Abstract
As an important mechanism of renal injury, oxidative stress (OS) is inseparable from the occurrence of renal fibrosis and the rapid progression of renal failure. However, the contribution of OS to IgA nephropathy (IgAN), the primary driver of chronic kidney disease remains uncertain. To investigate the effects of OS in IgAN, and identify the mechanisms of cell and tissue injury and protection, single-cell RNA sequencing (scRNA-seq) data and microarray data of IgAN were collected and analyzed. Through gene set variation analysis (GSVA), we identified significant alterations in the activity of multiple OS pathways within the proximal tubule cells (PTCs) of IgAN patients. Subsequent enrichment analysis revealed that the differentially expressed genes associated with OS in PTCs were primarily linked to the process of ferroptosis. Therefore, regulators of ferroptosis were collected to define the ferroptosis activity of PTCs in IgAN, and we found that the activity of suppressing ferroptosis was significantly enhanced. Moreover, being the central controller of ferroptosis, the expression of GPX4 in the PTCs of IgAN is extremely significant, which has been further verified by immunohistochemistry in kidney tissues of IgAN patients. Additionally, the GSVA of microarray data of IgAN indicated that the activity of driving ferroptosis and suppressing ferroptosis in tubulointerstitium were markedly decreased, however, the inhibition of ferroptosis in the tubulointerstitium of IgAN is relatively stronger. These findings demonstrate that ferroptosis inhibition may be a potential mechanism to alleviate OS injury in IgAN, and GPX4 could not only function as a specific marker for PTCs in IgAN but also represent a potential therapeutic target to halt the progression of the disease.
Similar content being viewed by others
Introduction
A growing body of evidence suggests that the kidney plays a pivotal role in generating reactive oxygen species (ROS)1. As a substantial contributing factor to the development of renal damage and the progression of chronic kidney disease (CKD)2, oxidative stress (OS) is involved in cell death, tissue inflammation and disease progression3. The vigorous metabolism of mitochondria in kidney makes it vulnerable to injury caused by OS4, but synchronously, the presence of endogenous antioxidant system protects the kidney from damage5. It is clear that the pathophysiology of kidney injury is inseparable from regulation of OS, whose development lead to CKD and have been a valuable target for the treatment and prevention of AKI progression6. As an important cause of CKD, researches on OS in the occurrence and development of glomerulonephritis (GN) have not been revealed clearly to date.
Undoubtedly, as the most common primary GN worldwide, most of IgA nephropathy (IgAN) patients are confronted with the risk of progressing to CKD7. The most prominent pathological manifestation of IgAN is IgA deposition in the mesenchyme of the glomeruli8, which results in the activation of various types of OS signaling pathways9. Furthermore, the relationship between OS and IgAN is not limited to mesangial cell damage, but also involved in endothelial cell apoptosis and macrophage-mediated inflammation10,11. Exploring variations in OS activity in various types of kidney cells may bring new perspectives to shed light on the mechanisms of cell death and tissue damage observed in IgAN, and provide potential therapeutic targets to prevent its progression.
Excessive OS is a critical driver of cellular damage and death, contributing to various forms of cell death, including ferroptosis, apoptosis, and necrosis. Among these, the relationship between ferroptosis and kidney pathology has garnered significant attention in recent years. Ferroptosis is a distinct form of cell death characterized by iron-dependent lipid peroxidation, driven by the accumulation of ROS through iron metabolism and lipid peroxidation pathways12. Given the kidney’s high sensitivity to oxidative damage, ferroptosis plays a pivotal role in promoting the death of tubular and glomerular cells under pathological conditions. The occurrence of ferroptosis is a key contributor to the injury and progression of IgAN13.
Consequently, single-cell RNA sequencing (scRNA-seq) data of IgAN were analyzed to explore the OS-related pathways activity of various cell types14, and multiple microarray datasets were collected and analyzed to validate the discovery in the scRNA-seq data. Finally, we verified the specific GPX4 expression on the proximal tubule cells (PTCs) of IgAN by immunohistochemistry (IHC). The design of this study is shown in Fig. 1.
Methods
Information sources and data manipulation
To investigate the differences in OS pathway activity across diverse cell types in IgAN, we obtained scRNA-seq data for kidney tissues from thirteen IgAN patients and six healthy controls via the GEO database. (GSE127136)15. Subsequently, we utilized the “Seurat” package (version 4.1.1) on the R platform (version 4.3.2) for data manipulation and analysis16. Cells with fewer than 200 genes detected were filtered out, and the functions “NormalizeData” and “ScaleData” were employed to standardize the gene expression levels. Then, PCA was performed based on the first 2000 variable genes screened by “FindVariableFeautres” function. The gene expression matrix was reduced to the first 11 principal components (PCs) and TSNE plot was used to visualize the clusters using a resolution of 0.2. After that, the marker genes were filtered out using the “FindAllMarkers” function according to average log2FC (Fold Change) greater than 0.25. In addition, heatmap was used to show the top five marker genes for each cell cluster.
Further, immunohistochemistry paraffin (IHC-P) of patients with IgAN diagnosed by renal biopsy at Shanxi Provincial People’s Hospital from January 2022 to September 2022 were collected for immunohistochemical analysis, and eight control samples were obtained from patients with renal cell carcinoma (RCC) undergoing nephrectomy at Shanxi Provincial People’s Hospital. (Table S1). To improve the accuracy of the study, patients who used immunosuppressants within 1 month before renal biopsy were excluded from this study.
Calculation and comparison of oxidative stress pathway activity
To calculate the OS score of different cell clusters, keyword “oxidative stress” was used to search for the oxidative stress-related genes (OSRGs) and pathways in the Gene Ontology (GO) database17, and “Homo sapiens” was served as the filter. Then, we calculated the enrichment score of each cell in the predefined pathways that consists of OSRGs using R software package “GSVA” (version 1.40.1)18. The OS scores of different renal cell clusters in IgAN patients and controls were compared by Wilcoxon Rank Sum test19, and a P-value less than 0.05 is deemed statistically significant.
Differentially expressed genes associated with oxidative stress
To explore the differentially expressed genes (DEGs) associated with OS (OS-DEGs) of PTCs between IgAN patients and controls, “limma” package (version 3.40.6) was used to screen the DEGs in the PTCs of IgAN under the criterion of P values < 0.0520, and the OS-DEGs were identified by intersecting DEGs with OSRGs.
Enrichment analysis
To uncover the mechanisms underlying disease pathogenesis, we conducted enrichment analysis on the OS-DEGs using the “clusterProfiler” package (Version 3.14.3), focusing on both GO and KEGG databases21,22,23.
Activity of ferroptosis in PTCs
In order to assess the changes in activity of ferroptosis in IgAN PTCs, thirty genes that regulate ferroptosis were downloaded from Tang et al.24, consisting of 18 drivers and 12 suppressors. Additionally, separate PPI networks for the 30 regulatory factors were built using the String database (see Table S2). Given that these regulatory factors have varying roles in ferroptosis, we utilized “Cytoscape” software (Version 3.9.1) along with the “MOCODE” plugin to sift through the networks for key regulators25, and the parameters were set as scores > 5.
Then, the gene sets composed of key drivers and suppressors were defined as two pathways, “driving ferroptosis” and “suppressing ferroptosis”, respectively. Similarly, the enrichment scores of each PTC in these two pathways were calculated using “GSVA” package.
Identification of oxidative stress related markers for PTCs of IgAN
The unique markers of PTCs in IgAN were selected from the up-regulated OS-DEGs. PTCs markers were screened out using the “FindAllMarkers” function. “FeaturePlots” function were used to show the gene expression in different cell clusters and different groups.
Immunohistochemistry
Each IHC-P result was verified in 53 patient samples and 8 controls, and antibody was performed in at least 3 slides. Tissue slices were first dewaxed in xylene for 40 min, then rehydrated with a solution of ethanol ranging from 100 to 70% and washed with distilled water. The system was allowed to cool to room temperature before being washed with TBS (Solarbio; T1080) after twice repairing the slices with sodium citrate (Proteinrech;PR30001) in the microwave over medium heat for 5 min.
The tissue in the goat serum (Biosharp; BL210A) was blocked at room temperature for an hour and then the slides were exposed at room temperature with anti-GPX4 (rabbit anti-human, Proteinrech; 67,763–1-Ig) for 90 min. Then, the tissue was then subjected to a 30-min incubation with secondary antibodies (Proteinrech; PK10006) at room temperature. Finally, the slides were stained with DAB and nucleated with haematoxylin.
Microarray data collection and analysis
To corroborate the insights gained from the scRNA-seq analysis, microarray datasets specific to the tubulointerstitium of IgAN were sourced from the GEO repository using the keyword “IgA Nephropathy”, and the information of datasets are shown in Table 1. The gene symbols are converted based on the platform information, and “inSilicoMerging” package (Version 1.0.8) was used to combine three datasets, while the “combat” function was devoted for batch effect removal to finally get the “merged data” 26. Then, same methods above were used to identify the OS-DEGs. To understand the function of OS-DEGs and the changes in the activity of pathways related to OS and ferroptosis between IgAN patients and controls in microarray data, enrichment analysis was performed. Linear regression models were constructed to compare the changes in the correlation between the two pathways of driving and suppressing ferroptosis.
Results
Identification of cell clusters
Quality control and dimensionality reduction were performed on the gene expression matrix of each cell in GSE127136. Subsequently, six distinct clusters of cells were identified using unsupervised clustering analysis, and specific marker genes of each cell clusters were calculated (Table S3). Then, the cell clusters were annotated based on published studies15 and CellMarker database (Table 2)30. The localization of various cell clusters within the kidney tissue from both control subjects and IgAN patients is illustrated in Fig. 2A. Figure 2B represents the top 5 specific marker genes expression of all cell clusters, while Fig. 2C indicates their annotation and expression in the 6 cell clusters.
Overview of ScRNA-seq Data Processing and Analysis. (A) Illustrates the cellular distribution within kidney tissue from both control subjects and IgAN patients, encompassing six distinct cell types: Proximal Tubular Cells, Mesangial Cells, T Cells, Macrophages, Endothelial Cells, and NKT Cells. (B) Showcases a heatmap of the top five marker genes across the six identified cell clusters; Cluster 0 corresponds to Proximal Tubular Cells, Cluster 1 to Mesangial Cells, Cluster 2 to T Cells, Cluster 3 to Macrophages, Cluster 4 to Endothelial Cells, and Cluster 5 to NKT Cells. (C) Displays a bubble chart illustrating cell-lineage-specific marker genes for each of the six identified cell clusters.
Oxidative stress pathway activity of cell clusters in scRNA-seq data
Four hundred and forty-four OSRGs and 8 important OS pathways were obtained from GO database (Table S4). The “GSVA” package was employed to compute the enrichment scores for individual cells across eight OS-related pathways, which are provided in Table S5 and could be considered as OS pathway activity. Bubble chart was applied to visualize the pathway activity in each cell cluster of IgAN and control groups (Fig. 3A). Subsequently, the Wilcoxon Rank Sum test was applied to evaluate the activity levels of each cell cluster within the OS pathways among both IgAN and control groups. Noteworthy differences were exclusively found in PTCs, as depicted in Fig. 3B–F. These differences encompassed various aspects like oxidative stress response, cellular adaptation to oxidative stress, modulation of cellular oxidative stress response, and control of oxidative stress-triggered cell death, and NKT cells were not analyzed as their number was insufficient in the control group to conduct the analysis.
Computation and Comparison of OS Pathway Activity in ScRNA-seq Data. (A) Depicts a Matrix-Bubble chart illustrating the activity of eight OS pathways across the six identified cell clusters in both IgAN patients and controls. (B) Presents a Bar graph showcasing the activity of eight OS pathways specifically in PTCs of IgAN patients compared to controls, with statistical significance denoted as * for P < 0.05, ** for P < 0.01. (C) Displays a Bar graph representing the activity of eight OS pathways in mesangial cells of IgAN patients and control group. (D) Shows a Bar graph representing the activity of eight OS pathways in endothelial cells of IgAN patients and control group. (E) Demonstrates a Bar graph representing the activity of eight OS pathways in T cells of IgAN patients and control group. (F) Exhibits a Bar graph representing the activity of eight OS pathways in macrophages of IgAN patients and controls. Annotation: A: regulation of oxidative stress-induced intrinsic apoptotic signaling pathway; B: intrinsic apoptotic signaling pathway in response to oxidative stress; C: regulation of oxidative stress-induced cell death; D: cell death in response to oxidative stress; E: regulation of cellular response to oxidative stress; F: cellular response to oxidative stress; G: regulation of response to oxidative stress; H: response to oxidative stress.
DEGs associated with oxidative stress in scRNA-seq data
To gain deeper insights into the regulatory dynamics of OSRGs in PTCs from IgAN patients, we isolated 920 DEGs from these cells, which included 507 up-regulated and 413 down-regulated DEGs, as presented in Fig. 4A and Table S6. Further analysis allowed us to identify 20 up-regulated and 27 down-regulated OS-DEGs, based on a cross-reference with 444 known OSRGs (Fig. 4B). A likert chart was employed to visually represent the quantity and relative prevalence of these OS-DEGs across eight OS pathways in PTCs (Fig. 4C).
Discovery of Differentially Expressed Genes Associated with Oxidative Stress in Proximal Tubular Cells. (A) Illustrates a volcano map displaying 920 DEGs from PTCs of IgAN patients, where red represents up-regulation and blue represents down-regulation, with a total of 507 up-regulated and 413 down-regulated DEGs identified. (B) Presents an UpSet-Topology diagram for the identification of OS-DEGs within PTCs of IgAN, revealing 20 up-regulated and 27 down-regulated OS-DEGs. (C) Depicts a Likert-Chart displaying the quantity and proportion of OS-DEGs across eight OS pathways in IgAN PTCs. Pathways with significant differences are indicated in red fonts, while the white fonts on the bar graph represent the number of OS-DEGs that are either upregulated or downregulated within each pathway.
Enrichment analysis of OS-DEGs in PTCs
For a more in-depth examination of the 47 OS-DEGs, both KEGG and GO enrichment analyses were carried out. 20 up-regulated OS-DEGs were mainly related to ferroptosis, autophagy-animal, Rap1 signaling pathway, and MAPK signaling pathway in KEGG database (Fig. 5A), these genes were aliso linked to processes such as the negative regulation of apoptosis, inhibition of macroautophagy, and GTPase-activating protein binding, according to the GO database (Fig. 5B). Simultaneously, 27 down-regulated OS-DEGs were associated with apoptosis-multiple species, ferroptosis, apelin signaling pathway in KEGG database and cellular response to hydrogen peroxide, antioxidant activity in GO database (Fig. 5C,D).
Enrichment Analysis for OS-DEGs in IgAN Proximal Tubular Cells. (A) Depicts KEGG enrichment analysis results for the 20 up-regulated OS-DEGs in IgAN PTCs. (B) Illustrates GO enrichment analysis findings for the 20 up-regulated OS-DEGs in IgAN PTCs. (C) Displays KEGG enrichment analysis outcomes for the 27 down-regulated OS-DEGs in IgAN PTCs. (D) Presents GO enrichment analysis results for the 27 down-regulated OS-DEGs in IgAN PTCs.
Ferroptosis activity in PTCs
Both up and down-regulated OS-DEGs are significantly enriched in the ferroptosis pathway. Thus, 18 drivers and 12 suppressors of ferroptosis were obtained from published studies, and PPI network was established to screen 12 key drivers and 5 suppressors which served as gene sets used for ferroptosis-related pathway enrichment analysis (Fig. 6A,B). Similarly, the ferroptosis-related pathway activity of PTCs were calculated and the activity of suppressing ferroptosis was significantly enhanced in the PTCs of IgAN (Fig. 6C and Table S7).
Identification of Ferroptosis Regulators and Evaluation of Ferroptosis Activity. (A) Displays the PPI network involving 18 drivers, with 12 key drivers highlighted in pink circles. (B) Illustrates the PPI network of 12 suppressors, with 5 suppressors identified. (C) Presents a Bar graph representing the activity levels of ferroptosis pathways in PTCs of IgAN patients and the control group, with statistical significance marked as * for P < 0.05.
The expression of ferroptosis core regulator GPX4 in IgAN
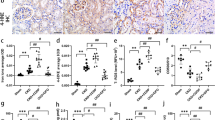
Given the differences in the expression of OS pathways and OSRGs between PTCs in IgAN and healthy controls, OSRGs may serve as potential cell markers for PTCs of IgAN. Hence, the 1194 cell markers of PTCs identified using “FindAllMarkers” function were compared with the 20 up-regulated OS-DEGs, and 5 potential marker genes for PTCs of IgAN were selected (Fig. 7A). The feature plot was used to represente the gene expression of the 5 potential marker genes in IgAN and control groups. The expression of these genes was significantly increased in PTCs of IgAN, especially GPX4 and AKT4 (Fig. 7B,C). Additionally, the elevated expression of GPX4, a central regulator of ferroptosis, suggests that the suppression of ferroptosis could be more pronounced in PTCs of IgAN. The GPX4 expression in IgAN was further evaluated by IHC-P using 53 IgAN and 8 normal tissues, the findings are consistent with the mRNA expression profiles derived from scRNA-seq data. In addition, GPX4 expression is already increased in PTC early stage in IgAN, and with the progression of CKD stages, the expression of GPX4 also becomes increasingly significant (Fig. 7D,E).
Discovery of Potential Oxidative Stress-Related Marker Genes for IgAN Proximal Tubular Cells. (A) Presents a Venn diagram used to identify potential OS-related marker genes for IgAN PTCs. (B) Shows the feature plot of the expression of the five OS-related marker genes across all PTCs from IgAN patients and the control group. (C) Displays the feature plot of the expression of the five oxidative stress-related marker genes in PTCs from IgAN patients and the control group separately. (D) Depicts IHC-Pverification of GPX4 expression in IgAN patients and normal controls, with scale bars at 60 mm and a magnification of 20 × . (E) Illustrates IHC-P verification of GPX4 expression in IgAN patients and normal controls, with scale bars at 30 mm and a magnification of 40 × .
Analysis of microarray data
Three datasets of tubulointerstitium in IgAN were combined to a new gene expression matrix “merged data” (Table S8), As previously demonstrated, an initial screening revealed 1254 up-regulated and 854 down-regulated DEGs in the tubulointerstitium of IgAN patients (Fig. 8A). Subsequent analysis further identified 41 up-regulated and 32 down-regulated OS-DEGs (Fig. 8B). Moreover, the enrichment analysis have shown that the 41 up-regulated OS-DEGs in tubulointerstitium of IgAN were involved in peroxisome, p53 signaling pathway, apoptosis, cellular response to hydrogen peroxide (Fig. 8C,D), while the 32 down-regulated OS-DEGs were mainly related to apoptosis, TNF signaling pathway, FoxO signaling pathway, and positive regulation of apoptotic process (Fig. 8E,F).
Identification and Enrichment Analysis of OS-DEGs in the Tubulointerstitium of IgAN. (A) Features a volcano map of 2,108 DEGs identified in the tubulointerstitium of IgAN patients, with red indicating up-regulation and blue denoting down-regulation; a total of 1,254 up-regulated and 854 down-regulated DEGs were found. (B) Displays an UpSet-Topology diagram for identifying the OS-DEGs in the tubulointerstitium of IgAN, revealing 41 up-regulated and 32 down-regulated OS-DEGs. (C) Presents the KEGG enrichment analysis for the 41 up-regulated OS-DEGs in the IgAN tubulointerstitium. (D) Illustrates the GO enrichment analysis for the 41 up-regulated OS-DEGs in the IgAN tubulointerstitium. (E) Depicts the KEGG enrichment analysis for the 32 down-regulated OS-DEGs in the IgAN tubulointerstitium. (F) Showcases the GO enrichment analysis for the 32 down-regulated OS-DEGs in the IgAN tubulointerstitium.
Additionally, similar results were obtained showing that the OS pathway activity in the tubulointerstitium of IgAN were significantly lower than control group (Fig. 9A and Table S9). In terms of ferroptosis, both the promotive and inhibitory activities were markedly reduced compared to those in the control group (Fig. 9B and Table S10). At this point, linear regression models were constructed to analyze the correlation between driving and suppressing ferroptosis in IgAN and control group, separately. Although the two pathways were positively correlated in the both groups, the correlation coefficient was higher in the tubulointerstitium of IgAN, suggesting a relative increase in the activity of suppressing ferroptosis in the tubulointerstitium of IgAN (Fig. 9C).
Assessment of Oxidative Stress and Ferroptosis Activity in IgAN Tubulointerstitium. (A) Features a bar graph depicting the activity levels of eight OS pathways in the tubulointerstitium of IgAN patients and control group; asterisks indicate statistical significance (*: P < 0.05, ***: P < 0.001). (B) Presents a bar graph illustrating the activity levels of ferroptosis pathways in the tubulointerstitium of IgAN patients and control group, with significance indicated (**: P < 0.01, ***: P < 0.001). (C) Displays linear regression models that examine the correlation between driving and suppressing ferroptosis; the x-axis represents the activity of driving ferroptosis, while the y-axis represents the activity of suppressing ferroptosis. Annotation: A: regulation of oxidative stress-induced intrinsic apoptotic signaling pathway; B: intrinsic apoptotic signaling pathway in response to oxidative stress; C: regulation of oxidative stress-induced cell death; D: cell death in response to oxidative stress; E: regulation of cellular response to oxidative stress; F: cellular response to oxidative stress; G: regulation of response to oxidative stress; H: response to oxidative stress.
Discussion
Reducing tissue damage caused by the deposits of Galactose-deficient IgA1 (Gd-IgA1), has become a major challenge in preventing renal function deterioration in IgAN patients31, especially in the situation of neither glucocorticoids nor immunosuppressants fail in halting or delay the progression to end-stage renal disease (ESRD)32. The nephrotoxicity of Gd-IgA1 in IgAN patients could be enhanced by OS33, and the variation in oxidation and antioxidant balance is closely linked to proteinuria levels and disease progression in IgAN patients34. Accumulating evidence revealed the unique importance of OS in mediating glomerular sclerosis and tubulointerstitial fibrosis35. OS may lead to irreversible damage to renal tissue in IgAN by regulating cell death and inflammation36.
OS has been extensively recognized as a critical driver of kidney injury in IgAN. Our study, leveraging both single-cell and bulk RNA sequencing data, provides a multi-dimensional characterization of OS and ferroptosis activity changes in PTCs of IgAN kidneys37. The deposition of Gd-IgA1 and subsequent formation of immune complexes can initiate an inflammatory cascade, leading to the establishment of an inflammatory milieu in the kidney. This microenvironment promotes the generation of excessive ROS, disrupts iron metabolism, and induces lipid peroxidation, ultimately triggering ferroptosis in renal cells. This process exacerbates kidney injury, creating a vicious cycle of damage and inflammation38.
To investigate the transformations of OS pathway activity in various cell types of IgAN, “seurat” package was used to identify the different cell clusters in the scRNA-seq data, and 6 cell clusters was annotated in the IgAN and control groups according to specific marker genes. The 8 OS pathways collected from GO database were considered as background gene sets, subsequently, enrichment scores of all cell clusters were calculated. Surprisingly, significant differences in OS pathway activity were found only between PTCs of IgAN and control groups. In principle, GSVA algorithm mainly calculates the enrichment score of a certain pathway according to the gene expression values16, which means that there is absolutely no difference in the OS pathway activity of other 5 cell clusters between IgAN patients and controls, but the activity of several important OS pathways in PTCs of IgAN was significantly altered. Recent research has established the pivotal influence of OS on the mobility, growth, and damage of PTCs39, which may be ascribed to the active mitochondrial and endoplasmic reticulum metabolism40. Sensitivity to OS is not only an explanation for PTCs death or injury, but also an emerging target of protecting renal function and delaying the progression of CKD41. Medications like dapagliflozin have demonstrated remarkable efficacy in counteracting OS-induced cell damage in human PTCs42, and previous studies have also indicated the protective effect of pharmaceutical molecules which could activate peroxisomes on PTCs in IgAN43.The selection and application of drugs to protect IgAN renal tissue damage are still limited. Drugs that modulate the cell injury caused by OS may be the key to breaking the deadlock that supportive and immunosuppressive therapies are not effective in preventing IgAN progression.
Upon identifying variations in OS pathway activity, we pinpointed 20 genes that were elevated and 27 that were reduced in the PTCs of those with IgAN. These up-regulated genes mainly contribute to Rap1 and MAPK signaling pathways, with Rap1 specifically helping to mitigate tubular damage primarily by regulating mitochondrial-originated OS44, Conversely, triggering the MAPK signaling pathway is associated with the onset of tubular fibrosis45. At the same time, several up-regulated genes related to OS were associated with biological activities like downregulating apoptosis and macroautophagy. Suppressing these pathways could minimize cell death in PTCs and decelerate kidney damage46. Notably, genes that were both elevated and reduced showed significant enrichment in the ferroptosis pathway according to KEGG analysis. The enrichment analysis suggested that PTCs in IgAN may have potential pathways to decrease OS damage. In view of that, drivers and suppressors of ferroptosis were collected and filtered to assess the ferroptosis activity in PTCs. Interestingly, both up-regulated and down-regulated genes were prominently represented in the KEGG ferroptosis pathway. As an important classifications of regulated necrosis47, ferroptosis is characterized by lipid peroxidation48. Accumulating evidence attributes the central pathophysiology of AKI, nephron loss, and acute tubular necrosis to ferroptosis and necrosis49. Particularly, cells could protect themselves by regulating ferroptosis when antioxidant mechanisms are ineffective50. As a consequence, PTCs of IgAN may reduce OS damage by suppressing ferroptosis.
Additionally, five OSRGs were further screened out as the potential marker genes, including GPX4, RBPMS, GSS, AK4, and SLC25A23. As the hub regulator of ferroptosis51, the GPX4 expression levels showed a notable rise in the PTCs of individuals with IgAN. Ample research has shown the significant correlation between GPX4 and renal tubular system, however, previous studies mainly focused on AKI and DN, and concluded that decreased GPX4 expression leads to increased ferroptosis and aggravate tubular injury in these diseases52. Moreover, GPX4 exerted its inhibiting ferroptosis role through regulating the GSS/GSR complex53, which is undoubtedly suggestive of the importance of ferroptosis activity in PTCs damage and protection in IgAN, and the IHC-P results shown that the high expression of GPX4 in PTCs of IgAN which further supports our findings and indicated the great potential of GPX4 for used as the specific marker of PTCs in IgAN as well as the therapeutic target for prevent the progression of IgAN.
Renal cells harbor a variety of antioxidant molecules, among which GPX4 plays a critical role. GPX4 functions to suppress lipid peroxidation and thereby inhibit ferroptosis. However, paradoxically, GPX4 is also recognized as a marker of ferroptotic activity54. The observed upregulation of GPX4 in PTCs likely represents an adaptive response to the OS environment, reflecting a cellular attempt to mitigate OS-induced damage. This regulatory mechanism underscores the cellular effort to counteract oxidative damage55. Nevertheless, the increased expression of GPX4 does not necessarily equate to the suppression of ferroptosis. The activity of ferroptosis is governed by a delicate balance between pro-oxidant and antioxidant pathways, and the disruption of this balance is a critical driver of cellular injury56. GPX4 represents just one component of this intricate network. Our integrated analysis of single-cell and bulk RNA sequencing data revealed a pronounced upregulation of pro-ferroptotic genes and heightened activity of ferroptosis-associated pathways in PTCs from IgAN kidneys. These findings highlight the complexity of ferroptosis regulation in IgAN and suggest that the increased GPX4 expression may reflect a compensatory, albeit insufficient, response to the OS-driven ferroptotic process.
The proximal tubule, rich in mitochondria and engaged in active oxidative phosphorylation, is especially susceptible to damage, potentially leading to cell death and the development of tubular glomeruli57, and serving as the monitor for both glomeruli and tubulointerstitium, the proximal tubule plays a critical role in the advancement of renal interstitial fibrosis and CKD58. Thus, to determine whether the changes of OS activity in PTCs and tubulointerstitium were synchronized, microarray datasets of IgAN tubulointerstitium were downloaded and analyzed, and 73 OS-DEGs were identified from the merged data. The 41 up-regulated OS-DEGs were mainly associated with peroxisome, apoptosis, cellular senescence and necroptosis, which are the common mechanisms of tubulointerstitium injury and fibrosis59. Furthermore, 32 down-regulated OS-DEGs were associated with apoptosis, TNF signaling pathway, and FoxO signaling pathway. TNF signaling pathway mediates renal interstitial injury and promote renal interstitial fibrosis60, while the activation of FoxO signaling pathway could protect tubulointerstitium through attenuating apoptosis61.
The findings of enrichment analysis and the differences of OS pathway activity between renal tubulointerstitium in IgAN and control groups indicated that there are also some mechanisms in the tubulointerstitium to alleviate the injury caused by OS. The tubulointerstitium may balance OS through multiple pathways, nevertheless, the relevant research in IgAN is critically insufficient, which may be one of the important reasons for the lack of effective treatment to delay the progression of IgAN. Considering the changes in ferroptosis activity of the PTCs in IgAN, the activity of driving ferroptosis and suppressing ferroptosis were also calculated and compared, and the activity of driving ferroptosis and suppressing ferroptosis were both significantly increased in the tubulointerstitium of IgAN patients. Moreover, regression models were established to reflect the correlation between pathway activity of driving and suppressing ferroptosis in IgAN and control groups. Although the activity of the two pathways was positively correlated in both groups, the correlation coefficient is higher in IgAN, which suggests that the inhibition of ferroptosis in the tubulointerstitium of IgAN is relatively more active than in controls.
Under these lights, suppressing ferroptosis may be a potential mechanism for PTCs or tubulointerstitium to alleviate OS injury in IgAN, which will enrich mechanisms of IgAN progression and inspire the research and development of drugs to protect renal functions of IgAN patients and prevent ESRD.
Conclusions
These findings is the first demonstrate that ferroptosis inhibition may be a potential mechanism to alleviate OS injury in IgAN, and GPX4 may act as both a unique identifier for PTCs in IgAN and a potential treatment focal point to halt the disease’s advancement.
Data availability
The foundational datasets underpinning the findings of this research are accessible through the GEO database, as well as in the supplementary material accompanying this article. For further in-depth data inquiries, please reach out to the corresponding author via email.
References
Coppolino, G., Leonardi, G., Andreucci, M. & Bolignano, D. Oxidative stress and kidney function: A brief update. Curr. Pharm. Des. 24(40), 4794–4799 (2018).
Tamay-Cach, F. et al. A review of the impact of oxidative stress and some antioxidant therapies on renal damage. Ren. Fail. 38(2), 171–175 (2016).
Chen, Q. et al. Astragalosides IV protected the renal tubular epithelial cells from free fatty acids-induced injury by reducing oxidative stress and apoptosis. Biomed. Pharmacother. 108, 679–686 (2018).
Daenen, K. et al. Oxidative stress in chronic kidney disease. Pediatr. Nephrol. 34(6), 975–991 (2019).
Marklund, S. L. Extracellular superoxide dismutase and other superoxide dismutase isoenzymes in tissues from nine mammalian species. Biochem. J. 222(3), 649–655 (1984).
He, L. et al. AKI on CKD: Heightened injury, suppressed repair, and the underlying mechanisms. Kidney Int. 92(5), 1071–1083 (2017).
Rodrigues, J. C., Haas, M. & Reich, H. N. IgA nephropathy. Clin. J Am. Soc. Nephrol.: CJASN. 12(4), 677–686 (2017).
Rajasekaran, A., Julian, B. A. & Rizk, D. V. IgA nephropathy: An interesting autoimmune kidney disease. Am. J. Med. Sci. 361(2), 176–194 (2021).
Coppo, R., Camilla, R., Amore, A. & Peruzzi, L. Oxidative stress in IgA nephropathy. Nephro. Clin. Pract. 116(3), c1996-8 (2010).
Zhang, Q. et al. Activation of Nrf2/HO-1 signaling: An important molecular mechanism of herbal medicine in the treatment of atherosclerosis via the protection of vascular endothelial cells from oxidative stress. J. Adv. Res. 34, 43–63 (2021).
Calle, P. & Hotter, G. Macrophage phenotype and fibrosis in diabetic nephropathy. Int. J. Mol. Sci. 21(8), 2806 (2020).
Ru, Q. et al. Iron homeostasis and ferroptosis in human diseases: Mechanisms and therapeutic prospects. Signal Transduct. Target. Ther. 9(1), 271 (2024).
Tian, Z. Y. et al. Iron metabolism and chronic inflammation in IgA nephropathy. Renal. Fail. 45(1), 2195012 (2023).
Clough, E. & Barrett, T. The gene expression omnibus database. Methods Mol. Biol. 1418, 93–110 (2016).
Zheng, Y. et al. Single-cell transcriptomics reveal immune mechanisms of the onset and progression of IgA nephropathy. Cell Rep. 33(12), 108525 (2020).
Pereira, W. J. et al. Asc-Seurat: Analytical single-cell seurat-based web application. BMC Bioinform. 22(1), 556 (2021).
Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–56.
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013).
Lin, T., Chen, T., Liu, J. & Tu, X. M. Extending the Mann-Whitney-Wilcoxon rank sum test to survey data for comparing mean ranks. Stat. Med. 40(7), 1705–1717 (2021).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43(7), e47 (2015).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.: A Publ. Protein Soc. 28(11), 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51(D1), D587–D592 (2023).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic acids research. 28(1), 27–30 (2000).
Tang, B. et al. Integrative analysis of the molecular mechanisms, immunological features and immunotherapy response of ferroptosis regulators across 33 cancer types. Int. J. Biol. Sci. 18(1), 180–198 (2022).
Qing, J., Song, W., Tian, L., Samuel, S. B. & Li, Y. Potential small molecules for therapy of lupus nephritis based on genetic effect and immune infiltration. Biomed. Res. Int. 2022, 2259164 (2022).
Taminau, J. et al. Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinform. 13, 335 (2012).
Shved, N. et al. Transcriptome-based network analysis reveals renal cell type-specific dysregulation of hypoxia-associated transcripts. Sci. Rep. 7(1), 8576 (2017).
Grayson, P. C. et al. Metabolic pathways and immunometabolism in rare kidney diseases. Ann. Rheum. Dis. 77(8), 1226–1233 (2018).
Reich, H. N. et al. A molecular signature of proteinuria in glomerulonephritis. PLoS One. 5(10), e13451 (2010).
Coppo, R., Feehally, J. & Glassock, R. J. IgA nephropathy at two score and one. Kidney Int. 77(3), 181–186 (2010).
Descamps-Latscha, B. et al. Early prediction of IgA nephropathy progression: proteinuria and AOPP are strong prognostic markers. Kidney Int. 66(4), 1606–1612 (2004).
Ji, L. et al. FOXO1 overexpression attenuates tubulointerstitial fibrosis and apoptosis in diabetic kidneys by ameliorating oxidative injury via TXNIP-TRX. Oxid. Med. Cell Longev. 2019, 3286928 (2019).
Nagata, M. Podocyte injury and its consequences. Kidney Int. 89(6), 1221–1230 (2016).
Duni, A., Liakopoulos, V., Roumeliotis, S., Peschos, D., Dounousi, E. Oxidative stress in the pathogenesis and evolution of chronic kidney disease: Untangling Ariadne’s thread. Int. J. Mol. Sci. 2019;20(15).
Han, Y. C. et al. AMPK agonist alleviate renal tubulointerstitial fibrosis via activating mitophagy in high fat and streptozotocin induced diabetic mice. Cell Death Dis. 12(10), 925 (2021).
Zaibi, N., Li, P. & Xu, S. Z. Protective effects of dapagliflozin against oxidative stress-induced cell injury in human proximal tubular cells. PLoS One. 16(2), e0247234 (2021).
Zhu, W. et al. Ferroptosis-related genes in IgA nephropathy: Screening for potential targets of the mechanism. Int. J. Genom. 2024, 8851124 (2024).
Wu, J. et al. Downregulation of PPARα mediates FABP1 expression, contributing to IgA nephropathy by stimulating ferroptosis in human mesangial cells. Int. J. Biol. Sci. 18(14), 5438–5458 (2022).
García-Pérez, E., Ryu, D., Kim, H. Y., Kim, H. D., Lee, H. J. Human proximal tubule epithelial cells (HK-2) as a sensitive in vitro system for ochratoxin a induced oxidative stress. Toxins. 2021;13(11).
Xiao, J., Leung, J. C., Chan, L. Y., Guo, H. & Lai, K. N. Protective effect of peroxisome proliferator-activated receptor-gamma agonists on activated renal proximal tubular epithelial cells in IgA nephropathy. Nephrol. Dial Transplant. 24(7), 2067–2077 (2009).
Xiao, L. et al. Rap1 ameliorates renal tubular injury in diabetic nephropathy. Diabetes. 63(4), 1366–1380 (2014).
Geng, X. Q. et al. Ganoderic acid hinders renal fibrosis via suppressing the TGF-β/Smad and MAPK signaling pathways. Acta Pharmacol. Sin. 41(5), 670–677 (2020).
Fujiki, K., Inamura, H., Sugaya, T. & Matsuoka, M. Blockade of ALK4/5 signaling suppresses cadmium- and erastin-induced cell death in renal proximal tubular epithelial cells via distinct signaling mechanisms. Cell Death Differ. 26(11), 2371–2385 (2019).
Sun, Y. et al. The emerging role of ferroptosis in inflammation. Biomed. Pharmacother. 127, 110108 (2020).
Su, L. J. et al. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid. Med. Cell Longev. 2019, 5080843 (2019).
Belavgeni, A., Meyer, C., Stumpf, J., Hugo, C. & Linkermann, A. Ferroptosis and necroptosis in the kidney. Cell Chem. Biol. 27(4), 448–462 (2020).
Chen, X., Li, J., Kang, R., Klionsky, D. J. & Tang, D. Ferroptosis: Machinery and regulation. Autophagy. 17(9), 2054–2081 (2021).
Bersuker, K. et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 575(7784), 688–692 (2019).
Tonnus, W. et al. Dysfunction of the key ferroptosis-surveilling systems hypersensitizes mice to tubular necrosis during acute kidney injury. Nat. Commun. 12(1), 4402 (2021).
Wu, W., Geng, Z., Bai, H., Liu, T. & Zhang, B. Ammonium ferric citrate induced ferroptosis in non-small-cell lung carcinoma through the inhibition of GPX4-GSS/GSR-GGT axis activity. Int. J. Med. Sci. 18(8), 1899–1909 (2021).
Zhang, Y. et al. Sirtuin 3 regulates mitochondrial protein acetylation and metabolism in tubular epithelial cells during renal fibrosis. Cell Death Dis. 12(9), 847 (2021).
Chevalier, R. L. The proximal tubule is the primary target of injury and progression of kidney disease: role of the glomerulotubular junction. Am. J. Physiol. Renal Physiol. 311(1), F145–F161 (2016).
Duffield, J. S. Cellular and molecular mechanisms in kidney fibrosis. J. Clin. Invest. 124(6), 2299–2306 (2014).
Feng, Q. et al. Broadening horizons: The multifaceted functions of ferroptosis in kidney diseases. Int. J. Biol. Sci. 19(12), 3726–3743 (2023).
Zhang, L. M., Liu, X. M., Guo, D. W., Li, F., Hao, J., Zhao, S. FBXW7-Mediated Downregulation of GPX4 Aggravates Acute Kidney Injury Following Ischemia‒Reperfusion. Inflammation. 2024.
Azuma, K. et al. Mitochondrial glutathione peroxidase 4 is indispensable for photoreceptor development and survival in mice. J. Biol. Chem. 298(4), 101824 (2022).
Shi, J., Xu, W., Zheng, R., Miao, H. & Hu, Q. Neuregulin 4 attenuate tubulointerstitial fibrosis and advanced glycosylation end products accumulation in diabetic nephropathy rats via regulating TNF-R1 signaling. Am. J. Transl. Res. 11(9), 5501–5513 (2019).
Huang, F. et al. FoxO1-mediated inhibition of STAT1 alleviates tubulointerstitial fibrosis and tubule apoptosis in diabetic kidney disease. EBioMedicine. 48, 491–504 (2019).
Uil, M. et al. Platelet inhibition by ticagrelor is protective against diabetic nephropathy in mice. Faseb J. 34(10), 13750–13761 (2020).
Zhang, C. et al. The HMGB1-RAGE/TLR-TNF-α signaling pathway may contribute to kidney injury induced by hypoxia. Exp. Ther. Med. 17(1), 17–26 (2019).
Zhang, M. et al. Angiotensin IV attenuates diabetic cardiomyopathy via suppressing FoxO1-induced excessive autophagy, apoptosis and fibrosis. Theranostics. 11(18), 8624–8639 (2021).
Acknowledgements
We express our sincere gratitude to those who helped us a lot during our writing process.
Funding
This study received financial support from multiple sources, including grants from the National Natural Science Foundation of China (Nos. 82170716 and 81870333), the National Key Research and Development Program of China (No. 2022YFC2505400), as well as Key Project of the Zhejiang Provincial Natural Science Foundation (491030-N22302ZJ).
Author information
Authors and Affiliations
Contributions
Conceptualization, J.Q. and L.Z.; methodology, J.Q. and C.L.; software, J.Q. and C.L.; validation, J.Q., L.Z., R.F., and H.Z.; formal analysis, J.Q..; investigation, J.Q. and Y.L.(Yaheng Li); writing—original draft preparation, J.Q. and J.W.; writing—review and editing, J.Q. and C.H.; visualization, C.Y. and J.W.; supervision, Y.L.(Yaheng Li); project administration, Y.L.(Yafeng Li); funding acquisition, Y.L.(Yafeng Li). All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
The research adhered to the ethical principles outlined in the Declaration of Helsinki and received approval from the Institutional Review Board at Shanxi Provincial People’s Hospital (Fifth Hospital) of Shanxi Medical University. The protocol code for this study is 2021–250, and it was approved in May 2021. In accordance with ethical standards, informed consent was obtained from all participants involved in the study. Our research did not include any individuals under the age of 18.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Qing, J., Zhang, L., Fan, R. et al. GPX4 expression changes in proximal tubule cells highlight the role of ferroptosis in IgAN. Sci Rep 15, 3886 (2025). https://doi.org/10.1038/s41598-025-87228-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-87228-9