Abstract
The resurgence of COVID-19 and the rise in severe outcomes emphasize the need for reliable prognostic markers to guide patient care and optimize ICU and hospital resources. This study investigates the potential of nasopharyngeal swabs to identify biomarkers that predict ICU admission or death in hospitalized COVID-19 patients. We analyzed nasopharyngeal exudates from 95 hospitalized patients in 2020 using high-plex RNA quantification on the NanoString® nCounter platform. Comparative analysis identified four genes, with KLRB1 (Killer cell lectin like receptor B1) (Odds Ratio OR 0.5, 95% CI: 0.27–0.96), along with age (OR 3.3, 95% CI: 1.25–8.93) emerging as independent prognostic markers in multivariate analysis. These findings were validated using qRT-PCR in an independent cohort of 168 patients hospitalized in 2022. While univariate analysis identified a significant association between KLRB1 expression and vaccination status (p < 0.05), only low KLRB1 expression (OR 1.135, 95% CI: 1.0-1.280), and age (OR 1.033, 95% CI: 1.006–1.061) were confirmed as independent risk factors for ICU admission or death, regardless of other studied variables such as comorbidities, vaccination status, or smoking habits. Our findings suggest that KLRB1 expression could improve prognostic tools by identifying patients at higher risk upon admission. Incorporating KLRB1 into multiplex diagnostic kits alongside SARS-CoV-2 detection could streamline prognostic assessment, providing a more comprehensive and efficient approach to patient management.
Similar content being viewed by others
Introduction
With an estimated 777 million reported cases and more than 7.1 million deaths as of December 2024, the Coronavirus Disease 2019 (COVID-19) pandemic has posed unprecedented health, social, and economic challenges worldwide. The disease manifests with great heterogeneity, ranging from mild (40%) to moderate symptoms (40%), to severe illness requiring oxygen support (15%) or critical disease (5%)1. Complications such as multiorgan failure and acute respiratory distress syndrome (ARDS), often triggered by the downregulation of natural killer (NK) cell numbers and function and elevated levels of pro-inflammatory cytokines (or cytokine storm syndrome), significantly contribute to patient mortality2,3,4. Although vaccination has had a substantial health impact by significantly reducing COVID-19 deaths5, challenges remain due to immune evasion by new strains and the waning of post-vaccine immunity, which promote periodic resurgences in infections and impose economic strains from prolonged hospitalizations and specialized care services6.
Identifying patients at risk of rapid progression and severe outcomes within hours is crucial for effective management and improved health outcomes. As patients with varying disease severity levels require different clinical management approaches, reliable prognostic markers are essential for ensuring the efficient allocation of medical resources and determining the appropriate level of care, such as hospitalization for moderate-to-severe cases and timely transfer to an intensive care unit (ICU) for the most critical patients. However, despite advancements, precise patient stratification at the time of admission remains challenging7.
Since the onset of the pandemic, different demographic and clinical variables have been identified as risk factors for severe COVID-19. Extensive evidence shows that older individuals, particularly those with pre-existing respiratory conditions and comorbidities, are at a heightened risk of adverse outcomes such as ICU admission and mortality8,9,10. Other factors, such as smoking habits and biological sex, have also been linked to severe infection11, although their reliability as standalone markers is insufficient to inform clinical decisions.
Regarding biological markers, different genetic variants have been associated with severe infection11, although their validation as prognostic biomarkers remains controversial. To support early triage, our group previously developed a machine-learning-based model integrating demographic data and blood parameters, including oxygen saturation and inflammation markers, to predict the risk of progressing to a score of 5 or higher on the WHO Clinical Progression Scale before requiring mechanical ventilation12. In recent years, inflammation biomarkers like C-reactive protein (CRP), interleukin-6 (IL-6), red cell distribution width (RDW), D-dimer, ferritin, and neutrophil-to-lymphocyte ratio (NLR) have been incorporated into clinical practice for stratifying patients based on expected disease progression. However, while these biomarkers show high sensitivity, their specificity is limited, as they can be elevated in various pathological conditions7. On the other hand, these biomarkers require blood sampling after patient admission and the confirmation of acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, followed by processing and analysis, which can delay critical decision-making.
Given the airborne transmission of SARS-CoV-2 and its primary impact on the mucosa of the upper respiratory tract, research has highlighted the role of local mucosal immunity in controlling viral spread within the nasopharynx13. Accordingly, our study focuses on the analysis of non-invasively obtained, widely available samples, such as surplus diagnostic nasopharyngeal swabs from hospitalized COVID-19 patients, to identify measurable, accurate, and reproducible biomarkers. Specifically, our aim was to identify biomarkers that can be analyzed concurrently with SARS-CoV-2 detection via reverse transcription polymerase chain reaction (qRT-PCR), the gold standard for early-stage COVID-19 diagnosis, to assist in the early prediction of disease progression and outcomes.
Gene expression analysis measures the activity levels of specific genes within a sample, providing critical insights into the biological pathways active in different conditions. Transcriptomics, the study of RNA molecules transcribed from DNA, reveals how cells respond to stimuli, such as viral infections, by regulating the production of mRNA transcripts.
Outputs from gene expression assays, including fold changes indicating upregulation or downregulation of specific genes, normalized expression values, and differential expression patterns between sample groups are used to infer biological pathways, identify disease markers, or assess treatment responses.
In this study, we employed two complementary methods for gene expression profiling: the NanoString nCounter ® platform, a high-throughput, fluorescence-based technology that provides quantitative and reproducible measurements of gene expression, even in challenging samples with degraded RNA, uses color-coded molecular barcodes to directly count mRNA transcripts, and provides reliable results without amplification bias. Additionally, qRT-PCR was used for validation, measuring cycle threshold (Ct) values, which are inversely proportional to RNA levels.
Gene expression analysis of nasopharyngeal exudates presents significant challenges, particularly because RNA from these samples is typically more degraded than that from other sources, such as blood. NanoString nCounter®technology, a high-throughput, multiplex, fluorescence-based digital hybridization method, is specifically designed for precise gene expression analysis in samples where RNA is substantially fragmented and degraded, delivering reliable data and facilitating the discovery of novel gene expression profiles14,15,16.
After optimizing the necessary protocol to analyze these complex samples17, we proceeded to identify potential genes related to immune and inflammatory responses in nasopharyngeal exudates from COVID-19 hospitalized patients, focusing on genes with differential expression between patients who were discharged and those who were admitted to the intensive care unit or succumbed to the disease.
Results
Discovery cohort
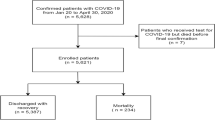
The discovery cohort included 95 hospitalized COVID-19 patients, representative of the affected population during the acute phase of the pandemic from March 7, 2020, to October 15, 2020. The average age of the patients was 62.9 years, with a slight predominance of females (68.4%). Upon admission, 43.2% of patients exhibited an increased oxygen demand (categorized as level 1: nasal cannula, level 2: Venturi mask; level 3: high-flow nasal cannula, level 4: non-invasive mechanical ventilation, level 5: invasive mechanical ventilation). On these patients, 71.6% were discharged without requiring ICU admission, while 28.4% experienced severe outcomes, including ICU admission or death. The socio-demographic and clinical characteristics of the discovery cohort are summarized in Table 1.
Data analysis using nSolver™ 4.0 revealed significant differences in gene expression between patients who required intensive care and those who were discharged without needing ICU support. Among the identified genes, KLRAP1 (Killer Cell Lectin Like Receptor A1) and KLRB1 (Killer Cell Lectin Like Receptor B1, CD161) exhibited significantly higher expression in patients discharged without requiring ICU admission, with Log Fold change (LogFC) values of 2.24 and 1.98, respectively (p < 0.001 for both). Conversely, IRF8 (Interferon Regulatory Factor 8) and TCF4 (Transcription Factor 4) showed significantly lower expression in these patients, as evidenced by LogFC values of −1.79 and − 1.59, respectively (p < 0.001 for both) (Table 2).
Following the individual gene analysis performed using nSolver™, we conducted a comprehensive pathway-based analysis through Gene Set Enrichment Analysis (GSEA) on patients from both groups. The results from GSEA revealed significant enrichment in the “regulation of natural killer (NK) cell-mediated immunity” pathway, which ranked among the top 20 up- and down-regulated pathways (Enrichment Score > 0.4, adjusted p-value < 0.05). This pathway-level analysis highlighted broader functional shifts that may not be captured by individual gene changes alone, emphasizing the coordinated regulation of NK cell-mediated immune responses and suggesting that alterations in NK cell activity may play a crucial role in disease progression (Fig. 1).
Gene Set Enrichment Analysis plot illustrating the enrichment of the gene set “Regulation of Natural Killer Cell Mediated Immunity” in the context of COVID-19 outcomes. The green curve represents the running enrichment score (ES), which reflects the degree to which the gene set is overrepresented at the top of the ranked gene list. A positive ES (ES > 0.4) indicates significant enrichment of these genes at the top of the list, suggesting their upregulation. The vertical black lines correspond to the positions of individual genes from the gene set in the ranked list. The concentration of these lines near the top indicates a strong gene set enrichment, suggesting an important role for natural killer cell regulation in the immune response of the studied patients.
In patients who were discharged without requiring ICU admission, GSEA revealed the upregulation of different genes including KLRAP, KLRB1, CXCL12 (C-X-C Motif Chemokine Ligand 12, a chemoattractant active on T-lymphocytes and monocytes), ZBTB16 (Zinc Finger and BTB Domain Containing 16, related to MHC-1 mediated antigen processing and presentation), TNFRSF11A (TNF Receptor Superfamily Member 11a, a regulator of interactions between T cells and dendritic cells), and IRAK2 (Interleukin 1 Receptor Associated Kinase 2, involved in IL-1 induced upregulation of NF-kappa B).
Conversely, in patients with severe COVID-19, downregulation was observed in interferon regulatory factors (IRF-8, IRF-7 and IRF-5, which are essential for coordinating the innate immune response), TCF4 (T-cell-specific transcription factor 4, involved in dendritic cell programming), ITGAX (Integrin Subunit Alpha X, involved in Th17 differentiation), BATF3 (Basic leucine zipper ATF-like transcription factor 3, with a role in repression of interleukin-2 and matrix metalloprotease-1 transcription), POLR2A (RNA polymerase II subunit A), SERPING1 (Serpin family G member 1, involved in the regulation of the complement cascade), CD3EAP (DNA-directed RNA polymerase I subunit RPA34), AICDA (Activation Induced Cytidine Deaminase) as well as components of the IL-1 receptor complex such as IL1RAP (Interleukin 1 Receptor Accessory Protein, necessary for early Th17 programming).
The volcano plot comparing patients who experienced severe outcomes with those who were discharged, highlights the differentially expressed genes between the study groups (Fig. 2).
Volcano plot depicting gene expression differences between COVID-19 patients with favorable outcomes (discharged without ICU admission) and adverse outcomes (requiring ICU admission or death). Log2 Fold Change (x-axis) indicates upregulation in favorable (positive) or adverse (negative) outcomes, while -log10(p-value) (y-axis) represents statistical significance. Relative to patients with favorable outcomes, significant genes are highlighted: upregulated (red), downregulated (green), and non-significant (gray).
After multivariate logistic regression analysis, only KLRB1 (p < 0.05, Odds Ratio OR 0.5, 95% CI: 0.27–0.96) and age (p < 0.02, OR 3.3, 95% CI: 1.25–8.93) were identified as independent prognostic markers for ICU admission or death, irrespective of sex, comorbidities, smoking habits or increased oxygen demand.
Validation cohort
The validation of these findings in a second cohort of 168 hospitalized patients recruited from August 2021 to November 2021 reinforced the importance of KLRB1 as a prognostic marker. Samples from this cohort were obtained and processed as previously described for the discovery cohort, but analyzed using qRT-PCR, with GAPDH as the housekeeping gene. Table 3 summarizes the socio-demographic and clinical characteristics, along with normalized KLRB1 expression (∆Ct) measured by qRT-PCR in these patients.
Regression analysis showed no significant correlation between age and KLRB1 expression, with Pearson’s Correlation Coefficient (PCC) = 0.202 (p < 0.05). A two-tailed t-test revealed significant associations between KLRB1 and vaccination status (p < 0.05) as well as ICU admittance or death (p < 0.05), but no significant association was observed with the remaining studied variables (Table 4).
Subsequent multivariate logistic regression analysis revealed that low KLRB1 expression, as indicated by a high ∆Ct value (Odds Ratio OR 1.135, 95% CI: 1.0–1.280), and older age (OR 1.034, 95% CI: 1.007–1.061), are independently associated with an increased risk of ICU admission or death (Table 5).
Discussion
Since the onset of the pandemic, different blood markers have been proposed as prognostic indicators for COVID-19, to assist in monitoring hospitalized patients18,19,20,21. Despite their utility, blood tests have limitations such as invasiveness, the need for specialized personnel and equipment, higher costs, and longer processing times compared to nasopharyngeal swabs. Moreover, blood samples may not accurately reflect the localized immune response in the respiratory tract, potentially limiting the effectiveness of blood-based biomarkers in predicting disease progression and outcomes specific to respiratory infections. We hypothesized that integrating a prognostic biomarker into nasopharyngeal swabs, a non-invasive test already recommended as the gold standard for COVID-19 diagnosis, could streamline prognostic assessment and provide a more efficient testing protocol.
Unlike previous studies focused on host response gene expression using shotgun RNAsequencing of nasopharyngeal swabs22, our study is the first to use the NanoString nCounter® platform for high-plex digital quantification of mRNA in diagnostic surplus samples from hospitalized COVID-19 patients. This approach led us to identify low KLRB1 (Killer Cell Lectin Like Receptor B1, CD161) RNA expression as a potential risk biomarker for adverse outcomes, such as ICU or mortality. Our exploratory study was based on samples collected during the early stages of the pandemic, before the introduction of vaccines and when variants such as A, B, B.1.177, Alpha (B.1.1.7), Beta (B.1.351) and Delta(P.1.617.2) were prevalent23.
To potentially translate our findings into clinical practice, we validated our results in an independent cohort of patients from 2022, during the prevalence of the Omicron variant. These samples were analyzed using conventional qRT-PCR, a technique renowned for its sensitivity, specificity, and reliability in quantifying gene expression, which facilitates the integration of KLRB1as a target in multiplex diagnostic kits to enable simultaneous assessment of a patient’s risk of deterioration and confirmation of SARS-CoV-2 infection24.
In this validation cohort we could control for potential confounding factors such as vaccination status, smoking, which is known to increase susceptibility to respiratory viruses25, as well as comorbidities and age, all known risk factors for severe COVID-19 outcomes26. Interestingly, the univariate analysis revealed a significant association between KLRB1 expression and vaccination status, with non-vaccinated individuals showing higher KLRB1 expression compared to vaccinated individuals, a finding suggesting that the adaptive immune response in the nasopharyngeal mucosa induced by vaccination differs from the innate response observed in non-vaccinated patients, warranting further investigation. In any case, multivariate analysis confirmed KLRB1 RNA expression and age as the only independent variables.
The association between low KLRB1 expression and adverse outcomes was also independent of other variables like smoking habit, medical conditions and sex. Although the prognostic significance of KLRB1 also remained robust across different viral variants, further research is needed to assess how emerging SARS-CoV-2 variants may influence its prognostic value.
Our findings show that elevated KLRB1RNA expression is a protective factor against COVID-19, suggesting a robust immune response in the nasopharyngeal mucosa capable of controlling viral replication and mitigating severe disease outcomes. However, the ongoing debate over the correlation between mRNA levels and protein abundance27 complicates the interpretation of the biological significance of our results. The KLRB1 gene, also known as CD161, CLEC5B, hNKR-PIA, NKR, NKR-P1 or NKR-P1A, encodes the CD161 receptor, a C-type lectin-like type-II transmembrane glycoprotein predominantly expressed on lymphocytes in both the innate and adaptive immune systems28. Consistent with the results of Gene Set Enrichment Analysis (GSEA), CD161 is particularly abundant on natural killer (NK) cells, marking a pro-inflammatory subset involved in early protective and inflammatory responses during infections and acknowledged within the immune cell population of the nasal mucosa for effectively limiting SARS-CoV-2 infection29,30,31.
Recent evidence suggests that SARS-CoV-2 can impair NK cell function11,32by upregulating the inhibitory ligand Lectin-Like-Transcript 1 (LLT1) on target cells31,33. This may lead to a reduction in both the proportion of plasma CD161 + NK cells and CD161 expression levels, along with impaired NK cell cytotoxicity31and decreased cytolytic activity34, potentially exacerbating the inflammatory response and contributing to the cytokine storm observed in severe COVID-19 cases35. Although the Human Immunology V2 CSO panel used in this study did not include the CLEC2D gene encoding the LLT-1 ligand, our findings demonstrate a clear association between elevated KLRB1 levels in the nasopharyngeal mucosa and favorable clinical outcomes, consistent with its protective role.
The dual role of CD161 as both an inhibitory receptor on NK cells and a co-activating factor influencing T-cell receptor-dependent responses28,36,37also suggests that the nasopharyngeal mucosa may be colonized by different immune cells as a result of an appropriate adaptive response. This is consistent with our GSEA findings, which revealed the upregulation of critical genes involved in T-cell activation, antigen presentation, and chemotaxis in patients who did not require ICU admission, indicating a robust immune response. In contrast, patients with severe outcomes showed significant downregulation of regulatory genes related to antiviral responses and T-cell differentiation, suggesting a compromised immune defense29,41.
The nasal mucosa, rich in epithelial cells and T cells, serves as a first line of defense against airborne pathogens, housing essential immune components for antiviral activity26,38. The importance of a well-functioning immune response in the nasopharyngeal region is emphasized by the dynamics of nasal immunity during SARS-CoV-2 infection, which involves increased levels of NK cells, granulocytes, and different T cell populations39. Consistent with our findings, the product of KLRB1gene is also abundant in subsets of CD4 + T memory cells, including T helper 17 cells (Th17), Mucosa-Associated Invariant T (MAITs) cells and non-MAIT CD8 + cells, serving as a marker for immune cell activation and response to infection28,40,41,42. Interestingly, several of these cells can also bear the immune inhibitory LLT143.
Indeed, the protective function of CD161 + T cells, particularly in mucosal tissues, is crucial in preventing disease progression in viral infections like HIV44as well as SARS-CoV-242, as evidenced by reduced counts of CD3+, CD4+, and CD8 + T cells in moderate and severe COVID-1945, and the robust activation and adaptive immune responses induced by COVID-19 vaccines46. Moreover, the expansion of tissue-resident cytotoxic CD8 + T cells and CCR6 + CD161 + CD4 + T cells in the nasal mucosa following mRNA COVID-19 vaccination48 further supports our results. Despite the strong association of KLRB1 expression with favorable outcomes in COVID-19, the involvement of this gene in antiviral responses across various infections suggests that it may not be entirely specific to SARS-CoV-2, thus justifying further research to analyze KLRB1 expression across a broader range of infections to clarify its specificity and assess potential cross-reactivity.
On the other hand, while qRT-PCR offers high sensitivity and specificity, reliable detection in low-expression samples requires high-quality specimens and strict protocol adherence. Future research should prioritize integrating KLRB1 into multiplex diagnostic panels and analyzing larger patient cohorts to establish standardized expression ranges, improving data interpretation and supporting informed therapeutic decisions for hospitalized COVID-19 patients.
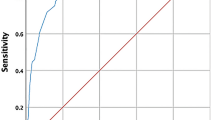
Building on this, our preliminary ROC (Receiver Operating Characteristic) curve analysis, based on the grouping of severe outcomes, demonstrated an AUC (Area Under the Curve) of 0.625 (data not shown), which highlights the potential of KLRB1 as a biomarker for risk stratification and underscores the need for further refinement and validation. Future studies should leverage larger and more diverse patient cohorts, as well as longitudinal designs, to confirm the sensitivity and specificity of KLRB1 as a prognostic marker. Additionally, exploring the integration of KLRB1 within a panel of complementary biomarkers could provide a more comprehensive tool for risk assessment, ultimately aiding in more precise and timely decision-making for COVID-19 patients.
Although our study primarily focused on hospitalized patients and did not address the long-term effects of KLRB1 expression, our results suggest practical implications for clinical care. Early identification of patients with low KLRB1 expression could allow for timely stratification, enabling healthcare providers to intensify monitoring and resource allocation for those at a greater risk of deterioration. The feasibility of detecting KLRB1 using Nanostring Platforms or qRT-PCR supports its integration into routine prognostic assessments.
The patient distribution in our study aligns with the epidemiological data reported by the WHO and previous literature1. Future research could build on our findings by including larger cohorts or focusing on specific subsets of patients, such as those with higher rates of respiratory compromise, to further validate the utility of KLRB1 as a prognostic biomarker in diverse clinical settings.
Methods
Study design
In this prospective observational study, we analyzed the RNA expression from a panel of genes associated with inflammatory and immune responses [NS_Immunology_v2_C2328]. The primary objective was to compare the gene expression patterns between COVID-19 patients who were discharged and those who required ICU admission or deceased.
Patient and samples
The study utilized diagnostic surplus samples for the confirmation of SARS-CoV-2 infection and included nasopharyngeal swabs, sputum or bronchial-aspirates, all obtained within the first 24 h of emergency room admission. Enrollment criteria required hospital admission, age over 18 years, a confirmed diagnosis of SARS-CoV-2 infection via qRT-PCR, the presence of compatible respiratory symptoms, and the provision of informed consent.
The study involved two patient cohorts:
-
Discovery cohort: This cohort consisted of 95 consecutively admitted COVID-19 patients at Costa del Sol University Hospital (Marbella, Spain) in 2020, with SARS-CoV-2 infection confirmed via qRT-PCR. Samples from this cohort were analyzed using the NanoString nCounter system to identify potential prognostic mRNA candidates.
-
Validation cohort: Validation of the candidate markers was performed using qRT-PCR on similar samples collected from an independent cohort of 168 COVID-19 patients admitted to Costa del Sol University Hospital in 2022. This cohort included 87 unvaccinated and 81 vaccinated patients, with confirmed SARS-CoV-2 infection.
Ethical approval
This study received approval from the Costa del Sol Ethics Committee (decision/file number: 003_jul20_PI-IMMU-COVID19 date: July 30, 2020). The study adhered to the Declaration of Helsinki and followed good clinical practice guidelines. Informed consent was obtained from all patients, who were briefed on the study’s objectives, and their diagnostic surpluses were analyzed in accordance with prevailing regulations. Approval for the analysis of diagnostic surplus samples from deceased patients was obtained from the same ethics committee.
Study variables
Patient information
Age at admission, biological sex, oxygen demand increase (level 1: nasal cannula, level 2: Venturi mask; level 3: high-flow nasal cannula, level 4: non-invasive mechanical ventilation, level 5: invasive mechanical ventilation), SARS-CoV-2 vaccination status, comorbidity (more than two pathologies recorded in patient’s medical history: arterial hypertension, obesity, angina, arrhythmia, atrial fibrillation, valvular disease, acute myocardial infarction, heart failure, cerebrovascular disease, peripheral vascular disease, mild chronic lung disease, moderate-severe chronic lung disease, dementia, hemiplegia/paraplegia, other neurological diseases, diabetes mellitus, diabetes mellitus with organ damage, other endocrine diseases, mild liver disease, moderate-severe liver disease, mild renal failure, moderate-severe renal failure, peptic ulcer, gastrointestinal bleeding, inflammatory bowel disease, malignant tumor, lymphoma, leukemia, metastatic tumor, lymphoma, AIDS, rheumatic disease, coagulopathy, history of deep vein thrombosis, pulmonary thromboembolism, obstructive sleep apnea syndrome, asthma, interstitial lung disease), smoking habit.
Outcome variables included hospital discharge, admission to the intensive care unit (ICU), or death during hospitalization. ICU admissions were determined based on the criteria outlined in the Documento Técnico Manejo del COVID-19: Atención Hospitalaria, published in June 2020 by the Spanish Ministry of Health. These criteria required the presence of at least one major criterion (need for invasive mechanical ventilation or shock requiring vasopressors) or three minor criteria (respiratory rate > 30 breaths per minute, PaO2/FiO2 < 250, multilobar infiltrates, confusion/disorientation, uremia with blood urea nitrogen BUN > 20 mg/dl, leukopenia < 4000 cells/mm3, hypothermia with central temperature < 36.8ºC, or hypotension requiring aggressive fluid resuscitation). Additionally, the Sequential Organ Failure Assessment (SOFA) Score was used to assess organ failure and monitor disease progression in cases of severe pneumonia and/or sepsis.
Molecular variables
NanoString nCounter® Human Immunology V2 Panel Gene [NS_Immunology_v2_C2328]: consists of gene-specific molecular barcoded reporter probes and biotin-labeled capture probes, that are analyzed for gene expression quantification using the NanoString nCounter® System.
qRT-PCR output: Relative fold gene expression of nasopharyngeal samples, measured as Ct (cycle threshold).
Methodology
High-throughput RNA analysis
We previously developed a protocol for multiplex gene expression analysis using the NanoString nCounter®platform in combination with a predesigned panel for the study of infectious disease immune response, the Human Immunology V2 panel [NS_Immunology_v2_C2328], which targets 579 genes associated with key pathways in immune and inflammatory responses, alongside 15 internal reference genes for data normalization47. Gene expression data were analyzed to identify differentially expressed genes among patient groups using nSolver TM 4.0 Analysis Software, which calculates the fold changes by comparing the means of log-transformed normalized data between groups. The statistical significance of these fold changes was determined using a two-tailed t-test with unequal variances (Welch’s t-test) and adjusted for multiple comparisons using the False Discovery Rate (FDR) method. The distribution of the t-statistic was estimated using the Welch-Satterthwaite equation to calculate degrees of freedom and determine 95% confidence intervals for the observed differential expression.
The protocol began with viral inactivation of diagnostic surplus samples at 98ºC for 2 min, followed by automated RNA extraction using the Viral Nucleic Acid Extraction Kit, Cartridge Code 203, with the MagCore®robot (Magcore Lamination India PvT. Ltd.). The initial sample volume was 400 µl, with an elution volume of 40 µl. Quality control measures included assessing the quality and concentration of the samples employing a NanoDrop™ spectrophotometer (Thermo Scientific™, Waltham, Massachusetts) to ensure that total RNA concentrations fell within the range of 100–130 ng, with A260/280 and A260/230 absorbance ratios between 1.80 and 2.00. Additionally, RNA concentration and integrity were evaluated using the Agilent 2100 Bioanalyzer (Agilent Scientific Instruments Inc. Santa Clara, California)17. Due to the limited RNA concentration from nasopharyngeal surpluses, we optimized the input concentration from the standard 200 ng/µl to 100 ng/µl. Samples were hybridized 22 h at + 65ºC, and results were obtained using the nCounter Prep Station and Digital Analyzer set at high sensitivity17.
qRT-PCR
For validation, we processed the diagnostic nasopharyngeal swabs from an independent cohort of 168 COVID-19 patients hospitalized in 2022. Virus inactivation and RNA extraction were performed according to the previously established protocol17. Gene expression was then assessed via multiplexed qRT-PCR reactions using the Bio-Rad CFX96 Real-Time PCR System (Bio-Rad Laboratories, Inc, California). After confirming that there were no statistically significant differences in the expression of the proposed housekeeping gene between the study groups (p = 0.61, t: −0.513), GAPDH (Glyceraldehyde-3-Phosphate Dehydrogenase) (PrimeTime Std®IDT Hs.PT.39a.22214836) was selected48,49. KLRB1 was targeted using the TaqMan Assay Hs00174469_m1 (ThermoFisher Scientific). The thermocycling scheme was as follows: 20 min at 50 °C and 15 min at 95 °C, followed by 44 cycles of 15 s at 94 °C and 30 s at 58 °C. For each sample, an amplification curve was obtained, with the cycle threshold (Ct) being inversely proportional to the initial RNA quantity. Data were normalized to the internal control as ΔCt to compare the relative expression levels of the target gene within each sample.
Data analysis
Descriptive analysis involved calculating measures of central tendency and dispersion for quantitative variables, while qualitative variables were summarized using frequency distribution. Bivariate analysis was conducted using Student’s t-test to compare qualitative variables, and Pearson’s correlation coefficient was employed for the quantitative variable of interest.
A multivariate logistic regression model was developed to examine factors associated with ICU admission or death during hospitalization. The primary variable of interest was initially included in the model, and additional independent variables were incorporated progressively. Variables were included in the model if they met the inclusion criterion of statistical significance (p < 0.05), and those that did not meet a higher threshold of significance (p < 0.01) during further testing were excluded. This stepwise approach ensured that only statistically relevant variables were retained, minimizing the potential for overfitting while enhancing the predictive accuracy of the model. Odds Ratio (OR) with 95% Confidence Intervals (95% CI) were reported. All analyses were conducted using SPSS v28 Statistical software.
To overcome the limitations of single-gene analysis, Gene Set Enrichment Analysis (GSEA) was employed to derive biological insights from the mRNA expression data and to identify groups of genes with shared biological functions, chromosomal locations, or regulatory mechanisms50. GSEA was conducted in R/Bioconductor51using the Fast Gene Set Enrichment Analysis (FGSEA)52package and the C5:GOBP collection from MSigDB53 as a reference.
Data availability
Research data that support the findings of this study have been deposited on Mendeley repository: García Aranda, Marilina; Martín García, Desirée; López Rodríguez, Inmaculada; Martínez, Beatriz; Hortas, María Luisa; Barragán-Mallofret, Isabel; Redondo, Maximino (2024), “Digital Quantification of RNA from nasopharyngeal exudates of COVID-19 patients obtained with the Nanostring nCounter platform”, Mendeley Data, V1, doi: 10.17632/xs7dnkffkv.1.
References
Wu, Z. & McGoogan, J. M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. jama, 323(13): pp. 1239–1242. (2020).
Yang, X. et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respiratory Med. 8 (5), 475–481 (2020).
Organization, W. H. Statement on the fifteenth meeting of the IHR (2005) Emergency Committee on the COVID-19 pandemic. [cited 2023 Jun 18, 2023]; (2023). Available from: https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic
Montazersaheb, S. et al. COVID-19 infection: an overview on cytokine storm and related interventions. Virol. J. 19 (1), 92 (2022).
Watson, O. J. et al. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet. Infect. Dis. 22 (9), 1293–1302 (2022).
Tatsis, F. et al. Economic Burden of ICU-Hospitalized COVID-19 patients: A systematic review and Meta-analysis. Cureus 15(7), e41802 (2023).
Rizzi, M. et al. COVID-19 biomarkers at the crossroad between patient stratification and targeted therapy: the role of validated and proposed parameters. Int. J. Mol. Sci., 24(8), 7099 (2023).
Vardavas, C. I. et al. Prognostic factors for mortality, intensive care unit and hospital admission due to SARS-CoV-2: a systematic review and meta-analysis of cohort studies in Europe. Eur. Respiratory Rev., 31(166), 220098 (2022).
Alizadehsani, R. et al. Risk factors prediction, clinical outcomes, and mortality in COVID-19 patients. J. Med. Virol. 93(4), 2307–2320 (2021).
Wolff, D. et al. Risk factors for Covid-19 severity and fatality: A structured literature review. Infection 49, 15–28/s15010-020-01509 (2021).
Ranjan, J., Ravindra, A. & Mishra, B. Gender and genetic factors impacting COVID-19 severity. J. Family Med. Prim. Care. 10(11), 3956–3963 (2021).
Garcia-Gutiérrez, S. et al. Machine learning-based model for prediction of clinical deterioration in hospitalized patients by COVID 19. Sci. Rep. 12 (1), 7097 (2022).
Flemming, A. Poor nasal immunity can lead to severe COVID-19. Nat. Rev. Immunol. 21 (9), 547–547 (2021).
NanosString Technologies, I.Using the nCounter®Analysis System with FFPE Samples for Gene Expression Analysis, inTech Note (2012).
Goytain, A. & Ng, T. NanoString nCounter Technology: High-throughput RNA validation. In Chimeric RNA: Methods and Protocols (eds. Li, H. & Elfman, J.) 125–139 (Springer US, New York, NY, 2020).
Veldman-Jones, M. H. et al. Evaluating robustness and sensitivity of the NanoString technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res. 75 (13), 2587–2593 (2015).
García Aranda, M. et al. Laboratory protocol for the digital multiplexed gene expression analysis of nasopharyngeal swab samples using the NanoString nCounter system. F1000Res 11, p133 (2022).
Kashyap, A. et al. Molecular markers for early stratification of disease severity and progression in COVID-19. Biol. Methods Protoc. 7(1), bpac028 (2022).
Osman, M. et al. Impaired natural killer cell counts and cytolytic activity in patients with severe COVID-19. Blood Adv. 4 (20), 5035–5039 (2020).
Hasan, M. Z. et al. SARS-CoV-2 infection initiates interleukin-17-enriched transcriptional response in different cells from multiple organs. Sci. Rep. 11 (1), 16814 (2021).
Sweet, D. R., Freeman, M. L. & Zidar, D. A. Immunohematologic biomarkers in COVID-19: insights into Pathogenesis, Prognosis, and Prevention. Pathog Immun. 8 (1), 17–50 (2023).
Lieberman, N. A. et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 18 (9), e3000849 (2020).
Troyano-Hernáez, P., Reinosa, R. & Holguín, Á. Evolution of SARS-CoV-2 in Spain during the first two years of the pandemic: circulating variants, amino acid conservation, and genetic variability in Structural, non-structural, and Accessory proteins. Int. J. Mol. Sci., 23(12), 6394 (2022).
García Aranda, M., Redondo, M., Barragán Mallofret, I. & Martínez Gálvez, B. Patent Application Number P202331044 - KLRB1 como biomarcador de pronóstico en pacientes con COVID-19 (KLRB1 as Prognostic Biomarker of COVID-19 Patients) (Spain, 2023). O. Madrid, Editor.
Horvath, K. M. et al. Nasal lavage natural killer cell function is suppressed in smokers after live attenuated influenza virus. Respir. Res. 12 (1), 102 (2011).
Winkley, K. et al. Immune cell residency in the nasal mucosa may partially explain respiratory disease severity across the age range. Sci. Rep. 11 (1), 15927 (2021).
Ponomarenko, E. A. et al. Workability of mRNA sequencing for Predicting protein abundance. Genes 14 (11), 2065 (2023).
Wyrożemski, Ł. & Qiao, S. W. Immunobiology and conflicting roles of the human CD161 receptor in T cells. Scand. J. Immunol. 94 (3), e13090 (2021).
Kurioka, A. et al. CD161 defines a functionally distinct subset of pro-inflammatory natural killer cells. Front. Immunol. 9, 486 (2018).
Lanier, L. L., Chang, C. & Phillips, J. H. Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. Journal of immunology (Baltimore, Md.: 1994. 153(6): pp. 2417–2428. (1950).
Lenart, M. et al. SARS-CoV-2 infection impairs NK cell functions via activation of the LLT1-CD161 axis. Front. Immunol. 14, 1123155 (2023).
Kirkham, C. L. & Carlyle, J. R. Complexity and diversity of the NKR-P1:Clr (Klrb1:Clec2) Recognition systems. Front. Immunol., 5, 214 (2014).
Braud, V. M. et al. LLT1-CD161 Interaction in Cancer: promises and challenges. Front. Immunol. 13, 847576 (2022).
Demaria, O. et al. Identification of druggable inhibitory immune checkpoints on natural killer cells in COVID-19. Cell Mol. Immunol. 17 (9), 995–997 (2020).
Ghasemzadeh, M., Ghasemzadeh, A. & Hosseini, E. Exhausted NK cells and cytokine storms in COVID-19: whether NK cell therapy could be a therapeutic choice. Hum. Immunol. 83 (1), 86–98 (2022).
Kamishikiryo, J. et al. Molecular basis for LLT1 protein recognition by human CD161 protein (NKRP1A/KLRB1). J. Biol. Chem. 286 (27), 23823–23830 (2011).
Fergusson, J. R., Fleming, V. M. & Klenerman, P. CD161-expressing human T cells. Front. Immunol. 2, 36 (2011).
He, X. et al. Breakthrough infection evokes the nasopharyngeal innate immune responses established by SARS-CoV-2–inactivated vaccine. Front. Immunol. 14, 1181121 (2023).
Roukens, A. H. E. et al. Prolonged activation of nasal immune cell populations and development of tissue-resident SARS-CoV-2-specific CD8 + T cell responses following COVID-19. Nat. Immunol. 23 (1), 23–32 (2022).
Karunathilaka, A. et al. CD161 expression defines new human γδ T cell subsets. Immun. Ageing. 19 (1), 11 (2022).
Truong, K. L. et al. Killer-like receptors and GPR56 progressive expression defines cytokine production of human CD4 + memory T cells. Nat. Commun. 10 (1), 2263 (2019).
Parrot, T. et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci. Immunol. 5 (51), eabe1670 (2020).
Germain, C. et al. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-γ contributes to modulate immune responses. J. Biol. Chem. 286 (44), 37964–37975 (2011).
McGary, C. S. et al. The loss of CCR6 + and CD161 + CD4 + T-cell homeostasis contributes to disease progression in SIV-infected rhesus macaques. Mucosal Immunol. 10 (4), 1082–1096 (2017).
Aljabr, W. et al. Evaluation of the levels of peripheral CD3(+), CD4(+), and CD8(+) T cells and IgG and IgM Antibodies in COVID-19 patients at different stages of infection. Microbiol. Spectr. 10 (1), e0084521 (2022).
Moss, P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 23 (2), 186–193 (2022).
Information, N. N. C. B. nCounter®Human Immunology V2 panel [NS_Immunology_v2_C2328]. [cited 2024 8 Aug. 2024]; (2023). Available from: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL32993
Amati, F. et al. Expression profiles of the SARS-CoV-2 host invasion genes in nasopharyngeal and oropharyngeal swabs of COVID-19 patients. Heliyon, 6(10), e05143 (2020).
Kuzan, A. et al. What to do if the qPCR test for SARS-CoV-2 or other Pathogen lacks endogenous Internal Control? A simple test on housekeeping genes. Biomedicines 11 (5), 1337 (2023).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 102 (43), 15545–15550 (2005).
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, 1–16 (2004).
Korotkevich, G. et al. Fast gene set enrichment analysis. bioRxiv, : p. 060012. (2021).
The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res., 47(D1), D330–D338 (2019).
Acknowledgements
The authors thank Alicia Aguilera for her technical support, and Martina Álvarez and Fernando Fernández for their advice on experimental design.
Funding
This work was supported by the [University of Malaga—Consejería de Transformación Económica, Industria, Conocimiento y Universidades—Junta de Andalucia] under Grant [CV20-76497]; [Fundación Fuerte and Isabel Luque Bardón]; [European Social Fund—Operational Program of Andalusia 2014–2020 - Consejería de Salud y Familias de la Junta de Andalucía] under Grant [RH-0055-2020]; [Sistema Andaluz de Salud – Nicolás Monardes] under Grant [RC-0009-2021]; [Gobierno de Navarra] under Grant [ANDIA 2021 and PORTTRAIT 0011-3947-2021-000023 and 0011-2750-2022-000000]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
MGA, IBM and MR: Conceived the study. MGA, MAO, RQ, AS: Initiated study design and DMG, TT, IL, BMG, MLH, MPR helped with implementation. FRR: Provided primary statistical analysis and AGG, JGG, RA provided statistical expertise in bioinformatics. All authors contributed to interpretation of data, drafting and reviewing and approved the final manuscript.All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
García-Aranda, M., Onieva, M.Á., Martín-García, D. et al. KLRB1 expression in nasopharyngeal mucosa as a prognostic biomarker in COVID-19 patients. Sci Rep 15, 3079 (2025). https://doi.org/10.1038/s41598-025-86846-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-86846-7