, C. et al. Climate change adaptation and sustainable regional development: a case study for the Federal State of Brandenburg, Germany. Reg. Envriron. Chang. 12, 523–542 (2012).
Steinfeld, J. P. et al. Identifying agroforestry characteristics for enhanced nutrient cycling potential in Brazil. Agric. Ecosyst. Environ. 362, 108828 (2024).
Harris, D., Horwáth, W. R. & Van Kessel, C. Acid fumigation of soils to remove carbonates prior to total organic carbon or carbon-13 isotopic analysis. Soil Sci. Soc. Am. J. 65, 1853–1856 (2001).
Beule, L. et al. Conversion of monoculture cropland and open grassland to agroforestry alters the abundance of soil bacteria, fungi and soil-N-cycling genes. PLOS ONE. 14, e0218779 (2019).
Guerra, V., Beule, L., Lehtsaar, E., Liao, H-L. & Karlovsky, P. Improved protocol for DNA extraction from Subsoils using phosphate lysis buffer. Microorganisms 8, 532 (2020).
Yu, Y., Lee, C., Kim, J. & Hwang, S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679 (2005).
Takai, K. & Horikoshi, K. Rapid Detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66, 5066–5072 (2000).
Frey, B. et al. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 92, fiw018 (2016).
Tedersoo, L. et al. Global diversity and geography of soil fungi. Science 346, 1256688 (2014).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1 (2013).
Toju, H., Tanabe, A. S., Yamamoto, S. & Sato, H. High-Coverage ITS primers for the DNA-Based identification of Ascomycetes and Basidiomycetes in Environmental samples. PLOS ONE. 7, e40863 (2012).
Vancov, T. & Keen, B. Amplification of soil fungal community DNA using the ITS86F and ITS4 primers. FEMS Microbiol. Lett. 296, 91–96 (2009).
Bednar, Z. et al. Earthworm and soil microbial communities in flower strip mixtures. Plant. Soil. 492, 209–227 (2023).
Beule, L. & Karlovsky, P. Tree rows in temperate agroforestry croplands alter the composition of soil bacterial communities. PLOS ONE. 16, e0246919 (2021).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 13, 581–583 (2016).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Abarenkov, K. et al. UNITE QIIME release for Fungi 2. UNITE Community. https://doi.org/10.15156/BIO/1264763 (2021).
Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90 (2018).
R Core Team. : A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (2024). https://www.R-project.org/
Liu, C., Cui, Y., Li, X. & Yao, M. Microeco: an R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 97, fiaa255 (2021).
Beule, L. & Karlovsky, P. Improved normalization of species count data in ecology by scaling with ranked subsampling (SRS): application to microbial communities. PeerJ 8, e9593 (2020).
Heidrich, V., Karlovsky, P. & Beule, L. SRS’R Package and ‘q2-srs’ QIIME 2 Plugin: normalization of microbiome data using scaling with ranked subsampling (SRS). Appl. Sci. 11, 11473 (2021).
Zhou, H., He, K., Chen, J. & Zhang, X. LinDA: linear models for differential abundance analysis of microbiome compositional data. Genome Biol. 23, 95 (2022).
Ivezić, V., Lorenz, K. & Lal, R. Soil Organic Carbon in Alley Cropping systems: a Meta-analysis. Sustainability 14, 1296 (2022).
Ramírez, P. B., Fuentes-Alburquenque, S., Díez, B., Vargas, I. & Bonilla, C. A. Soil microbial community responses to labile organic carbon fractions in relation to soil type and land use along a climate gradient. Soil Biol. Biochem. 141, 107692 (2020).
Vos, C., Jaconi, A., Jacobs, A. & Don, A. Hot regions of labile and stable soil organic carbon in Germany – spatial variability and driving factors. SOIL 4, 153–167 (2018).
Cardinael, R. et al. Impact of alley cropping agroforestry on stocks, forms and spatial distribution of soil organic carbon — a case study in a Mediterranean context. Geoderma 259–260, 288–299 (2015).
Hao, J. et al. The effects of Soil depth on the structure of Microbial communities in Agricultural soils in Iowa (United States). Appl. Environ. Microbiol. 87, e02673–e02620 (2021).
Kramer, S., Marhan, S., Haslwimmer, H., Ruess, L. & Kandeler, E. Temporal variation in surface and subsoil abundance and function of the soil microbial community in an arable soil. Soil Biol. Biochem. 61, 76–85 (2013).
Yuan, C-L. et al. Limited effects of depth (0–80 cm) on communities of archaea, bacteria and fungi in paddy soil profiles. Eur. J. Soil. Sci. 71, 955–966 (2020).
Fierer, N., Schimel, J. P. & Holden, P. A. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 35, 167–176 (2003).
Dragone, N. B., Hoffert, M., Strickland, M. S. & Fierer, N. Taxonomic and genomic attributes of oligotrophic soil bacteria. ISME Commun. 4, ycae081 (2024).
Stone, B. W. G. et al. Life history strategies among soil bacteria—dichotomy for few, continuum for many. ISME J. 17, 611–619 (2023).
Aguado-Norese, C. et al. Topsoil and subsoil bacterial community assemblies across different drainage conditions in a mountain environment. Biol. Res. 56, 35 (2023).
Lopes, L. D. et al. Soil depth and geographic distance modulate bacterial β-diversity in deep soil profiles throughout the U.S. Corn Belt. Mol. Ecol. 32, 3718–3732 (2023).
Brewer, T. E., Handley, K. M., Carini, P., Gilbert, J. A. & Fierer, N. Genome reduction in an abundant and ubiquitous soil bacterium ‘Candidatus Udaeobacter copiosus’. Nat. Microbiol. 2, 16198 (2017).
Navarrete, A. A. et al. Verrucomicrobial community structure and abundance as indicators for changes in chemical factors linked to soil fertility. Antonie Van Leeuwenhoek. 108, 741–752 (2015).
Ozimek, E. & Hanaka, A. Mortierella Species as the plant growth-promoting Fungi present in the Agricultural soils. Agriculture 11, 7 (2021).
Li, F. et al. Mortierella Elongata’s roles in organic agriculture and crop growth promotion in a mineral soil. Land. Degrad. Dev. 29, 1642–1651 (2018).
Botha, A. Yeasts in soil. In: Biodiversity and Ecophysiology of Yeasts. The Yeast Handbook. (eds (eds Péter, G. & Rosa, C.) Springer, Berlin, Heidelberg. (2006).
Cordovez, V. et al. Priming of Plant Growth Promotion by volatiles of Root-Associated Microbacterium spp. Appl. Environ. Microbiol. 84, e01865–e01818 (2018).
Olanrewaju, O. S. & Babalola, O. O. Streptomyces: implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 103, 1179–1188 (2019).
Asaf, S., Numan, M., Khan, A. L. & Al-Harrasi, A. Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 40, 138–152 (2020).
Luo, Y. et al. Sphingomonas sp. Cra20 increases Plant Growth Rate and alters Rhizosphere Microbial Community structure of Arabidopsis thaliana under Drought stress. Front. Microbiol. 10, 1221 (2019).
Yang, F. et al. Effects of Rhizosphere Microbial communities on Cucumber Fusarium wilt Disease suppression. Microorganisms 11, 1576 (2023).
Vaupel, A. et al. Tree-distance and tree-species effects on soil biota in a temperate agroforestry system. Plant. Soil. 487, 355–372 (2023).
Jose, S. Agroforestry for Ecosystem Services and Environmental Benefits: An Overview (Springer, 2009).
Yang, Y-J. et al. Effect of organic mulches on soil bacterial communities one year after application. Biol. Fertil. Soils. 38, 273–281 (2003).
Zhang, S. et al. Organic mulching positively regulates the soil microbial communities and ecosystem functions in tea plantation. BMC Microbiol. 20, 103 (2020).
Peng, Z. et al. Land conversion to agriculture induces taxonomic homogenization of soil microbial communities globally. Nat. Commun. 15, 3624 (2024).
Funding
Open Access funding enabled and organized by Projekt DEAL.
This study was financed by funds of the Federal Ministry of Food and Agriculture (BMEL) based on a decision of the Parliament of the Federal Republic of Germany via the Federal Office for Agriculture and Food (BLE), grant number 28DE204D21 (DaVaSus project). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Open access funding enabled and organized by Project DEAL.
Author information
Authors and Affiliations
Contributions
A.V., M.K., J.T., B.B., and L.B. contributed to the conception and design of the study. A.V. and L.B. performed the fieldwork. A.V., N.H., and L.B. coordinated the laboratory work. A.V. and L.B. performed the statistical analyses. A.V., J.T., and L.B. wrote the first draft of the manuscript. M.K., N.H. and B.B. critically revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vaupel, A., Küsters, M., Toups, J. et al. Trees shape the soil microbiome of a temperate agrosilvopastoral and syntropic agroforestry system. Sci Rep 15, 1550 (2025). https://doi.org/10.1038/s41598-025-85556-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-85556-4
Keywords
This article is cited by
-
Data-driven scenario analysis supports the revival of historic silvoarable systems for carbon smart rural landscapes
Scientific Reports (2025)
-
Assessment of pea (Pisum sativum L.) and cauliflower (Brassica oleracea L. var. botrytis) productivity in Mulberry (Morus alba L.) based agroforestry system in mid-hills of the north-western Himalayas: effects of tree canopy, fertiliser and bio-stimulants
Agroforestry Systems (2025)
-
South African Myxococcota: an untapped resource for microbial ecolo gy and biotechnology
Applied Microbiology and Biotechnology (2025)
-
Guidance on spatial sampling designs and data reusability in agroforestry research
Agroforestry Systems (2025)