Abstract
To investigate the mechanism of ultra-high dose rate pulsed radiation in radiation-induced lung injury (RILI), providing an experimental and theoretical basis for the application of FLASH-RT (FLASH-radiotherapy) in radiotherapy. C57BL/6J mice were randomly divided into three groups: a sham group, a FLASH-irradiated group, and a CONV-irradiated group. A whole-body irradiation with a single dose of 3 Gy (200 Gy/s for FLASH group and 0.3 Gy/s for CONV group) using electron rays was used to establish models of lung injury. After 3 months, lung tissues were stained with HE and Masson stains to observe pathological changes in lung tissue and subjected to 4D-Fast DIA quantitative proteomics, with the sequencing data validated by Western blot and immunofluorescence. The mice in FLASH group had less lung tissue damage and lower levels of fibrosis compared to the CONV group. Proteomic sequencing showed significant differences in CCT6b protein expression between the two irradiation groups. As verified by the WB and immunofluorescence assays, the expression level of CCT6b was significantly reduced in the CONV group of mice compared with the SHAM and FLASH groups. With the down-regulation of CCT6b, there was a notable decrease in the expression of E-cadherin, accompanied by an increase in the expression of α-smooth muscle actin and Vimentin. The differential response in the level of lung fibrosis caused by the two types of radiation may be related to the level of CCT6b expression, but the specific mechanism of action needs to be further investigated.
Similar content being viewed by others
Introduction
Radiotherapy is one of the most important means of treating thoracic malignant tumors such as lung cancer, esophageal cancer, breast cancer, etc. While it enhances patient survival rates and extends lifespan, those with poor prognoses often suffer from radiation-related complications. Radiation-induced lung injury (RILI) stands as a major complications of radiation sickness, particularly prevalent in lung cancer radiotherapy, and is a key area of research in radiotherapy1,2,3. Ionizing rays aims to eradicate tumor cells but concurrently exposes the paraneoplastic and adjacent normal tissues to potentially damaging levels of radiation, significantly compromising lung function in treated patients and impacting the efficacy of thoracic tumor treatments. More serious patients may also develop symptoms of chronic pulmonary heart disease, such as dyspnea and cyanosis, leading to a progressive decrease in lung function until respiratory failure4,5. Therefore, RILI significantly impairs the therapeutic outcome of thoracic tumor treatment and restricts advancements in the field of thoracic tumor therapy. At present, various precision radiotherapy techniques, such as intensity-modulated radiation therapy, have been widely used in the clinic, but the tumor irradiation dose is still limited by the tolerance dose of normal tissues6. Additionally, recent research has revealed that FLASH-radiotherapy (FLASH-RT), as an emerging technology, holds great promise in tumor control and protection of normal tissues7,8,9,10.
FLASH-RT is a novel type of radiotherapy employing ultra-high dose rate (UHDR) pulsed radiation, with an average dose rate typically exceeding 40 Gy/s, which is significantly higher by several orders of magnitude compared to the conventional dose-rates (CONV) utilized in clinical settings11,12. UHDR pulsed radiation can enhance the tolerance of normal tissues to the radiation dose, enabling the administration of higher radiation dose specifically to tumors, which is considered a promising method for tumor radiotherapy13. In recent years, several studies have revealed that mice irradiated with UHDR pulsed radiation exhibit significant lower lung tissue fibrosis and higher survival rates compared to those irradiated with CONV radiation during the same period14,15. However, the underlying biological mechanism responsible for this effect remains elusive and necessitates further in-depth research.
We employed FLASH and CONV electron rays to administer 3 Gy of whole-body irradiation to mice. Subsequently, we harvested lung tissues from these mice at one, two and three months after irradiation for pathological sectioning. Our findings revealed that the severity of lung damage progressively escalated with time in both irradiation groups, with a significant difference between the two groups after 3 months (Supplementary Fig. 1). Specially, the extent of lung damage in the FLASH group was significantly lower compared to the CONV group. In order to analyze the biological mechanisms of FLASH effects at the molecular level, we performed a 4D-FastDIA quantitative proteomics study on lung tissues from mice 3 months after irradiation. Unlike the traditional DDA (data-dependent acquisition mode) technique, the DIA (Data-independent Acquisition) technique divides the entire full scanning range of the mass spectrum into a number of windows, and selects, fragments, and detects all ions in each window at high speed and in a cyclic manner. Thus, full fragmentation information for all ions in the sample can be obtained without omission, with much better data utilization and fewer missing values. Our objective was to identify the key differential molecules in mouse lung tissue, which we subsequently experimentally validated, aiming to furnish a theoretical cornerstone and empirical foundation for implementation of FLASH-RT in radiotherapy.
Results
The pathological changes in mouse lung tissue induced by FLASH radiation milder than those induced by CONV radiation
The HE analysis showed that the lung tissue of the sham mice was structurally normal, characterized by intact alveolar walls, absence of exudates within the alveolar lumen, and no inflammatory cell infiltration in the alveolar septa. However, the lung tissue of mice in the CONV group exhibited a significant aggregation of inflammatory cells (As indicated by the arrows in Fig. 1A), accompanied by evident thickening of the alveolar septa and alveolar collapse (As indicated by the asterisk in Fig. 1A). In contrast, the lung tissue of mice in the FLASH group demonstrated only minimal inflammatory cell infiltration and partial thickening of the alveolar septum (Fig. 1A). Furthermore, the pathological damage score in the lung tissue of the CONV group was markedly elevated compared to that of the FLASH group (Fig. 1B). Masson staining showed prominent collagen deposition in the alveolar and peribronchiolar regions in the CONV group, whereas collagen deposition was significantly reduced in the FLASH group compared to the CONV group (Fig. 1A,C). So, the degree of lung injury and fibrosis were notably lower in the FLASH group compared to the CONV group at the three-month point following radiation exposure.
Comparative analysis of HE staining and Masson staining in mouse lung tissues at three-month point after FLASH and CONV 3 Gy whole-body radiation (n = 3). (A) HE staining of mouse lung tissues displayed on the left side and Masson staining displayed on the right side. (B) Pathological damage score of lung tissues. (C) Results of Masson staining analysis.
Overview of proteomics testing
To explore the potential biological mechanism of FLASH electron radiation in mice, we performed 4D-DIA quantitative proteomics sequencing on individual sets of lung tissue samples. The sample quality was evaluated, revealing that the molecular weight of all detected proteins and the lengths of all peptides were within normal ranges, indicative of a stable overall sample condition (Supplementary Fig. 2). A Principal component analysis (PCA) was conducted on the protein quantification data, and the resulting PCA plots were visualized. These plots revealed significant intergroup differences and intragroup correlations for each sample group (Fig. 2A), conforming the good reproducibility and reliability of our proteomics data.
(A) The PCA plot of protein expression correlation for all samples displays the degrees of interpretation of PC1 and PC2 on the horizontal and vertical axes, respectively. A larger value indicates a higher degree of interpretation. The degree of clustering within each group reflects the similarity or reproducibility of the samples within that group, with duplicates tending to cluster together, indicating good duplication or reproducibility among the grouped samples (n = 6). (B) Volcano plots showcasing differential protein expression between the FLASH and CONV groups. In this plot, each point represents an individual gene. The horizontal coordinate signifies the logarithm of the fold change in gene expression between the two groups, while the vertical coordinate represents the negative logarithm of the statistical significance associated with the observed change in gene expression. Blue dots signify proteins that are down-regulated and differentially expressed, while red dots represent proteins that are up-regulated and differentially expressed, on the other hand, grey dots represent genes with no change. (C) Venn diagram illustrating overlapping differentially expressed proteins. The Venn diagram shows the differentially expressed proteins in different comparative combinations, and the number on each region represents the number of proteins under the corresponding classification, where the overlapping region indicates the number of differentially expressed proteins shared between related combinations in the region. (D) The heatmap displays the clustering of differentially expressed protein. The horizontal coordinates correspond to the names of the samples along with their clustering results. The vertical coordinates represent the clustering results of differentially expressed proteins. Within the heatmap, red color indicates high expression levels, blue color represents low expression levels, and gray signifies where quantification is not possible for the corresponding samples.
Identification of differentially expressed proteins
The sequencing outcomes were analyzed in order to investigate the effects of two types of rays on the mouse lung tissue. A comprehensive identification was made, yielding a total of 8192 quantitative proteins and 66,315 unique peptides. Differential proteins were screened through intergroup comparisons by comparing the CONV group with the SHAM group, the FLASH group with the SHAM group, and the FLASH group with the CONV group, respectively. A total of 254 differential proteins were screened for relative quantification in the CONV group compared with the SHAM group, with 214 being up-regulated and 40 being down-regulated. In the FLASH group versus SHAM group comparison, 70 differential proteins were screened, comprising 54 up-regulated and 16 were down-regulated. Furthermore, when comparing the FLASH group to CONV group, 154 differential proteins were relatively quantified, of which 37 were up-regulated and 117 were down-regulated. The up and down-regulated differentially expressed proteins (DEPs) were presented as volcano plots (Supplementary Fig. 3, Fig. 2B). In addition, we evaluated the expression patterns of DEPs in the samples by cluster analysis. Figure 2C shows the Venn diagram of protein expression in the SHAM, CONV, and FLASH groups. The cluster analysis depicted in Fig. 2D shows the different expression patterns among the three groups. These findings suggest that the two types of rays significantly affected the protein expression patterns in mouse lung tissues, potentially modulating their biological functions. Furthermore, in the comparison between the FLASH group and the CONV group, we found a notable differential expression of a protein closely related to fibrosis, CCT6b.
FLASH and CONV radiation affect the expression of fibrosis-related proteins in mouse lung tissues
Based on the above sequencing results, we found that the expression of CCT-6b was significantly reduced in the CONV group (Fig. 3A). The down-regulation of CCT-6b expression could serve as a key indicator of the activation process in fibroblasts, suggesting that CCT-6b may be involved in the fibrotic process of fibroblasts and potentially function as an antifibrotic factor. The WB results (Fig. 3B) showed that compared with mice in the SHAM group and FLASH group, the expression level of CCT-6b was significantly down-regulated in mice in the CONV group (Fig. 3C). At the same time, the expression of E-cadherin was significantly reduced in mice in the CONV group (Fig. 3D), and the expression of Vimentin (Fig. 3E) and α-SMA (Fig. 3F) was significantly increased. Immunofluorescence results (Fig. 3G) also confirmed a significant decrease in E-cadherin expression (Fig. 3H) and a significant increase in Vimentin expression (Fig. 3I) in the CONV group compared to the other two groups. These results suggest that FLASH-RT reduces the down-regulation of CCT6b compared with CONV-RT, which attenuates epithelial-mesenchymal transition (EMT) and thus reduces the degree of lung fibrosis in mice.
Western blotting detection and quantification of CCT-6b, E-cadherin, α-SMA and Vimentin in mouse lung tissues after 3 months of FLASH-RT and CONV-RT irradiation. Immunofluorescence staining results of E-cadherin, Vimentin. (A) Box plots showing the expression of CCT-6b in the three groups. (B) Western blotting results of CCT-6b, E-cadherin, α-SMA and Vimentin western blot results (n = 6). (C) Relative expression of CCT-6b protein, (D) Relative expression of E-cadherin protein. (E) Relative expression of Vimentin protein. (F) Relative expression of α-SMA protein. (G) Immunofluorescence results for E-cadherin and Vimentin (n = 3). (H) Semi-quantitative analytical results of E-cadherin. (I) Semi-quantitative analytical results of Vimentin.
Enrichment analysis of differential proteins
The GO secondary classification was performed for the screened differential proteins (Supplementary Fig. 4). The different proteins between the CONV and SHAM groups were mainly enriched in ribosome biosynthesis, vesicle transport, and electron transfer in the respiratory chain (Supplementary Fig. 5A); the different proteins between the FLASH and SHAM groups were mainly related to angiogenesis and ribosome composition (Supplementary Fig. 5B); the different proteins between the FLASH and CONV groups were mainly related to mitochondrial composition, assembly of respiratory chain complexes, and mitochondrial transport (Fig. 4A). Protein-protein interaction enrichment analysis of differential proteins between the two irradiated groups was performed through the Metascape website using the STRING, BioGrid, OmniPath, and InWeb_IM databases, and the results showed that there were four main types of protein-interacting clusters, among which the proteins in the MCODE2 cluster were mainly related to mitochondrial function (Fig. 4B).
(A) Metascape clustering annotation of differentially expressed proteins between FLASH and CONV groups is shown in color code, where nodes sharing the same code are close to each other. (B) Differential protein interaction network and MCODE components between FLASH and CONV groups. (C) KEGG analysis of differentially expressed proteins in FLASH and CONV groups. The vertical axis is the KEGG pathway description information, and the horizontal axis is the degree of functional enrichment (Fold enrichment) after Log2 transformation, the larger the value indicates the higher degree of enrichment; the color of the point indicates the P value of enrichment significance, the bluer the color represents the stronger the significance of the enrichment; the size of the point indicates the number of differentially expressed proteins in the KEGG pathway, and the larger the point indicates that the more differentially expressed proteins are involved. The KEGG-related access points in this article are copyrighted.
KEGG enrichment analysis of differentially expressed proteins in lung tissue samples from mice in the FLASH, CONV and SHAM groups showed that the different proteins between the CONV and SHAM groups were mainly involved in IL-17 signaling pathway, autophagy-related pathway, retrograde endogenous cannabinoid signaling, and autoimmune-related pathway (Supplementary Fig. 6A); the different proteins between FLASH and SHAM groups were mainly involved in PPAR signaling pathway, lipolysis regulation-related pathway, oxytocin signaling pathway (Supplementary Fig. 6B); the different proteins between FLASH and CONV groups were mainly involved in sphingolipid metabolism, ret rograde endogenous cannabinoid signaling pathway, etc. (Fig. 4C).
Discussion
Flash-RT is an innovative radiation therapy technique that boasts promising clinical application prospects, mainly characterized by its capability to deliver ultra-high dose rate irradiation exceeding 40 Gy/s. Preclinical data have demonstrated that FLASH-RT significantly enhances the tolerance of normal tissues to radiation compared with CONV-RT. This effect significantly reduces radiation-induced toxicity in normal tissues without affecting the tumor-killing effect16,17,18. Although this new technology holds great promise, the radiobiological mechanism of FLASH-RT remains unclear to date, significantly hindering its clinical translation.
In this study, a radiation damage model was established using FLASH and CONV radiations with a whole-body irradiation at 3 Gy. After 3 months, lung tissues from the mice were harvested, and histomorphology was observed using HE staining and Matson’s trichrome staining. These analyses revealed that the degree of fibrosis in lung tissues was significantly lower in the FLASH group compared to the CONV irradiation group. Additionally, by analyzing sequencing results, we observed a significantly lower expression level of CCT6b in the CONV group compared to both the FLASH and SHAM groups. To further validate the role of CCT6b in radiological lung injury, the expression level of CCT6b protein was further verified using the WB method.
CCT (TCP-1 ring complex), also known as molecular chaperone TRiC, is a type II molecular chaperonin present in eukaryotic cells. It is a cylindrical structural body consisting of two rings stacked back-to-back, each containing eight homologous subunits (CCT1-8) with similar three structural domains19. CCT acts as a molecular chaperone, using actin, microtubule proteins and cell cycle regulatory proteins as specific substrates, promoting the correct folding of about 10% of proteins in eukaryotic cells in an ATP-dependent manner and participating in many biological processes20,21,22. Some studies have confirmed that CCT7 is significantly elevated in fibrotic diseases, and cellular fibrosis indexes are significantly decreased after interfering with CCT7 expression23. CCT6b is a subunit of CCT6, and some scholars have found that CCT6b is significantly elevated in mucosal wounds, which may affect fibroblast function24. Other researchers have found low CCT6b protein and gene expression in arthrofibrosis models, and CCT6b may be involved in the fibrotic process as an antifibrotic factor25. However, the role of CCT6b in pulmonary fibrosis, especially radiographic pulmonary fibrosis, has not been studied. In lung fibrosis, alveolar type II epithelial cells serve as a pivotal source of myofibroblasts, a transformation known as epithelial-mesenchymal transition (EMT). This EMT process can be induced by the unfolded protein response, thereby accelerating the progression of fibrosis26,27,28. Whereas the most important function of the CCT family is to facilitate proteins folding, we hypothesized that CCT6b might potentially influence the process of EMT. EMT occurs with loss of intercellular tight junctions, downregulation of epithelial-specific markers such as E-cadherin, and increased expression of mesenchymal markers including α-smooth muscle actin (α-SMA) and Vimentin. Therefore, we examined the expression levels of CCT6b, α-SMA, E-cadherin, and Vimentin in mouse lung tissues and found that down-regulation of CCT6b led to decreased expression of E-cadherin and enhanced expression of α-SMA and Vimentin29. These findings suggest that FLASH radiation, in comparison to CONV radiation, mitigates the down-regulation of CCT6b, thereby inhibiting the irradiation-induced EMT process and consequently reducing the severity of radiation-induced lung fibrosis.
Furthermore, we found that the main enrichment pathways of differential proteins between the two irradiation groups were mostly related to the respiratory chain which is located on the inner mitochondrial membrane as a role of representing the most basic function of mitochondria. The respiratory chain is an important component of the mitochondria, and is also the biological basis of the electron transport chain30. Impairment of the assembly and delivery of the respiratory chain will affect the function of the electron transport chain, increase the level of intracellular free radicals, and exacerbate the damage of the respiratory chain, which will ultimately affect the structure and function of the mitochondria31,32,33. Impairment of mitochondrial structure and function is often accompanied by an abnormal increase in reactive oxygen species (ROS)34,35. Excessive ROS can damage cellular biomolecules such as lipids and DNA36, induce oxidative stress and lead to apoptosis, senescence, and gene mutation, and activate excessive inflammatory mediators, ultimately leading to pulmonary fibrosis37,38. We hypothesize that the rapid oxygen depletion effect of FLASH-RT can limit the generation of ROS, thereby mitigating damage to the respiratory chain. This, in turn, means reduced impact on the structure and function of mitochondria, potentially resulting in a milder level of lung fibrosis in mice. However, further experimental investigations are essential to establish a robust foundation for this hypothesis.
Conclusion
The present study demonstrated that FLASH radiation caused less damage to the lung tissue of mice compared with CONV radiation. And the differential response in the level of lung fibrosis caused by the two rays may be related to the expression level of CCT6b, which seems to be a key molecule in the lung fibrosis caused by CONV radiation, and FLASH radiation may lightly cause lung fibrosis by maintaining the expression level of CCT6b. The proteomics results also showed that the effects of the two rays on mouse lung tissues were mainly related to the mitochondrial respiratory chain, and the specific mechanism of their effects on mitochondrial function will be a key issue for further exploration and study in the future.
Materials and methods
Animals
Ethical approval for all experiments in this study was obtained from the Ethics Committee of the Laboratory Animal Center of the Military Medical Research Institute under the approval number IACUC-DWZX-2024-520.All experiments were conducted in accordance with ARRIVE guidelines and regulations. All methods were performed in accordance with the relevant guidelines and regulations.
Eight-week-old male C57BL/ 6 J mice, with an average body weight of 20 ± 0.9 g, were purchased from SPF (Beijing) Biotechnology Co Ltd (Beijing, China). All procedures for use of animals and their care were approved by the Animal Laboratory Review Board of the Center for Laboratory Animals, Military Medical Research Institute. The mice were housed in standard conditions, five per cage, at a room temperature of 22 ± 2 °C, with a 12-hour light-dark cycle and a relative humidity maintained between 50 and 60%. The animals had ad libitum to standard food (SPF Beijing) Biotechnology Co., Ltd., Beijing, China) and water. Following a 7-day acclimatization, 60 mice were randomly divided into three groups: a sham group (n = 20), a FLASH-irradiated group (n = 20), and a CONV-irradiated group (n = 20).
Irradiation systems and dosimetry
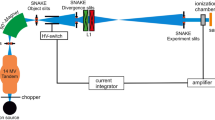
The CONV and UHDR (UHDR standing for FLASH-irradiated group) electron pulse radiation was administered to mice utilizing the electron line UHDR vertical test platform, situated within the Department of Engineering Physics, Tsinghua University (Fig. 5). The electron energy employed was 6 MeV, and both types of rays were administered at 3 Gy, with an average dose rate of 0.3 Gy/s for CONV and 200 Gy/s for UHDR respectively. To ascertain the precise radiation dose delivered and ensure the overall dose accuracy, the Gafchromic TM EBT3 film from Ashland Company, USA, was utilized for dosimetry measurements (Table 1). The sham group was treated in the same way, but without radiation. The radiation device, irradiation environment and specific experimental workflow arrangement are shown in (Fig. 5).
Lung HE staining and Masson’s trichrome staining
After anesthetizing mice using 1% concentration of sodium pentobarbital, mice were euthanized by cervical dislocation. Mouse lung tissue was then taken, and the left lung was fixed, dehydrated, and paraffin-embedded. Then, the tissue sections were sliced into 4 μm-thick slices, which underwent both hematoxylin-eosin staining and Masson’s trichrome staining. The morphology of the lung tissue was then observed under a 400 × microscope (OLYMPUS BX41; Olympus Corporation, Japan). A semi-quantitative analysis of the lung tissue inflammation was performed according to the Szapiel scoring criteria39,40, detailed as follows: score 0: absence of alveolitis. Alveolitis. score 1: mild alveolitis, mononuclear cell infiltration was seen locally and in the near thorax, with an area of less than 20% of the whole lung, and the alveolar structure was more or less normal. score 2: moderate alveolitis, with the lesion area accounting for 20-50% of the whole lung. score 3: severe alveolitis and pulmonary fibrosis, with the area of the lesion being greater than 50% of the whole lung, with occasional mononuclear cells and/or hemorrhage in the alveolar lumen causing solid lesions.
4D-DIA proteomics analysis
Protein extraction
The samples, retrieved from a temperature of −80℃, were weighed and transferred into a pre-cooled mortar liquid nitrogen. Subsequently, they were ground into a fine powder using liquid nitrogen. To each group of samples, four volumes of lysis buffer (8 M urea, 1% protease inhibitor) were added, and the mixture was subjected to ultrasonic lysis for complete disruption of the cellular structure. The samples were centrifuged at 4℃ and 12,000 g for 10 min to remove the cellular debris. Following this, the clarified supernatant was transferred to a fresh centrifuge tube, and the protein concentration was quantified using the BCA (Beyotime Biotechnology, China) assay kit, adhering to the manufacturer’s protocol.
Trypsin digestion
The protein sample was added with 1 volume of pre-cooled acetone, vortexed to mix, and added with 4 volumes of pre-cooled acetone, precipitated at -20℃ for 2 h. The precipitate was washed 2 ~ 3 times with the pre-cooled acetone. The protein sample was then redissolved in 200 mM TEAB and ultrasonically dispersed. Trypsin was added at 1:50 trypsin-to-protein mass ratio for the first digestion overnight. The sample was reduced with 5 mM dithiothreitol for 30 min at 56 ℃ and alkylated with 11 mM iodoacetamide for 15 min at room temperature in darkness. Finally, the peptides were desalted by Strata X SPE column.
Liquid chromatography-mass spectrometry analysis
The peptides were separated using a NanoElute ultra-high performance liquid chromatography (UHPLC) system after being solubilized with liquid chromatography mobile phase (A) Home-made analytical column with integrated spray tip (100 μm i.d. × 25 cm) packed with 1.9 μm/120 Å ReproSil-PurC18 resins (Dr. Maisch GmbH, Ammerbuch, Germany). Mobile phase A was an aqueous solution containing 0.1% formic acid and 2% acetonitrile; mobile phase B was an acetonitrile-water solution containing 0.1% formic acid. The liquid-phase gradient was set at 0–14 min, 6–24% B; 14–16 min, 24–35% B; 16–18 min, 35–80% B; and 18–20 min, 80% (B) The flow rate was maintained at 500 nl/min. The peptides were separated by an UHPLC system and injected into a Capillary ion source for ionization and then into a timsTOF Pro mass spectrometer for data acquisition. The ion source voltage was set to 1.75 kV, and both the peptide parent ion and its secondary fragments were detected and analyzed using TOF. The data acquisition mode used was the data-independent parallel accumulation serial fragmentation (dia-PASEF) mode, with the primary MS scan range set to 300–1500 m/z, one primary MS acquisition followed by 20 PASEF mode acquisitions, and the secondary MS scans in the interval of 400–850, with a window of 7 m/z each.
Database search
The DIA data were processed using DIA-NN search engine (v.1.8). Tandem mass spectra were searched against Mus_musculus_10090_SP_20231220.fasta (17191 entries) concatenated with reverse decoy database. Trypsin/P was specified as cleavage enzyme allowing up to 1 missing cleavages. Excision on N-term Met and carbamidomethyl on Cys were specified as fixed modification. FDR was adjusted to < 1%.
Bioinformatics analysis
Gene ontology (GO) analysis, the identified proteins were analyzed by using eggnog-mapper software to extract the GO ID of each protein based on the EggNOG database, and then the proteins were categorized and annotated according to the cellular components, molecular functions, and bioprocesses. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis41,42,43 integrates currently known information on protein interaction networks, such as pathways and related complexes (Pathway database), genes and gene products (Gene database), and intrabiotic complexes and related reactions (Compound and Reaction database). The protein pathways were annotated based on the KEGG pathway database, and the identified proteins were subjected to BLAST comparison (blastp, evalue ≤ 1e-4), and for the BLAST comparison results of each sequence, the one with the highest comparison score was selected for annotation.
Western blot
All samples were lysed with RIPA buffer to obtain protein extracts, and then the samples were centrifuged at 4 °C, 12,000 g for 15 min. The protein content of the supernatant was determined using a BCA protein assay kit (Thermo Fisher Scientific Inc, USA). Equal amounts of protein were diluted in RIPA lysis buffer and then boiled in 1× super sampling buffer. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and after electrophoresis, they were transferred to PVDF membranes, which were sealed with 5% skimmed milk at room temperature for 1 h. The membranes were infiltrated with specific primary antibodies overnight at 4 °C, shaken at room temperature for 1 h, and then incubated with secondary antibodies the next day for 1 h. The membrane was incubated with a secondary antibody using an ECL 1228 Plus (Beijing Lablead Biotechnology Co, China) and Fluor Chem R Western blot detection system (Protein Simple, USA) and the bands were quantified using Image J software. Primary and secondary antibodies for Western blot are listed in Table 2.
IF and antibodies
Mouse lung tissue was embedded with OCT and then cut into 8 µ m thick sections using a freezing microtome (CM 1950, Leica, Germany). Sections were rinsed three times with 1xPBS buffer at 4 °C for 5 min each. Sections were closed with 10% bovine serum albumin (BSA) for an additional 1 h at room temperature protected from light, followed by staining with 3% bovine serum albumin diluted primary antibody (rabbit anti-E-cadherin 1:200, rabbit anti- Vimentin 1:200) for 10 h at 4 °C. After one hour of rewarming at room temperature the sections were rinsed three times with 1xPBS buffer at 4 °C for 5 min each time. Immunofluorescent secondary antibody was added and the sections were restained for 1 h at room temperature. Finally, the sections were fixed with a sealer containing DAPI. The sections were observed with a confocal microscope (Nikon, Japan) and images were collected using NikonMosaic 1.6 software and correlation analysed using ImageJ software. The following experimental antibodies were used: rabbit E-cadherin antibody (10366-1-AP, Proteintech Group, China), rabbit Vimentin antibody (80541-7-RR, Proteintech Group, China), goat anti-rabbit IgG (ab150077, Abcam, USA).
Statistical analysis
All data are presented as mean ± standard error of the mean and all graphs were generated using GraphPad Prism 8.0 software (San Diego, CA, USA). Normality of the data was first tested using the Shapiro-Wilk test. Normally distributed data were analyzed using a two-tailed Student’s t-test or one-way analysis of variance (ANOVA), and multiple comparisons were performed using the Tukey test. For non-normally distributed data, non-parametric analysis with a p < 0.05 was considered statistically significant. Significant statistical differences were set at p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD057784.
References
An, N. et al. Inhibition of Rac1 attenuates radiation-induced lung injury while suppresses lung tumor in mice. Cell. Death Discov. 8, 26 (2022).
Guo, H. et al. Activation of Nrf2/ARE pathway by anisodamine (654-2) for inhibition of cellular aging and alleviation of radiation-induced lung injury. Int. Immunopharmacol. 124, 110864 (2023).
Li, L. et al. NVP-AUY922 alleviates radiation-induced lung injury via inhibition of autophagy-dependent ferroptosis. Cell. Death Discov. 8, 86 (2022).
Rabbani, Z. N. et al. Overexpression of extracellular superoxide dismutase reduces acute radiation induced lung toxicity. BMC Cancer 5, 59 (2005).
Wei, L., Zhang, J., Yang, Z. L. & You, H. Extracellular superoxide dismutase increased the therapeutic potential of human mesenchymal stromal cells in radiation pulmonary fibrosis. Cytotherapy 19, 586–602 (2017).
Lühr, A. et al. Radiobiology of proton therapy: results of an international expert workshop. Radiother Oncol. 128, 56–67 (2018).
Montay-Gruel, P. et al. Hypofractionated FLASH-RT as an effective treatment against glioblastoma that reduces neurocognitive side effects in mice. Clin. Cancer Res. 27, 775–784 (2021).
Okoro, C. M., Schüler, E. & Taniguchi, C. M. The therapeutic potential of FLASH-RT for pancreatic cancer. Cancers (Basel) 14, 1167 (2022).
Limoli, C. L. et al. The sparing effect of FLASH-RT on synaptic plasticity is maintained in mice with standard fractionation. Radiother. Oncol. 186, 109767 (2023).
Iturri, L. et al. Proton FLASH radiation therapy and immune infiltration: evaluation in an orthotopic glioma rat model. Int. J. Radiat. Oncol. Biol. Phys. 116, 655–665 (2023).
Zhou, S. et al. Minimum dose rate estimation for pulsed FLASH radiotherapy: a dimensional analysis. Med. Phys. 47, 3243–3249 (2020).
Schüler, E. et al. Ultra-high dose rate electron beams and the FLASH effect: from preclinical evidence to a new radiotherapy paradigm. Med. Phys. 49, 2082–2095 (2022).
Bourhis, J. et al. Clinical translation of FLASH radiotherapy: why and how. Radiother Oncol. 139, 11–17 (2019).
Dai, Y. et al. Fractionated FLASH radiation in xenografted lung tumors induced FLASH effect at a split dose of 2 gy. Int. J. Radiat. Biol. 99, 1542–1549 (2023).
Favaudon, V. et al. Ultrahigh dose-rate FLASH irradiation increases the differential response between normal and tumor tissue in mice. Sci. Transl. Med. 6, 245ra93 (2014).
Buonanno, M., Grilj, V. & Brenner, D. J. Biological effects in normal cells exposed to FLASH dose rate protons. Radiother. Oncol. 139, 51–55 (2019).
Fouillade, C. et al. FLASH irradiation spares lung progenitor cells and limits the incidence of radio-induced senescence. Clin. Cancer Res. 26, 1497–1506 (2020).
Shi, X. et al. FLASH X-ray spares intestinal crypts from pyroptosis initiated by cGAS-STING activation upon radioimmunotherapy. Proc. Natl. Acad. Sci. USA 119, e2208506119 (2022).
Liu, W. et al. Current understanding on the role of CCT3 in cancer research. Front. Oncol. 12, 961733 (2022).
Yam, A. Y. et al. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 15, 1255–1262 (2008).
Meyer, A. S. et al. Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell 113, 369–381 (2003).
Joachimiak, L. A., Walzthoeni, T., Liu, C. W., Aebersold, R. & Frydman, J. The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell 159, 1042–1055 (2014).
He, R. et al. Chaperonin containing T-complex polypeptide subunit eta is a potential marker of joint contracture: an experimental study in the rat. Cell. Stress Chaperones 20, 959–966 (2015).
Satish, L. et al. Chaperonin containing T-complex polypeptide (CCT) subunit expression in oral mucosal wounds and fibroblasts. Cell. Stress Chaperones 16, 675–680 (2011).
Yi, X. et al. Overexpression of chaperonin containing T-complex polypeptide subunit zeta 2 (CCT6b) suppresses the functions of active fibroblasts in a rat model of joint contracture. J. Orthop. Surg. Res. 14, 125 (2019).
Bartis, D., Mise, N., Mahida, R. Y., Eickelberg, O. & Thickett, D. R. Epithelial-mesenchymal transition in lung development and disease: does it exist and is it important. Thorax 69, 760–765 (2014).
Hewlett, J. C., Kropski, J. A. & Blackwell, T. S. Idiopathic pulmonary fibrosis: epithelial-mesenchymal interactions and emerging therapeutic targets. Matrix Biol. 71–72, 112–127 (2018).
Nagaraja, S. S. & Nagarajan, D. Radiation-induced pulmonary epithelial-mesenchymal transition: a review on targeting molecular pathways and mediators. Curr. Drug Targ. 19, 1191–1204 (2018).
Kim, S. Y. et al. Efferocytosis and enhanced FPR2 expression following apoptotic cell instillation attenuate radiation-induced lung inflammation and fibrosis. Biochem. Biophys. Res. Commun. 601, 38–44 (2022).
Zeng, M. et al. Domesticated and optimized mitochondria: mitochondrial modifications based on energetic status and cellular stress. Life Sci. 265, 118766 (2021).
Chenna, S., Koopman, W., Prehn, J. & Connolly, N. Mechanisms and mathematical modeling of ROS production by the mitochondrial electron transport chain. Am. J. Physiol. Cell. Physiol. 323, C69–C83 (2022).
Cadenas, E. & Davies, K. J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol. Med. 29, 222–230 (2000).
Murphy, M. P. Understanding and preventing mitochondrial oxidative damage. Biochem. Soc. Trans. 44, 1219–1226 (2016).
Cao, Z., Lindsay, J. G. & Isaacs, N. W. Mitochondrial peroxiredoxins. Subcell. Biochem. 44, 295–315 (2007).
Selivanov, V. A. et al. Reactive oxygen species production by forward and reverse electron fluxes in the mitochondrial respiratory chain. PLoS Comput. Biol. 7, e1001115 (2011).
Indo, H. P. et al. Evidence of ROS generation by mitochondria in cells with impaired electron transport chain and mitochondrial DNA damage. Mitochondrion 7, 106–118 (2007).
Chistiakov, D. A., Shkurat, T. P., Melnichenko, A. A., Grechko, A. V. & Orekhov, A. N. The role of mitochondrial dysfunction in cardiovascular disease: a brief review. Ann. Med. 50, 121–127 (2018).
Liu, X. & Chen, Z. The pathophysiological role of mitochondrial oxidative stress in lung diseases. J. Transl. Med. 15, 207 (2017).
Yang, D. et al. The histone methyltransferase DOT1L is a new epigenetic regulator of pulmonary fibrosis. Cell. Death Dis. 13, 60 (2022).
Ma, J. et al. Neutralization of interleukin-11 attenuates silica particles-induced pulmonary inflammation and fibrosis in vivo. J. Environ. Sci. (China) 126, 772–783 (2023).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y. & Kawashima, M. Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Acknowledgements
Thanks to Professor Hao Zha from Tsinghua University for his help and support in irradiation and dose calculation.
Author information
Authors and Affiliations
Contributions
MHL, GFD, and CZW designed experiments. MHL, LHZ, JX, XYW, and XML performed experiments. MHL, ANG, DFY, DFZ, SW, and BS analyzed data.MHL wrote the manuscript. MHL, GFD, and CZW participated in the writing and subsequent revision of the article.All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, M., Zhang, L., Gao, A. et al. Molecular mechanisms of lung injury from ultra high and conventional dose rate pulsed radiation based on 4D DIA proteomics study. Sci Rep 15, 5425 (2025). https://doi.org/10.1038/s41598-025-87247-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-87247-6