Abstract
Feedlot cattle may be subjected to digestive disorders, including ruminal acidosis, due to high concentration of grain in their diet. Therefore, novel feeding strategies are required to maximize animal performance and mitigate economic losses in the operation. This study employed a two-period crossover design to assess the effect of direct ruminal administration of native rumen microorganisms (NRM) inoculation on cattle that underwent a high-grain challenge. The NRM inoculation consisted of six microorganisms (1.70 M CFU /day/animal) isolated from the rumen of healthy feedlot cattle: Succinivibrio dextrinosolvens ASCUSBF53, Prevotella albensis ASCUSBF41, Chordicoccus furentiruminis ASCUSBF65, Bacteroides xylanisolvens ASCUSBF52, Clostridium beijerinckii ASCUSBF26, and Syntrophococcus sp. ASCUSBF60. The trial consisted of 16 Angus heifers receiving NRM (n = 8) or a CON (CON = Carrier Buffer; n = 8) inoculation daily for 14-days as pre-challenge while on a high-grain diet and continued daily for a 21-day treatment period. The combined 35 days of microbial supplementation resulted in an improved average daily gain (ADG) of 29% (P = 0.037) and a tendency toward a 19% decrease in the feed efficiency metric, gain to feed ratio (G: F) (P = 0.055). Additionally, administration of NRM to animals on a high-grain diet, improved ruminal microbiome stability (P < 0.001), potentially encouraging the conversion of rumen lactate to propionate over time via the succinate pathway and alleviating metabolic stress.
Similar content being viewed by others
Introduction
Diets in modern cattle finishing programs have been designed to encourage accelerated animal growth. Due to the shift in feed nutrient density, feedlot cattle are particularly prone to diet induced rumen microbial dysbiosis which can lead to rumen acidosis, a serious metabolic disease that negatively impacts animal health and productivity1,2. Among the many variables that influence rumen microbial communities (e.g., season, host genetics, and management), diet plays a pivotal role in shaping the rumen microbiome and its function and is also directly involved in the development of subacute ruminal acidosis (SARA)3,4. The transition to a high-grain diet and feeding mismanagement promotes rapid digestion and fermentation by rumen microbes and leads to the accumulation of organic acids and reduction in ruminal pH, which can cause metabolic diseases5,6,7,8.
Diet-induced microbial dysbiosis can be detrimental to rumen function. Accompanying the reduced rumen pH, studies have reported the accumulation of lactic acid in rumen fluid and its association with contributing to the development of ruminal acidosis1,9. Rapid rumen fermentation also results in the production of a large quantity of CO2. Laporte-Uribe (2016)10 theorized that under rapid fermentation, a large amount of dissolved CO2 (dCO2) in rumen fluid would remain undissociated and contribute to the reduced rumen pH and rumen fluid buffer capacity, thus interfering with host metabolic functions10,11. Therefore, optimizing the rumen microbial interactions (e.g., competition and cooperation) that could reduce the production of lactic acid and dCO2 may be able to stabilize rumen pH and aid the adaptation of the rumen microbiome to high-grain diets, improving ruminant digestive health and animal productivity.
Compositional variations of diverse microbial populations in a healthy rumen have been linked to feed efficiency, methane production, and other production traits12,13,14. Supplementing microorganisms associated with these compositional variations may be a viable mechanism to influence the ruminal microbiome and positively impact animal production traits. A large survey experiment was conducted to identify strains of rumen microorganisms strongly associated with high performance in feedlot cattle13. Based on the significance of their associations to animal efficiency (e.g., feed efficiency, ADG, and ADFI) and impacts on the rumen microbiome stability, six strains of Native Rumen Microorganisms (NRM) (Succinivibrio dextrinosolvens ASCUSBF53, Prevotella albensis ASCUSBF41, Chordicoccus furentiruminis ASCUSBF65, Bacteroides xylanisolvens ASCUSBF52, Clostridium beijerinckii ASCUSBF26, and Syntrophococcus sp. ASCUSBF60) were selected to examine their ability at maintaining healthy ruminal fermentation and improving animal performance on a high-grain diet15,16.
Four species have been previously studied in context of rumen fermentation: S. dextrinosolvens, P. albensis, B. xylanisolvens, and C. beijerinckii. S. dextrinosolvens, members of Gammaproteobacteria, can ferment an array of carbohydrates, including glucose and xylose, and requires exogenous CO2 to produce succinate, acetate, formate, and lactate17,18,19. Studies have also shown that other members of succinate producing Gamma-proteobacteria are able to generate additional ATP from CO2 fixation via phosphoenolpyruvate (PEP) carboxykinase, suggesting that succinate production may play a role in microbial carbon capture and energy conservation in the rumen20,21. P. albensis has previously been noted for its ability to degrade starches and xylan, suggesting that it may play a significant role in feed digestion in the rumen22,23,24. Studies have also suggested that P. albensis may play a protective role in an acidotic rumen environment25. Also, B. xylanisolvens has been shown to degrade many plant-derived polysaccharides, including xylan, to produce acetate, propionate, and succinate26. In addition, C. beijerinckii, has previously shown to work in conjunction with other microorganisms in the rumen to increase acetate and butyrate production, positively influencing animal metabolism27,28. The remaining two, C. furentiruminis and Syntrophococcus sp., are novel microbes with little literature documentation. Most notably, C. furentiruminis is a novel acetogenic species that can convert CO2 to acetate via the Wood-Ljungdahl pathway29. Both species are prevalent in ruminants based on 16 S rRNA gene sequencing (> 97% sequence identity) and have been found in different ruminant species globally based on the National Center for Biotechnology Information (NCBI) sequence repository.
Building on literature and previous findings, it was hypothesized that supplementing these NRM may modify overall rumen microbial interactions, alleviate negative ruminal conditions, and sustain cattle performance while experiencing ruminal grain challenge. The objective of the current study was to determine the effect of administering the previously discussed NRM in Angus cattle during grain challenge on rumen microbial populations, rumen microbial interactions, ruminal conditions, and performance.
Materials and methods
This study was approved and carried out in accordance with the recommendations of the Institutional Animal Care and Use Committee at the University of Tennessee, Knoxville (Protocol Number 2570 − 1217). The study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Experimental design
A total of 16 cannulated, registered purebred Angus heifers from the University of Tennessee East Tennessee Research and Education Center (ETREC) were used in the study. Heifers were utilized due to the impact and longevity of females in the herd. Sample size to detect differences was determined based on previous research7,12. Animals were housed in individual outdoor covered pens (pen size: 10.97 m x 3.66 m) with ad libitum access to water. Animals were individually housed at the ETREC Blount Unit, in Louisville, TN. At the beginning of the study, the animals were an average of 458 days old and weighed approximately 385±30 kg. Cattle received a total mixed ration (TMR; Table 1) daily ad libitum but were managed on a slick-bunk basis to achieve little to no feed refusals. The TMR was delivered directly to feed bunks once daily using a mixer wagon (WIC MDR-48; Wickham, Canada) at 0700 h. Feed refusals were collected and weighed (Adams CPWplus; Oxford, CT) immediately prior to the following morning’s feeding.
The project consisted of an acclimation phase and an experimental phase with 2 × 2 (AB/BA) crossover design (Fig. 1). During the acclimation phase, all animals were transitioned and acclimated from a 100% hay diet to a finishing ration (pre-challenge ration) in 3 steps over 20 days (Table 1) prior to being fed the finishing ration, and no NRM were administered. At the end of the acclimation phase, animals were blocked by body weight (BW) into two groups for the experimental phase. The experimental phase consisted of two crossover periods, with a 7-day wash-out period between each phase. NRM (carrier medium of 6 mL phosphate buffer solution containing S. dextrinosolvens ASCUSBF53, P. albensis ASCUSBF41, C. furentiruminis ASCUSBF65, B. xylanisolvens ASCUSBF52, C. beijerinckii ASCUSBF26, and Syntrophococcus sp. ASCUSBF60, each at 1.7 × 108 CFU/mL) were administered to treatment animals (n = 8) daily through rumen cannula during each crossover period and control animals (CON; n = 8) received the same NRM carrier buffer (6 mL phosphate buffer solution) but without microorganisms. No NRM or carrier buffer was administered during the wash-out period. Within each crossover period, animals remained on the pre-challenge ration for 14 days (pre-challenge dosing period) followed by a 21-day grain challenge period (Fig. 1). Beginning on day 1 of each challenge period, approximately 50% of the roughage dry matter was replaced with dry-rolled corn, and TMR offerings were increased daily by approximately 0.57 kg of DM per day until feed refusals were greater than or equal to 10% of the previous day’s offerings. All animals were returned to the herd after study completion.
Schematic of the 2 × 2 cross-over trial design. Each group consisted of 8 cannulated heifers. Cattle in the treatment group received daily dosing of native rumen microorganism (NRM) supplement suspended in a carrier medium via cannula while cattle in the control group received a blank carrier medium via cannula. The “Washout” period refers to “washing” the supplement out of the rumen by a period of non-supplementation. Pj and Ck represent the study periods and the crossover periods in the statistical model, respectively.
Sample collection and analysis
Throughout the study, BW was measured weekly along with daily feed intake and refusal. Rumen content samples were collected via cannula 1 day before the start of the trial, then every 7 days during the acclimation period, and every 3 to 4 days for the remainder of each challenge. All rumen content samples were collected in the morning prior to feeding. To account for the heterogeneity within the rumen, rumen content from the ventral, central, anterior, and posterior regions of the rumen were mixed. Each rumen content sample consisted of approximately 3:1 of rumen fluid: rumen solids. Rumen fluid pH was measured using an UltraBasic pH probe (Denver Instruments, Arvada, CO, United States), and rumen temperature and rumen fluid dCO2 were measured using the Inpro 5000 CO2 sensor (Mettler Toledo, Greifensee, Switzerland) on the composited rumen contents at the time of sampling. Rumen content samples were stored at − 80 °C until DNA extraction and amplicon sequencing were performed.
An electronic bolus (ebolus by eCow limited, Glasgow, UK) was also placed in the rumen of each animal to monitor rumen pH in real-time with pH measurements taken every 15 min. Throughout the study, 3 eboluses failed to operate and 4 were replaced due to mechanical malfunction and failure to calibrate. These interruptions led to missed periods of pH measurements and sometimes mismatched baselines. Additionally, since the rumen content samples were collected as a composite to accurately represent the entire rumen, associating the above-mentioned manual pH measurements with rumen chemistry and microbiome becomes more appropriate. Therefore, these ebolus pH measurements were used to monitor the real-time rumen pH changes due to the grain-challenges but not for statistical analyses (Figure S1)30,31). Henceforth, the term “rumen pH” or “rumen fluid pH” pertains to the measurement derived from the composite rumen content samples unless specified otherwise.
A subsample of rumen contents was aliquoted from each rumen content sample and prepared for organic acid analysis, including volatile fatty acid (VFA) and lactic acid. Strained samples were centrifuged at 10,000 RCF for 10 min at 4 °C. A mixture was then prepared consisting of 5 mL of rumen fluid supernatant and 1 mL of 25% (w/v) meta-phosphoric acid, 25mM2ethyl butyric acid solution. This mixture was stored on ice for 30 min, then centrifuged for 10 min at 10,000 RCF and 4 °C. Using the method described by Erwin et al. (1961)32, the samples were analyzed via gas chromatography (Agilent 7890B, Agilent Technologies, Inc., Santa Clara, CA, USA). The gas chromatograph was equipped with an FID detector, Nukol fused silica capillary column (Supelco, Sigma-Aldrich Co., LLC, Bellefonte, PA, USA), and utilized helium as the carrier gas.
A weekly blood sample (10 mL; Corvac serum separator, Kendall Health Care, St. Louis, MO) was collected via coccygeal venipuncture, which aligns with rumen sampling at approximately every 3–4 days. Immediately following blood collection, a 1 mL aliquot of blood was analyzed using the VetScan i-STAT (Abbott, Princeton, New Jersey, USA) and the CG4 + cartridges. The remainder of each blood sample was cooled and centrifuged at 2000 RCF at 4 °C for 20 min. Serum was separated and stored at −80 °C until further processing and analysis. Parameters analyzed using the VetScan i-STAT included L-lactate (mmol/L), pH, PCO2 (mmHg), PO2 (mmHg), TCO2 (mmol/L), HCO3 (mmol/L), base excess in the extracellular fluid compartment (BEEcf; mmol/L), and sO2 (%).
DNA extraction and sequencing
To concentrate the sample, 1.5 ml of the rumen samples were centrifuged at 4000 RCF at 4 °C for 15 min and the supernatant was discarded. The resultant pellet was resuspended in 0.5 ml of the appropriate fluid from the PowerMag DNA Isolation Kit (Qiagen, Hilden, Germany). DNA extraction was carried out following the commercial protocol provided in the kit.
Microbial 16 S rRNA gene amplicon sequences were used to evaluate the rumen microbiome. Specifically, the bacterial 16 S rRNA gene was amplified following standard protocols33 (9Q5 High-Fidelity DNA Polymerase; New England Biolabs, Inc., Ipswich, MA, USA) using 27 F and 534R34 primers modified for Illumina sequencing. Standard gel electrophoresis (2% agarose) was used to verify the size of PCR fragments. The amplicon products were then purified using AMPure XP bead (Beckman Coulter, Brea, CA, USA), followed by quantification using the PicoGreen dsDNA quantification assay. Final pooled amplicon products were sequenced on the MiSeq Platform (Illumina, San Diego, CA, USA) according to standard protocols using a 1 × 300 v3, 600-cycle kit. Raw sequence reads were de-multiplexed on the MiSeq Platform using the Illumina factory software.
Rumen bacterial cell counts
The number of bacterial cells in each rumen content sample was determined using a Sony SH3000 Flow Cytometer (Sony Biotechnology, San Jose, CA, USA). Specifically, approximately 0.5 g of rumen sample was homogenized and then centrifuged to remove the debris. SYBR Green I (Invitrogen, ThermoFisher, Waltham, MA) was used to measure all particles containing nucleic acids and Calcofluor-White (MilliporeSigma, Burlington, MA) was used to stain 1,4-β-glucans, which is a common component of fungal cell wall. SYBR green I and Calcofluor-White solution were mixed with an aliquot of sample supernatant for a final volume of 500 µL35. Samples mixed with dyes were incubated in the dark at room temperature for 20 min. Prior to counting, Spherotech AccuCount Particles (7–7.9 μm; Spherotech, Lake Forest, IL) were added to each sample. SYBR Green I was measured at excitation/emission (Ex/Em) of 497/520 nm and Calcofluor-White was measured at Ex/Em of 380/475 nm. Cells detected by both SYBR Green I and Calcofluor-White were considered the fungal cells and the bacterial cell counts were determined by subtracting the fungal cell counts from the total SYBR Green I counts.
Statistical analyses for animal performance, blood, and rumen fluid measurements
A 2 × 2 crossover design could not differentiate the carry-over effect, treatment sequence effect36. Because the treatment groups in this study were with NRM and CON, it is reasonable to assume that treatment sequence effect is negligible and only the carry-over effect and the crossover period specific treatment effect need to be evaluated. A statistical pre-test was carried out to specifically evaluate the carry-over effect37,38. Specifically, for each sequence group, the sum of squares between two crossover periods of each subject was calculated. An un-paired t-test between the two sequence groups was conducted using the formula:
Where m is the sample size of treatment sequence 1 (AB), n is the sample size of treatment sequence 2 (BA), \(\:{\overline{C}}_{AB}\) is the mean of within-subject crossover period sum of sequence 1, \(\:{\overline{C}}_{BA}\) is the mean of within-subject crossover period sum of sequence 2, SQAB is the total sum of squares of sequence 1 within-subject crossover period sum, and SQBA is the total sum of squares of sequence 2 within-subject crossover period sum. Significance was declared at P ≤ 0.1 and no carryover effect was observed for any of the measurements (Table S1).
Once the carryover effect was eliminated, a linear mixed effect (LME) with residual maximum likelihood (REML) regression model was used to evaluate the animal performance, blood, and rumen content measurements in R. The model formula is shown as follows:
Yijklmno = µ + Ti + Pj + Ck + Sl + Hm(l) + eijklmno.
Where Yijklr is the performance response (box-cox transformed to ensure normality), µ represents the average, Ti is the treatment effect, Pj is the study period representing the pre-challenge dosing and grain challenge period (Fig. 1), Ck is the crossover period effect (Fig. 1), Sl represents the crossover period specific treatment effect, individual heifer (Hm(l)) is included as the random effect, and eijklr is the residual error. The interaction effect of treatment and study period (Ti x Pj), baseline value for each response (the acclimation period), and baseline trajectory (linear regression slope of the baseline values for each response) was also evaluated. If these effects exist (P ≤ 0.1), the terms were added to the model above. The final model P-values are reported in Supplementary Table S1. Significance for interaction, baseline, baseline trajectory was declared at P ≤ 0.10. For main effects and post-hoc analysis, significance was declared at P ≤ 0.05 and a tendency toward significance was declared at 0.05 < P ≤ 0.10.
Rumen microbiome analysis
Considering the abundance of rumen microorganisms are in constant flux, the abundance of administered NRM strains between CON and NRM group was evaluated on each sampling day using a linear mixed effect model with individual heifer as the random effect. Significance was declared at P ≤ 0.05 and a tendency toward significance was declared at 0.05 < P ≤ 0.10.
The de-multiplexed raw amplicon sequence reads were processed to ensure accurate microbial inference. Specifically, the adapter sequences were removed using Trim Galore31. The reads were trimmed to 250 bases. USEARCH was used in downstream read analysis, where PhiX control reads and low complexity reads were removed39. Sequences were quality filtered with a maximum estimated error of 140. Sequencing errors, including chimeras, were identified and removed using the USEARCH UNOISE3 command41. The final zero-radius operational taxonomic units (zOTUs) were defined by clustering at zero-radius and their taxonomies were defined using the USEARCH SINTAX command with RDP 16 S rRNA database at a confidence threshold of 0.542. The presence of inoculated microorganisms was confirmed by comparing their 16 S rRNA gene sequences with the final zOTU sequences at 97% identity. The absolute abundance of each zOTU in each sample was estimated based on the sample bacterial cell counts. The abundance of the NRM in the samples was determined by sequence alignment using USEARCH with a minimum of 97% 16 S rRNA gene identity.
Samples with a low sequencing depth and coverage were excluded from the downstream microbiome analysis (35 out of 462 samples). Based on the sequencing depth and Good’s coverage plot, a sample was considered adequately sequenced if it had at least 10,000 quality sequence reads and a minimum Good’s coverage of 0.95. For samples with fewer than 10,000 quality sequences, if their bacterial cell counts were also fewer than median across all samples, the sequencing depth and coverage were likely influenced by the sample cell counts. In this case, a sample was also considered adequately sequenced if its Good’s coverage is greater than 0.9. Shannon’s diversity index and Pielou evenness were also calculated to provide an overview of the rumen microbial diversity. Both Good’s coverage and Shannon’s diversity index were calculated using python package scikit-bio (v0.5.9).
Core microbiome for control and treatment group was identified if a zOTU was detected in all animals of the specific experimental group. Correlations among the core zOTUs, diet components (i.e., percent of hay, percent of corn silage, percent of dry rolled corn), and rumen measurements (i.e., rumen fluid pH, dCO2, acetate concentration, butyrate concentration, propionate concentration, and lactate concentration) using both monotonic (Spearman’s rank-order correlation) and non-monotonic (Hoeffding’s dependence) relationships was conducted. To eliminate the compositional bias introduced by the amplicon sequencing, the absolute abundance of each core zOTU was used in the correlation analysis. A correlation was considered significant if the false discovery rate (FDR) adjusted P-value was less than 0.05.
For all significant correlations, the node degrees and betweenness centrality for each zOTU were calculated using Python (networkx). The relationship between node degrees and betweenness centrality for CON and NRM was modeled using a generalized additive model with the following formula in R (mgcv):
Where y represents betweenness centrality, T is treatment, \(\:{f}_{1}\left(N\right)\bullet\:T\) is the smoothing term of node degrees constrained by the treatment effect, and \(\varepsilon\)is the residual.
An impact score (I) for each core zOTU was calculated based on its node degree and betweenness centrality:
Where A represents Node Degree, B represents Betweenness Centrality, and \(\:\theta\:=\:{\text{tan}}^{-1}\left(\frac{B}{A}\right)\). The mean of impact scores of CON and NRM group was compared using one-way ANOVA in R.
Results and discussion
Grain challenges reduced ruminal pH in real-time
The real-time measurement of ruminal pH using eCow eboluses indicated a reduction in pH correlating with an escalation in grain intake (Figure S1). Across the grain challenge phases, the daily median ebolus ruminal pH consistently fell from a daily peak of approximately 7 to within the range of 5 to 5.5, aligning with expectations of SARA and acute ruminal acidosis43. This suggests that the grain challenge effectively induced ruminal acidosis.
Animal performance
The administration of NRM to Angus heifers improved ADG and tended to significantly improve animal G: F (Table 2). Compared to when animals were receiving no NRM (CON group), NRM animals were gaining 0.41 kg/d (29%) on average when NRM was administered (P = 0.037). No difference in dry matter intake (DMI) was observed between CON and NRM group (P = 0.98). This led to a tendency toward a significant improvement (24%; P = 0.055) in G: F, which was driven by the improvement in ADG. The ADG and G: F improvements observed while animals were administered with NRM suggest that NRM may have assisted the rumen microbiome’s adaptation to the presence of highly fermentable dietary ingredients and hence improved rumen function throughout the grain challenge period44,45.
Due to the experimental design, the overall endpoint BW was similar between CON and NRM (P = 0.94; Table 2) and a less relevant measurement comparing to ADG in the current study. This is consistent with the significant crossover (Ck) effect observed in the BW (P < 0.001; Table S1), where the animals were 63 kg heavier at the end of crossover period 2 than the end of crossover period 1 (P < 0.001; Table S3). During the washout period, although no microorganisms were administered, animals of CON and NRM group continued to gain weight at their respective rate and the NRM group weighed marginally more than CON group (P = 0.12; Table 2). This suggests that administrating NRM likely influenced the rumen microbiome and led to an extended weight gain, despite the effect diminishes within 7 days44,45,46,47,48. After the washout period, the animal treatment groups were swapped, meaning the NRM group started with a marginally lower BW than CON group during crossover period 2. At the end of the crossover period 2, the BW difference between the two treatment groups was smaller (P = 0.36; Table 2). Because ADG measures the growth rate of the animals and were not influenced by the crossover model (Ck: P = 0.20; Table S1), this led to the observation of the significant improvement in ADG but not in the overall endpoint BW (Table 2).
Blood measurements
Rumen acidosis leads to physiological changes in animals and blood chemistry often reflects these changes10,49,50. It was hypothesized that the alleviation of rumen acidotic condition would alter animal blood chemistry as well. Blood temperature, pH, base excess in extracellular fluid (BEEcf), carbon dioxide measurement (TCO2, pCO2), bicarbonate concentration (HCO3), oxygen measurements (sO2, pO2), and lactate concentrations were not affected by administrating NRM, suggesting that blood chemistry may not be sensitive enough to detect the effects of NRM supplementation in the rumen (P ≥ 0.26; Table 3)51,52.
Pereira et al. (2021)53 reported a range of blood chemistry measurements in high-grain fed feedlot beef (Angus cross and Nellore). All blood measurements were within the reported range of previous mentioned study, except for the blood pCO2 measurements53,54. In the current study, these measurements were less than the minimum value reported by Pereira et al. (2021)53. Further, a low blood pCO2 value indicates an imbalance of serum bicarbonate buffer system and the blood HCO3 concentration is expected to decrease as well55. However, the blood HCO3 concentration is similar to the average of those reported in the previous mentioned study, suggesting an increase in HCO3 flux may be compensating for the acidotic condition in the rumen. This is further supported by an elevated value of BEEcf being detected in this study53. This finding agrees with the hypothesis that CO2 species play an important role in rumen acidosis10 and cattle adapt to high-grain diets through rumen and blood acid-base manipulations.
Rumen fluid chemistry, bacterial load, and diversity
Using the composite rumen content samples, rumen fluid temperature, pH, carbon dioxide measurements (pCO2, dCO2), lactate and VFA concentration (total VFA, acetate, propionate, butyrate), and bacterial load were measured to evaluate the NRM effect on rumen fermentation products. Although likely not biologically relevant, rumen temperature significantly decreased by 0.13ºC (P = 0.047) while cattle received NRM in this study. Further, a greater evenness was observed in the rumen bacterial communities (Pielou’s Evenness) during the pre-challenge dosing period (P = 0.039; Table 3), suggesting a more balanced microbiome. No other significant differences were observed in rumen chemistry measurements between the treatment groups (P ≥ 0.29; Table 3).
It is important to highlight the discrepancy between the rumen pH values obtained from composited rumen content samples and the real-time measurements using eboluses. This variance can be attributed to two factors: (1) the collection of composite rumen content samples before morning feeding, contrasting with the continuous real-time pH measurements occurring every 15 min; (2) the comprehensive nature of the composite samples, drawn from four quadrants of the rumen contents to more accurately represent the overall rumen condition, as opposed to focusing on a singular region. Hence, the limitation of the rumen pH measurements obtained from the composited rumen content may not comprehensively reflect the occurrence of ruminal acidosis, as the diurnal pH pattern relative to morning feeding would be missed, but is supported by the representative eBolus data (Figure S1)30,31).
The effect of grain challenged induced ruminal acidosis on rumen temperature has been previously reported56. It was found that beef cattle experiencing ruminal acidosis had a higher rumen temperature than those that did not undergo the challenge. Wahrmund et al. (2012)56 and Sato (2016)57 also reported a negative correlation between rumen pH and rumen temperature, suggesting that a rise in rumen temperature is likely related to rapid rumen fermentation. The significant decrease in rumen temperature while animals received NRM in this study may indicate that NRM can potentially influence the rate of rumen fermentation and prevent the undesirable rapid pH decrease. This is also in line with the observation of a more even bacterial community in during the pre-challenge dosing period while cattle received NRM, where a greater initial community evenness has been associated with improved community function stability58,59. It is also worth mentioning that rumen fermentation is in constant flux and the host animal also plays a significant role in removing the fermentation products from rumen60,61. Further investment of the effect of NRM on rumen chemistry is needed.
Crossover period specific treatment effects
A tendency toward significance was detected in G: F (P = 0.10), blood pCO2b (P = 0.084), blood HCO3− (P = 0.065), and rumen lactate (P = 0.062; Table S1). Further examination of treatment effect within each crossover period revealed that blood pCO2 and HCO3 concentration were trending significantly (P = 0.054 and P = 0.085, respectively) greater in cattle that received NRM during crossover period 1. No statistical differences were observed (P ≥ 0.18) in G: F and rumen lactate during this period. During crossover period 2, G: F was significantly (P = 0.05) greater in cattle receiving NRM, while marginally less blood HCO3 (P = 0.12) and rumen lactate (P = 0.11) were associated with NRM animals and no difference was observed in blood pCO2 (Table S2).
These opposite trends of treatment effects during each crossover period may be linked to various stages of rumen microbiome adaptation and fermentation. Clemmons et al. (2019)7 previously observed that cattle adaptation to high-grain diets can take much longer than a traditional step-up period, suggesting rumen microbiome turnover occurs in stages. It is plausible that NRM may be able to promote beneficial microbial interactions that achieve temporal rumen fluid and blood acid-base balance during each stage7,62. Further studies are necessary to specifically examine this hypothesis.
Crossover period and study period effects
Significant differences and tendencies between two crossover periods (crossover period 1 VS crossover period 2) and two study periods (pre-challenge dosing VS grain challenge) were detected in animal performance and several blood and rumen measurements (Table S1). While the crossover period specifically evaluated the effect of time, the study period evaluated both the effect of time and grain challenge.
Among the animal performance measurements, endpoint BW was significantly greater in crossover period 2 compared to crossover period 1 (P < 0.001; Table S3). Because DMI is sensitive to both animal size and diet change, it is not surprising that animals ingested significantly more during both crossover period 2 and grain challenge period (P < 0.001; Tables S3 and S4). Compared to pre-challenge dosing period, a greater intake on a high energy diet (Table 1) during grain challenge period was also consistent with a significantly greater ADG (P = 0.002) (Table S4).
Rumen pH significantly decreased in crossover period 2 (P = 0.033; Table S3) and grain challenge period (P < 0.001; Table S4), suggesting that diet inducing acidosis was a major factor. The rumen CO2 measurements (dCO2, mg/L and pCO2, mmHg) were significantly greater in crossover period 2 (P ≤ 0.004; Table S3) but decreased during grain challenge period (P < 0.001; Table S4). While this opposite of trends may seem contradicting, they are consistent with each other when evaluated as unit of CO2 per unit of pH [U = log10(rumen CO2 measurements) / pH]. The calculated U(dCO2 mg/L) and U(pCO2 mmHg) were greater in both crossover period 2 (U(dCO2 mg/L): crossover period 1 = 0.41 ± 0.003, crossover period 2 = 0.42 ± 0.004, P < 0.001; U(pCO2 mmHg): crossover period 1 = 0.39 ± 0.003, crossover period 2 = 0.40 ± 0.003, P < 0.001) and grain challenge period (U(dCO2 mg/L): pre-challenge dosing = 0.41 ± 0.004, grain challenge = 0.42 ± 0.003, P < 0.001; U(pCO2 mmHg): pre-challenge dosing = 0.39 ± 0.003, grain challenge = 0.4 ± 0.003, P < 0.001). This is consistent with the observation that the accumulation of dissolved CO2 in rumen fluid is associated with reduced rumen pH10.
Rumen lactate concentration was significantly greater in crossover period 2 compared to crossover period 1 (P < 0.001; Table S3) and no study period effect was observed (P = 0.88; Table S1). However, the concentration of lactate in blood decreased in both crossover period 2 and grain challenge period (P < 0.007; Table S3 and S4). This suggests that the concentration of lactate in rumen fluid does not always translate into lactate concentration in blood63. Lactate concentrations in the blood are largely influenced by time instead of the induced acidosis events in this study.
The increase in rumen VFA production has been documented in cattle fed a high concentrate diet64. As expected, the VFA concentrations in rumen fluid increased from crossover period 1 to crossover period 2 (P < 0.001; Table S3), as well as from pre-challenge dosing to grain challenger period (P ≤ 0.065; Table S4). The number of bacterial cells were greater in crossover period 2 compared to crossover period 1 (P < 0.001; Table S3), agreeing with the literature that rumen VFA production is associated with ruminal bacterial growth2,3. However, fewer bacteria cells were detected during grain challenge than pre-challenge dosing period (P < 0.001; Table S4), indicating that diet induced acidosis can lead to rumen microbial dysbiosis by encouraging the growth of some VFA producing bacteria while inhibiting the growth of some other bacteria25. Overcoming this imbalance in rumen microbiome is the main objective in alleviating acidosis effects in feedlot cattle.
Bacterial growth and rumen fermentation
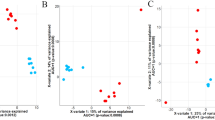
Rapid microbial growth from highly fermentable dietary ingredients drives the changes observed in rumen fermentation, including reduced rumen pH, increases in dCO2, and the accumulation of lactate and VFA in rumen fluid2,10. Consistent with the literature, upon the transition to the high-grain diets, the concentration of ruminal bacteria (cells/mL) increased while the bacterial community diversity decreased throughout the study (Fig. 2A)15,57. Rumen fluid pH decreased over the course of the study, with the least pH recorded during the first grain challenge period (Fig. 2B). As expected, the concentration of propionate, lactate, and butyrate in rumen fluid increased continuously throughout the study, while the concentration of acetate decreased (Fig. 2C).
The data presented in this figure includes both the control (no microbial supplementation) and the treatment (with microbial supplementation) groups to display the effect that time had on (A) the ruminal bacterial diversity and abundance throughout the study, (B) the rumen pH throughout the study and (C) the ruminal concentration of fatty acids throughout the trial. The vertical lines separate the experimental stages, from left to right, acclimation (no microbe supplementation), pre-challenge dosing 1, grain challenge 1, washout (no microbe supplementation), pre-challenge dosing 2, grain challenge 2.
The accumulation of lactate in the rumen has been linked to rumen acidosis2,65. Literature suggests that the production of microbial lactic acid was the primary drive of the decrease in rumen pH2,65. However, it has been shown that a greater amount of lactic acid was produced when the surrounding environment was acidic66,67. Thus, the accumulation of ruminal lactic acid may not be the main cause of decreasing rumen pH and other factors may be contributing to the changes of pH in rumen10,11.
Rumen propionate contributes directly to animal’s gluconeogenesis68,69. Three main pathways for bacterial propionate production have been identified and lactate is a key substrate in two of them, the succinate and acrylate pathways70. An in vitro isotopic enrichment study found that lactate was converted to propionate preferentially through the acrylate pathway at neutral pH. However, with increased dCO2 and reduced pH in rumen fluid, the succinate pathway, which depends on exogenous CO2 fixation to generate energy, may be favored in the rumen of grain fed beef cattle20,21. Literature reported the accumulation of succinate in lactating dairy cows that were experiencing SARA71. Further, succinate has also been identified as a key metabolite associated with highly efficient beef steers72, suggesting that the succinate pathway could play a major role in rumen fermentation of highly fermentable dietary ingredients. Unfortunately, succinate was not measured in this study. However, a strong and significant correlation was observed between the ruminal lactate and propionate concentrations (Spearman’s coefficient = 0.90, false discovery rate adjusted P < 0.05). Hypothesizing from the literature and the observation of this study, the conversion from lactate to propionate via succinate pathway may help fuel the animal glucose needs in beef cattle. Additional experiments are required to further investigate this hypothesis.
Abundance of administered microorganisms
Investigating the microbial composition of the rumen using 16 S rRNA gene sequencing revealed that all 6 strains administered were present in rumen samples of both treatment and control animals’ rumen samples (Fig. 3). This is expected as all 6 strains are commensal rumen microbes. The abundance of all six microbes fluctuated throughout the study, largely following the changes in diet. However, S. dextrinosolvens ASCUSBF53 was more abundant in NRM during the first pre-challenge dosing period and C. furentiruminis ASCUSBF65 were more abundant in NRM at the beginning of both grain challenge periods, suggesting that these four microbes may be contributing to the rumen microbiome modification that led to the observed improvements in animal performance. The other 4 microorganisms were found more abundant or tended to be more abundant in CON groups on different days throughout the study (Fig. 3).
The observed abundance of the microorganisms supplemented in the NRM in rumen throughout the study. Control animals did not receive additional microbe supplementation. Treatement animals received 1.7 × 108 CFU/mL per day supplementation of each S. dextrinosolvens ASCUSBF53, P. albensis ASCUSBF41, C. furentiruminis ASCUSBF65, B. xylanisolvens ASCUSBF52, C. beijerinckii ASCUSBF26, and Syntrophococcus sp. ASCUSBF60. The vertical lines separate the experimental periods, from left to right, acclimation (no microbe supplementation), pre-challenge dosing 1, grain challenge 1, wash-out (no microbe supplementation), pre-challenge dosing 2, grain challenge 2. Symbol “*” and “◎” denote significant (P ≤ 0.05) and trending significant (0.05 < P ≤ 0.1) differences in abundance between CON and NRM, respectively.
Two of the four microorganisms, S. dextrinosolvens and C. furentiruminis can utilize CO2 in the rumen for VFA and lactate production18,19. Numerically lower but not significant rumen dCO2 was observed to be reduced in the Treatment animals in this study (Table 3). Due to the heterogeneity of rumen fluid, a larger sample size is likely needed to detect significance for rumen dCO2. This observation is consistent with the hypothesis and observations of Laporte-Uribe (2016 & 2019)10,11 that rumen dCO2 accumulation can exacerbate rumen acidosis and microbial activities that reduce the dCO2 in rumen fluid can alleviate metabolic stress. Promoting the growth of rumen microorganisms that can utilize dCO2 in a reduced pH environment could minimize the effects of rumen acidosis and potentially improve animal performance. B. xylanisolvens can degrade xylan under a wide pH range25,73,74 observed that P. albensis was highly abundant in cases of alfalfa-induced SARA that did not elicit a host immune response. These studies corroborate observations in the current study and suggest that both B. xylanisolvens and P. albensis likely assist in stabilizing rumen function under reduced rumen pH.
The effect of NRM on the rumen microbiome
It is reported that the abundance of microorganisms in rumen microbiome does not consistently reflect their significance in the microbial community. Instead, the functional outcomes of the microbiome are defined by the compositional state of individual microorganism14. Correlation analysis can be used to identify potential relationships and influences of microorganisms in a community. Furthermore, these potential relationships can be associated with microbiome stability and functional resilience59. Hence, while B. xylanisolvens, C. beijerinckii, P. albensis, and Syntrophococcus sp. were more abundant in the CON group, the administration of these NRM may exert influences on the rumen microbiome, extending beyond mere abundance measurements.
Given the unique roles of the administered NRM, it is plausible that these microorganisms can increase the production of substrates (e.g., simple saccharides, VFA) or other types of small molecules that can be used by other members of the microbial community and increase the overall rumen microbiome connectivity and stability. In this study, the stability of the core rumen microbiome while undergoing and not undergoing daily NRM administration was evaluated based on individual microbial connectivity (node degree) and the influence of particular microorganisms in the community (betweenness centrality). The core microbiome correlation revealed that the rumen microbiome of animals that were being administered NRM contained more highly connected zOTUs (node degree) and for zOTUs with the same degree of connectivity, greater influences (betweenness centrality) were observed in the NRM group (Fig. 4A). The relationship between node degree and betweenness centrality was significantly strengthened (40.2 vs. 59.7 for CON vs. NRM, respectively; P < 0.001), suggesting that NRM administration improved the overall rumen microbiome stability of beef cattle that underwent a grain challenge.
The stability of CON (no microbial administration) and NRM (with microbial administration) rumen microbiome measured based on microbial network connectivity. (A) The number of edges connected to a node (Node Degree) and the amount of influence a node has (Betweenness Centrality) of all significant correlations identified in control and treatment group. Each point (including grey colored points) represents a node (i.e., a core zOTU) in CON (l) and NRM (p) group. The regression lines were fitted using LOESS for CON (dashed line) and NRM (solid line) group separately. The zOTUs matched to administered NRM were highlighted in colors. (B) The changes in impact of NRM species in rumen microbiome upon the supplementation of NRM strains, shown as the difference between CON (l) and NRM (p) group. Each zOTU matched to NRM species is shown on the y-axis. X-axis are the one-dimensional impact number calculated from “Node Degree” and “Betweenness Centrality”. The connectivity identified in control group and treatment group are shown in circular and triangle points, respectively. Solid lines represent the difference between CON and NRM group. Dash lines indicate the zOTU with significant impact was found in NRM group only.
The network analysis also revealed that five out of six NRM strains, B. xylanisolvens ASCUSBF52, C. furentiruminis ASCUSBF65, C. beijerinckii ASCUSBF26, P. albensis ASCUSBF41, and Syntrophococcus sp. ASCUSBF60, became more impactful upon the administration of NRM, despite four of which were less abundant in the NRM group than the CON group (Fig. 4B). P. albensis ASCUSBF41 and C. furentiruminis ASCUSBF65 were the two most impactful rumen bacterial species. S. dextrinosolven ASCUSBF53 was not detected in the network due to its brief presence during pre-challenge dosing period 1, implying a potential pivotal role in the early stages of microbiome transition induced by grain feeding, though its significance may diminish as animals transition into a grain-based diet.
Besides the administered NRM strains, sixteen zOTUs with betweenness centrality greater than 550 were found to be highly impactful (I > 145) in animal’s core microbiome while being administered NRM. All these zOTUs were present in the core microbiome of heifers not receiving NRM, albeit with impact scores below 105. Five of these were identified as S. dextrinosolvens, one as unclassified Prevotella, and the remaining ten zOTUs were members of unclassified Clostridia or Clostridiales. Further examination using BLAST against indexed nucleotides75,76 revealed that these microorganisms may contribute to three rumen niche environments. The S. dextrinosolvens zOTUs were similar to those found in grain fed beef cattle or calves, suggesting a strong association with rumen fermentation conditions induced by grains77,78. This is also consistent with our hypothesis that the succinate pathway may play an important role in dCO2 removal and lactate to propionate conversion in NRM group. The unclassified Clostridia or Clostridiales zOTUs were similar to sequences of rumen epithelial associated microorganisms79,80. Members of Clostria and Clostridiales are known butyrate producers and butyrate plays an important role in rumen epithelial development70,81,82. Lastly, the unclassified Prevotella zOTU was similar to sequences of uncultured Prevotella bacterium that were isolated from the rumen content of sheep and bovine (GenBank ID: LT975539.1, KC338612.1) of two unpublished studies. While no detailed information from literature is available on this specific unclassified Prevotella, Rodriguez (2003)83 has identified strains of Prevotella that consume lactate and produce succinate and propionate. This is in line with the observed increase on impact score of the unclassified Prevotella zOTU while cattle were administered NRM and further supports that members of Prevotella may play a protective role in rumen acidosis25. These findings suggest that the above-mentioned 3 groups of rumen microorganisms may play important roles in alleviating dCO2 and promoting microbe-host interaction via VFA production in rumen.
Conclusion
The current study employed a 2 × 2 crossover design to evaluate the influence of NRM on the beef cattle rumen microbiome, particularly under a diet-induced grain challenge. It was found that NRM supplementation increased rumen microbial connectivity and centrality and improved animal performance, suggesting that NRM enhanced rumen microbiome stability in the presence of highly fermentable dietary ingredients. Supporting existing literature where lactate accumulation may not always be the etiology of ruminal acidosis in all cases, the current study also finds that rumen lactate accumulation may not be the primary cause of rumen acidosis. Instead, rumen lactate may be fueling the production of propionate through the succinate pathway and contributing to animal weight gain. However, additional experiments are necessary to support this hypothesis. In conclusion, while additional studies are needed to further understand the roles of these NRM on rumen fermentation and their influences on beef cattle physiology, NRM supplementation may be a consistent and effective way to improve feedlot cattle production.
Data availability
The data from this study are available at the NCBI Sequence Read Archive, accession number PRJNA1173199.
References
Kleen, J. L., Hooijer, G. A., Rehage, J. & Noordhuizen, J. P. T. M. Subacute Ruminal acidosis (SARA): a review. J. Vet. Med. Ser. A 50, 406–414 (2003).
Nagaraja, T. G. & Titgemeyer, E. C. Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J. Dairy Sci. 90, E17–E38 (2007).
Gruninger, R. J., Ribeiro, G. O., Cameron, A. & McAllister, T. A. Invited review: application of meta-omics to understand the dynamic nature of the rumen microbiome and how it responds to diet in ruminants. Animal 13, 1843–1854 (2019).
Henderson, G. et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5, 14567 (2015).
Petri, R. M. et al. Characterization of the core Rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE 8, e83424 (2013).
Mccann, J. C., Wickersham, T. A. & Loor, J. J. High-throughput methods redefine the Rumen microbiome and its relationship with nutrition and metabolism. Bioinforma Biol. Insights 8, (2014).
Clemmons, B. A. et al. Temporal stability of the Ruminal bacterial communities in beef steers. Sci. Rep. 9, 9522 (2019).
Stecher, B., Maier, L. & Hardt, W. D. Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 11, 277–284 (2013).
Calsamiglia, S., Blanch, M., Ferret, A. & Moya, D. Is subacute ruminal acidosis a pH related problem? Causes and tools for its control. Anim. Feed Sci. Technol. 172, 42–50 (2012).
Laporte-Uribe, J. A. The role of dissolved carbon dioxide in both the decline in Rumen pH and nutritional diseases in ruminants. Anim. Feed Sci. Technol. 219, 268–279 (2016).
Laporte-Uribe, J. A. Rumen CO2 species equilibrium might influence performance and be a factor in the pathogenesis of subacute ruminal acidosis. Transl. Anim. Sci. 3, 1081–1098 (2019).
Myer, P. R., Smith, T. P. L., Wells, J. E., Kuehn, L. A. & Freetly, H. C. Rumen Microbiome from steers differing in feed efficiency. PLoS ONE 10, e0129174 (2015).
Myer, P. R., Freetly, H. C., Wells, J. E., Smith, T. P. L. & Kuehn, L. A. Analysis of the gut bacterial communities in beef cattle and their association with feed intake, growth, and efficiency1,2,3. J. Anim. Sci. 95, 3215–3224 (2017).
Mizrahi, I., Wallace, R. J. & Moraïs, S. The rumen microbiome: balancing food security and environmental impacts. Nat. Rev. Microbiol. 19, 553–566 (2021).
Clemmons, B. A., Voy, B. H. & Myer, P. R. Altering the gut microbiome of cattle: considerations of host-microbiome interactions for persistent microbiome manipulation. Microb. Ecol. 77, 523–536 (2019).
Zengler, K. & EMBREE, M. Methods, apparatuses, and systems for analyzing microorganism strains from complex heterogeneous communities, predicting and identifying functional relationships and interactions thereof, and selecting and synthesizing microbial ensembles based thereon. (2017).
Dehority, B. A. Carbon dioxide requirement of various species of Rumen bacteria. J. Bacteriol. 105, 70–76 (1971).
O’Herrin, S. M. & Kenealy, W. R. Glucose and carbon dioxide metabolism by Succinivibrio dextrinosolvens. Appl. Environ. Microbiol. 59, 748–755 (1993).
Bryant, M. P. Succinivibrio. Bergey’s Manual of Systematics of Archaea and Bacteria (ed Whitman, W. B.) 1–3. https://doi.org/10.1002/9781118960608.gbm01087. (Wiley, 2015).
Samuelov, N. S., Lamed, R., Lowe, S. & Zeikus, J. G. Influence of CO 2 -HCO 3 – levels and pH on growth, Succinate production, and enzyme activities of Anaerobiospirillum succiniciproducens. Appl. Environ. Microbiol. 57, 3013–3019 (1991).
Song, H. et al. Effects of dissolved CO2 levels on the growth ofMannheimia succiniciproducens and succinic acid production. Biotechnol. Bioeng. 98, 1296–1304 (2007).
Avgustin, G., Wallace, R. J. & Flint, H. J. Phenotypic diversity among Ruminal isolates of Prevotella ruminicola: proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int. J. Syst. Bacteriol. 47, 284–288 (1997).
Matsui, H. et al. Phenotypic characterization of polysaccharidases produced by four Prevotella type strains. Curr. Microbiol. 41, 45–49 (2000).
Bandarupalli, V. V. K. & St-Pierre, B. Identification of a candidate starch utilizing strain of Prevotella albensis from Bovine Rumen. Microorganisms 8, 2005 (2020).
Khafipour, E., Li, S., Plaizier, J. C. & Krause, D. O. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl. Environ. Microbiol. 75, 7115–7124 (2009).
Chassard, C., Delmas, E., Lawson, P. A. & Bernalier-Donadille, A. Bacteroides xylanisolvens sp. nov., a xylan-degrading bacterium isolated from human faeces. Int. J. Syst. Evol. Microbiol. 58, 1008–1013 (2008).
Gomez-Flores, M., Nakhla, G. & Hafez, H. Hydrogen production and microbial kinetics of Clostridium termitidis in mono-culture and co-culture with Clostridium beijerinckii on cellulose. AMB Express 7, 84 (2017).
Dickerson, A. M., Yang, F., Green, H. B., Embree, M. M. & Drackley, J. K. Feeding native rumen microbial supplements increases energy-corrected milk production and feed efficiency by Holstein cows. JDS Commun. 3, 239–244 (2022).
Ragsdale, S. W. Enzymology of the wood–Ljungdahl pathway of acetogenesis. Ann. N. Y. Acad. Sci. 1125, 129–136 (2008).
Falk, M., Münger, A. & Dohme-Meier, F. Technical aote: A comparison of reticular and ruminal pH monitored continuously with 2 measurement systems at different weeks of early lactation. J. Dairy Sci. 99, 1951–1955 (2016).
Silberberg, M., Mialon, M. M., Meunier, B. & Veissier, I. Sensor-captured modifications in cow behaviour under subacute ruminal acidosis. Anim. Open Space 3, 100063 (2024).
Erwin, E. S., Marco, G. J. & Emery, E. M. Volatile fatty acid analyses of blood and Rumen fluid by gas chromatography. J. Dairy Sci. 44, 1768–1771 (1961).
Weinroth, M. D. et al. Considerations and best practices in animal science 16S ribosomal RNA gene sequencing microbiome studies. J. Anim. Sci. 100, skab346 (2022).
Muyzer, G., De Waal, E. C. & Uitterlinden, A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700 (1993).
Berglund, D. L., Taffs, R. E. & Robertson, N. P. A rapid analytical technique for flow cytometric analysis of cell viability using calcofluor white M2R. Cytometry 8, 421–426 (1987).
Hills, M., Kenward, G., Chapman & Hall Design and analysis of cross-over trials. Byron Jones Michael and No. of pages: 340. Price: £D27.50. Stat. Med. 9, 1007–1007 (1990).
Grizzle, J. E. The two-period change-over design and its use in clinical trials. Biometrics 21, 467 (1965).
Wellek, S. & Blettner, M. On the proper use of the crossover design in clinical trials. Dtsch. Ärztebl Int. https://doi.org/10.3238/arztebl.2012.0276 (2012).
Edgar, R. Usearch.https://www.osti.gov/sciencecinema/biblio/1137186 (2010).
Edgar, R. C. & Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482 (2015).
Edgar, R. C. SINTAX: a simple non-bayesian taxonomy classifier for 16S and ITS sequences. https://doi.org/10.1101/074161 (2016).
Cole, J. R. et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2014).
Hernández, J., Benedito, J. L., Abuelo, A. & Castillo, C. Ruminal acidosis in feedlot: from aetiology to prevention. Sci. World J. 2014, 1–8 (2014).
Steele, M. A. et al. Bovine rumen epithelium undergoes rapid structural adaptations during grain-induced subacute ruminal acidosis. Am. J. Physiol. Regul Integr. Comp. Physiol. 300, R1515–R1523 (2011).
Bevans, D. W., Beauchemin, K. A., Schwartzkopf-Genswein, K. S., McKinnon, J. J. & McAllister, T. A. Effect of rapid or gradual grain adaptation on subacute acidosis and feed intake by feedlot cattle1,2. J. Anim. Sci. 83, 1116–1132 (2005).
Chiquette, J. Evaluation of the protective effect of probiotics fed to dairy cows during a subacute ruminal acidosis challenge. Anim. Feed Sci. Technol. 153, 278–291 (2009).
Goto, H. et al. Effects of a bacterial probiotic on ruminal pH and volatile fatty acids during subacute ruminal acidosis (SARA) in cattle. J. Vet. Med. Sci. 78, 1595–1600 (2016).
Mansilla, F. I. et al. Effect of probiotics on the growth, blood profile, and nutritional-metabolic profile of feedlot cattle - Academia.edu.
Mackenzie, D. D. S. Production and utilization of lactic acid by the ruminant. A review. J. Dairy Sci. 50, 1772–1786 (1967).
Minuti, A. et al. Experimental acute rumen acidosis in sheep: consequences on clinical, rumen, and gastrointestinal permeability conditions and blood chemistry1. J. Anim. Sci. 92, 3966–3977 (2014).
Brown, M. S. et al. Evaluation of models of acute and subacute acidosis on dry matter intake, ruminal fermentation, blood chemistry, and endocrine profiles of beef steers. J. Anim. Sci. 78, 3155 (2000).
Antanaitis, R., Juozaitienė, V., Malašauskienė, D. & Televičius, M. Can rumination time and some blood biochemical parameters be used as biomarkers for the diagnosis of subclinical acidosis and subclinical ketosis? Vet. Anim. Sci. 8, 100077 (2019).
Pereira, I. C. et al. Voluntary daily fluctuation in dry matter intake is associated to feedlot performance, feeding behavior and rumen morphometrics in beef cattle. Livest. Sci. 250, 104565 (2021).
De Nardi, R. et al. Blood parameters modification at different ruminal acidosis conditions. Agric. Conspec. Sci. 78, 259–262 (2013).
Messina, Z. & Patrick, H. Partial pressure of carbon dioxide. In StatPearls (StatPearls Publishing, Treasure Island (FL), 2024).
Wahrmund, J. L. et al. Ruminal acidosis challenge impact on ruminal temperature in feedlot cattle1. J. Anim. Sci. 90, 2794–2801 (2012).
Sato, S. Pathophysiological evaluation of subacute ruminal acidosis (SARA) by continuous ruminal pH monitoring. Anim. Sci. J. 87, 168–177 (2016).
Wittebolle, L. et al. Initial community evenness favours functionality under selective stress. Nature 458, 623–626 (2009).
Shade, A. et al. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3, (2012).
Dijkstra, J., Boer, H., Van Bruchem, J., Bruining, M. & Tamminga, S. Absorption of volatile fatty acids from the rumen of lactating dairy cows as influenced by volatile fatty acid concentration, pH and rumen liquid volume. Br. J. Nutr. 69, 385–396 (1993).
Dijkstra, J. Production and absorption of volatile fatty acids in the rumen. Livest. Prod. Sci. 39, 61–69 (1994).
Clemmons, B. A. et al. Blood parameters associated with residual feed intake in beef heifers. BMC Res. Notes 16, 177 (2023).
He, B., Fan, Y. & Wang, H. Lactate uptake in the rumen and its contributions to subacute rumen acidosis of goats induced by high-grain diets. Front. Vet. Sci. 9, 964027 (2022).
Wang, L., Zhang, G., Li, Y. & Zhang, Y. Effects of high forage/concentrate Diet on volatile fatty acid production and the microorganisms involved in VFA production in cow Rumen. Animals 10, 223 (2020).
Nocek, J. E. Bovine acidosis: implications on laminitis. J. Dairy Sci. 80, 1005–1028 (1997).
Fu, W. & Mathews, A. P. Lactic acid production from lactose by Lactobacillus plantarum: kinetic model and effects of pH, substrate, and oxygen. Biochem. Eng. J. 3, 163–170 (1999).
Tang, J., Wang, X. C., Hu, Y., Zhang, Y. & Li, Y. Effect of pH on lactic acid production from acidogenic fermentation of food waste with different types of inocula. Bioresour. Technol. 224, 544–552 (2017).
Van Gylswyk, N. O. Succiniclasticum ruminis gen. nov., sp. nov., a Ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int. J. Syst. Bacteriol. 45, 297–300 (1995).
Yost, W. M., Young, J. W., Schmidt, S. P. & McGilliard, A. D. Gluconeogenesis in ruminants: Propionic acid production from a high-grain diet fed to cattle. J. Nutr. 107, 2036–2043 (1977).
Reichardt, N. et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 8, 1323–1335 (2014).
Krause, K. M. & Oetzel, G. R. Inducing subacute Ruminal acidosis in lactating dairy cows. J. Dairy Sci. 88, 3633–3639 (2005).
Clemmons, B. A. et al. Rumen fluid metabolomics of beef steers differing in feed efficiency. Metabolomics 16, 23 (2020).
Gong, X., Gruniniger, R. J., Forster, R. J., Teather, R. M. & McAllister, T. A. Biochemical analysis of a highly specific, pH stable xylanase gene identified from a bovine rumen-derived metagenomic library. Appl. Microbiol. Biotechnol. 97, 2423–2431 (2013).
Mirande, C., Mosoni, P., Béra-Maillet, C., Bernalier-Donadille, A. & Forano, E. Characterization of Xyn10A, a highly active xylanase from the human gut bacterium Bacteroides xylanisolvens XB1A. Appl. Microbiol. Biotechnol. 87, 2097–2105 (2010).
Zhang, Z., Schwartz, S., Wagner, L. & Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214 (2000).
Morgulis, A. et al. Database indexing for production MegaBLAST searches. Bioinformatics 24, 1757–1764 (2008).
Durso, L. M. et al. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl. Environ. Microbiol. 76, 4858–4862 (2010).
Fernando, S. C. et al. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 76, 7482–7490 (2010).
Li, M., Zhou, M., Adamowicz, E., Basarab, J. A. & Guan, L. L. Characterization of bovine ruminal epithelial bacterial communities using 16S rRNA sequencing, PCR-DGGE, and qRT-PCR analysis. Vet. Microbiol. 155, 72–80 (2012).
Santos, T. M. A. & Bicalho, R. C. Diversity and succession of bacterial communities in the uterine fluid of Postpartum Metritic, Endometritic and healthy dairy cows. PLoS ONE 7, e53048 (2012).
Malhi, M. et al. Increased papillae growth and enhanced short-chain fatty acid absorption in the rumen of goats are associated with transient increases in cyclin D1 expression after ruminal butyrate infusion. J. Dairy. Sci. 96, 7603–7616 (2013).
Shen, H., Xu, Z., Shen, Z. & Lu, Z. The regulation of Ruminal short-chain fatty acids on the functions of Rumen barriers. Front. Physiol. 10, 1305 (2019).
Rodriguez, F. Control of lactate accumulation in ruminants using Prevotella bryantii. (Iowa State University, United States – Iowa).
Acknowledgements
This project was funded by Native Microbials. Inc. The authors would like to thank Brandon Beavers and the staff at the East Tennessee Research and Education Center for their work and support throughout this project.
Author information
Authors and Affiliations
Contributions
PRM and MME contributed to the conception of the study. All authors contributed with design, data acquisition, interpretation of data, data analyses, and preparation of manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
MTH, EAM, BAC, TBA, PM, JFC, DEA, and PRM have no conflicts of interest to declare. FY, ASI, JRG, CM, MLS, JJE, and MME are employed by Native Microbials, Inc, the company that supported this experiment.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, F., Henniger, M.T., Izzo, A.S. et al. Performance improvements and increased ruminal microbial interactions in Angus heifers via supplementation with native rumen bacteria during high-grain challenge. Sci Rep 15, 2289 (2025). https://doi.org/10.1038/s41598-025-86331-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-86331-1