Abstract
Isoliquiritigenin (ISL), a flavone isolated from licorice, has been demonstrated to exhibit anti-inflammatory and antioxidant properties in the treatment of Alzheimer’s disease (AD). However, the molecular details of the contribution of ISL to AD remain largely elusive. The present study aimed to investigate the molecular mechanisms of ISL against AD. In this study, AD targets and ISL targets were collected via different databases. The overlapped targets between AD and ISL were generated with Venny. Then we performed Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analyses on these common targets. The protein-protein interaction (PPI) network was constructed and clusters were obtained using the Molecular Complex Detection (MCODE) and the Cytohubba plugins. Further, molecular docking study was performed for these core targets. Subsequently, the receiver operating characteristic (ROC) curve analysis and the assessment of hub gene expression levels between AD and healthy individuals were used to estimate a possible link between target genes in AD. Finally, experiments were conducted to verify the therapeutic mechanism of ISL in lipopolysaccharide (LPS)-induced BV2 microglial cells. GO and KEGG pathway analysis found that ISL was significantly enriched in regulation of mitogen-activated protein kinase (MAPK) signaling pathway. The PPI network manifested 7 key targets including albumin (ALB), epidermal growth factor receptor (EGFR), solute carrier family 2 member 1 (SLC2A1), insulin-like growth factor 1 (IGF1), mitogen-activated protein kinase 1 (MAPK1), peroxisome proliferator activated receptor alpha (PPARA) and peroxisome proliferator activated receptor gamma (PPAR-γ, PPARG). Molecular docking showed that ISL had high binding affinity with these key targets. The experimental results revealed that ISL decreased extracellular signal-regulated kinase 1/2 (ERK1/2) phosphorylation and increased the expression of PPAR-γ, and suppressed the production of proinflammatory mediators. Our work revealed that ISL might be an effective treatment strategy in the treatment of AD by its anti-inflammatory effect towards microglia through the ERK/PPAR-γ pathway.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is the most common age-related neurodegenerative disorder, characterized by progressive cognitive impairment and abnormal behavior. It is estimated to be approximately 47 million people worldwide suffer from this disease1. Despite considerable progress in AD research in the past decade, there is still no effective treatments to cure or prevent the disease2. Thus, exploring the biological mechanisms of AD and finding successful therapeutic and preventive interventions are the focus of current research.
Although the pathogenic mechanisms that ultimately cause AD are still unknown, the widely recognized central process of two hallmark pathological mechanisms in AD are extracellular abnormal amyloid-beta (Aβ) deposits and intracellular neurofibrillary tangles (NFTs)3. In addition, neuroinflammation has been considered as a crucial factor of AD pathology4. Increasing evidence reveals that inflammation can be routinely observed in preclinical model systems of AD before the appearance of Aβ deposition5,6. As the main factor leading to neuroinflammation of the central nervous system (CNS), microglia have long been implicated in the development and progression of AD pathophysiology5,7. Activated microglia are known to produce pro-inflammatory factors including cyclooxygenase-2 (COX-2), nitric oxide synthase (iNOS), nitric oxide (NO), interleukin-1β (IL-1β), interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α), which in turn triggers neuroinflammation, resulting in CNS damage4. Inflammatory factors, released from activated microglia and overexpressed in proximity to the Aβ plaques8, appears to be protective by increasing Aβ uptake and clearance9. However, as the disease progresses, excessive and chronic inflammatory responses can affect amyloid precursor protein (APP) processing through β-secretase and thus contribute Aβ deposition, which exacerbates neurodegenerative processes10. In contrast, inhibition of microglial activation prevents Aβ accumulation, neuronal death and synaptic impairment4,11. As a consequence, intervention of microglia activation and neuroinflammation may be an effective therapeutic target to restrain the further progression of AD.
Licorice (Glycyrrhiza uralensis Fisch), a widely used herb in traditional Chinese medicine (TCM) as well as food additive12, has broad biological application prospects. It has been found that their active monomers and derivatives play a significant role in improving neuropsychiatric and neurodegenerative diseases, such as anti-oxidation, anti-inflammation, neuroprotection and mitochondrial protection13,14. Isoliquiritigenin (ISL) is a natural flavone found in licorice with good ability to penetrate the blood-brain barrier15. Previous studies have shown that ISL can obviously prevent neuroinflammation and oxidative stress in neurodegenerative diseases such as AD and Parkinson’s disease (PD)16. Although growing evidence indicates a protective role in the treatment of AD, its exact mechanism and molecular targets underlying remain unclear.
With the development of bioinformatics, network pharmacology has been successfully applied to discovery new drug candidates. It is widely accepted that the application of network pharmacology can assist to effectively and systematically explore the drug targets and molecular mechanisms underlying complex diseases17. More and more natural product studies are including this approach with a view to combat problems such as improving therapeutic efficacy and reducing the toxic and side effects18. The network pharmacology is considered as an effective approach to find therapeutic targets of ISL.
In this study, we used network pharmacology, molecular docking and experimental validation to explore the underlying mechanisms of ISL against AD. First, the potential targets of ISL in the treatment of AD were predicted by network pharmacology and the targets were validated through molecular docking. Then, experiments were conducted to prove the regulation of targets by ISL in lipopolysaccharide (LPS)- stimulated BV2 microglial cells. Our results suggested that ISL considerably ameliorated microglial inflammatory response, which may be linked with regulation of the extracellular signal-regulated kinase/peroxisome proliferator activated-receptor gamma (ERK/PPAR-γ) signaling pathway. This study provides a basis for ISL in treating AD.
Materials and methods
Screening of ISL targets
The potential targets of ISL were manually acquired from public databases such as Traditional Chinese Medicine Systems Pharmacology (TCMSP, https://old.tcmsp-e.com/tcmsp.php, Version 2.3, accessed on 20 November 2022) with oral bioavailability (OB) ≥ 30% and drug likeness (DL) ≥ 0.1819, BATMAN-TCM (http://bionet.ncpsb.org.cn/batman-tcm/index.php, version 2.0, accessed on 20 November 2022)20, PharmMapper (http://www.lilab-ecust.cn/pharmmapper/index.html, accessed on 20 November 2022)21 and Swiss target prediction (https://www.swisstargetprediction.ch/, accessed on 20 November 2022)22. Above databases species were limited to Homo Sapiens once the target had been predicted. After merging the results obtained from the 4 databases and excluding nonhuman genes, the rest genes were regarded as potential targets of ISL. Uniprot protein database (https://www.uniprot.org/) was used to retrieve the predicted targets23, and the protein targets and genes of ISL were standardized and integrated.
Collection of AD targets
Identification of critical genes in AD from disease databases
AD targets were collected via different databases to prepare for subsequent analysis. The keywords “Alzheimer’s Disease” were input to public databases such as Online Mendelian Inheritance in Man (OMIM, https://omim.org/, accessed on 20 November 2022)24, PharmGkb (https://www.pharmgkb.org/, accessed on 20 November 2022)25, DrugBank (https://go.drugbank.com/, accessed on 20 November 2022)26, Therapeutic Target Database (TTD, http://db.idrblab.net/ttd/, accessed on 20 November 2022)27 and Genecards (https://www.genecards.org/, accessed on 20 November 2022) to obtain targets for AD28. Then, the results of the five databases were summarized and the duplicates were deleted. All targets were input to Uniprot protein database (https://www.uniprot.org/) to get corresponding gene symbols23.
Identification of critical genes in AD from expression datasets
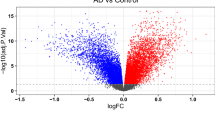
The gene expression omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) is an international public repository that distributes high-throughput gene expression datasets29. Expression profiling data from GSE5281 were then downloaded from the GEO database based on the microarray platform GPL570 (Affymetrix Human Gene Expression Array), which contains 74 samples from healthy individuals and 87 AD samples. GEO2R, an online tool incorporating R/Bioconductor and the Limma package 3.26.8 was used to analyze the raw gene expression data. We used the built-in methods in GEO2R to determine the differentially expressed genes (DEGs) between patients with AD and healthy individuals. Volcano plot was plotted using Bioinformatics platform (https://www.bioinformatics.com.cn/) for AD vs. healthy individuals to represent significant up and down regulated genes. The genes with adjusted p-value < 0.01 and a |log2FC| ≥ 1 were filtered and ranked to obtain the top 100 up and down regulated DEGs. The network of the top 100 DEGs was constructed using STRING database (https://cn.string-db.org/, version 12.0, accessed on 20 November 2022) with interaction scores > 0.4 30 and plotted using Cytoscape software (version 3.7.2, accessed on 20 November 2022) and the heat map was plotted using bioinformatics platform31.
GO and KEGG pathway enrichment analyses
The overlaps between ISL and AD targets generated with Venny (https://bioinfogp.cnb.csic.es/tools/venny/index.html, version 2.1) were then subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. GO, a functional system, is designed to expound gene functions and properties of gene products32, and KEGG pathway analysis designed to systematically analyze gene function links gene lists to higher-order functional information to receive significantly enriched biological pathways33. To explore the biological function of these ISL potential targets in AD, the GO enrichment analysis which consists of BP (biological process) and KEGG pathway analysis were conducted by the Metascape database (https://metascape.org/, version 3.5, accessed on 10 December 2024)34 with the species limited to “Homo sapiens” and p-value < 0.05. The top 20 terms were then selected and further presented visually. The results of GO and KEGG enrichment analyses from Metascape database were visualized by Sangerbox platform (http://vip.sangerbox.com/, version 3.0).
Protein-protein Interaction network construction
The protein-protein interaction (PPI) of the top 100 up and down DEGs from the GSE5281 dataset and the shared targets between ISL and AD targets were retrieved from the STRING database30 with a medium confidence, and the PPI network was then constructed with the use of Cytoscape 3.7.2 software31. To investigate node composition in the PPI network, the cluster analysis was calculated by the Molecular Complex Detection (MCODE) algorithm with default parameters in Cytoscape software. Proteins were annotated using GO terms in the Metascape database34 with the species limited to “Homo sapiens” and p-value < 0.01 to study identified ISL targets at the functional level. To further identify the hub genes with high degree of connectivity for ISL against AD, we used a Cytohubba tool (a plug-in of Cytoscape) to analyze the topological properties of targets based on four parameters of BottleNeck, Betweenness, Radiality and Stress35. Top ten genes of every sub-net were searched, and overlapping genes generated with Venny were chosen as hub genes.
Molecular docking
Molecular docking is a computer structure-based method widely used in drug discovery, which can predict ligand target interactions at molecular level and identify new compounds of therapeutic significance36. To predict the interaction between hub genes and ISL, molecular docking was performed37. Structures of hub genes were retrieved from the RCSB Protein Data Bank (PDB, http://www.rcsb.org, accessed on 20 November 2022) (ALB: 6YG9, EGFR: 1XKK, SLC2A1: 6THA, IGF1: 1TGR, MAPK1: 4QTA, PPARA: 6KAX and PPARG: 7AWC)38. Structure of ISL was retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/)39. The SDF format was converted into PDB format using Open Babel. During the docking process, the water molecules and native ligand groups from the protein structure were removed using the PyMOL 2.6 software, which then predicted the active site for most efficient ligand binding. Further processing of proteins and docking of proteins (10 docking runs were performed for each protein) were performed using AutoDock Tools 1.5.7. The interaction between ISL and the hub genes was evaluated by means of binding energy scores. The obtained docking protein structures were further optimized using PyMOL software.
Analysis of the hub gene expression in GSE5281 dataset and ROC curve
In order to analyze whether the hub genes were dysregulated in AD, the expression level of these DEGs was preliminarily validated in the GSE5281 dataset, which covered the gene expression profiling of brain tissues generated from 74 healthy individuals and 87 AD patients. Normalization and log2 transformation were performed for the raw data. GraphPad Prism 9.0 software was used for graphical visualization. In order to detect the diagnostic value of the hub genes in AD, receiver operating characteristic (ROC) curves were analyzed and plotted using GraphPad Prism software. The area under the curve (AUC) of ROC curves was calculated to assess the performance of each gene.
Cell culture and drug treatment
The immortalized BV2 murine microglial cell line was obtained from the Cell Culture Center of the Chinese Academy of Medical Sciences (China). BV2 microglial cells were cultured in complete medium, containing 90% Dulbecco’s modified Eagle’s medium (DMEM; HyClone, USA), 10% fetal bovine serum (FBS, Adamas life, China) and 1% penicillin/streptomycin (Gibco, USA). All cells grew in a humidified incubator (Thermo Fisher Scientific, USA) at 37 ℃ with 5% CO2. Cells were seeded into 6-well plates (2 × 106 cells/well) and pretreated with 5 µM ISL (Tauto Biotech, China) for 1 h, and then injured with 100 ng/ml LPS (Sigma-Aldrich, USA) for 5 h. The cellular morphology of BV2 microglial cells were captured under microscope (Olympus, Japan). The cell experiment was divided into 4 groups: Control group (Ctrl), Control + ISL group (Ctrl + ISL), LPS group (LPS), LPS + ISL group (LPS + ISL).
Gene quantification by quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from harvested BV2 microglial cells using TRIzol Reagent (Thermo Fisher Scientific, USA), and cDNA was synthesized by reverse transcription of total RNA using the PrimescriptTMRT reagent kit with gDNA Eraser (Takara Bio, Japan) according to the manufacturer’s instructions. The Real-time PCR assay was conducted using TB Green Premix Ex Taq™ (Takara Bio, Japan) with the LightCycler 96 detection system (Roche, Switzerland). The relative expression of mRNA (fold change) compared to the control group was calculated after normalization to β-actin.
The primer sequences used in PCR were provided: PPAR-γ, forward 5′-TTTTCAAGGGTGCCAGTTTC-3′, Reverse 5′-AATCCTTGGCCCTCTGAGAT-3′; MAPK1, forward 5′-TTGCTTTCTCTCCCGCACAAA-3′, Reverse 5′-AGAGCCTGTTCAACTTCAATCC-3′; IGF-1, forward 5′-CTCAGAGCATA CCTGCCTGG-3′, Reverse 5′-GGTACTATGAGGCCGAGGTG-3′; SLC2A1, forward 5′-CCCCGTCCTGCTGCTATTG-3′, Reverse 5′-GCACCGTGAAGATGATGAAGAC-3′; IL-1β, forward 5′-CAGGCAGGCAGTATCACTCA-3′, Reverse 5′-AGCTCATATGGGTCCGACAG-3′; IL-6, forward 5′-CCACTTCACAAGTCGGAGGCTTA-3′, Reverse 5′-GCAAGTGCATCATCGTTGTTCATAC-3′; TNF-α, forward 5′-GAACTGGCAGAAGAGAGGCACT-3′, Reverse 5′-AGGGTCTGGGCCATAGAACT-3′; β-actin, forward 5′-AGCCATGTACGTAGCCATCC-3′, Reverse 5′-TCTCAGCTGTGGTGGTGAAG-3′.
Western blot (WB) analysis
After treatment, BV2 microglial cells were harvested, washed with ice-cold phosphate buffered solution (PBS), lysed with RIPA buffer (CST, USA) supplemented with phenylmethanesulfonylfluoride (PMSF), protease inhibitor and phosphatase inhibitor (Well Biotech, China) on ice for 5 min. The extracts were subsequently centrifuged at 12,000 g for 10 min. Protein concentration was quantified by a Bicinchoninic acid (BCA) assay kit (Beyotime Biotechnology, China). Aliquots of protein (40 µg/lane) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose (NC) filter membranes (Pall, USA). The membranes were blocked in Tris-buffered saline/Tween-20 (TBST) containing 5% non-fat milk for 1 h at room temperature, and incubated with primary antibodies overnight at 4 °C. After washing three times with TBST, membranes were incubated with secondary horseradish peroxidase (HRP)-conjugated IgG for 1 h at room temperature. The bands were visualized with SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, USA), and the optical density of the bands was quantitatively analyzed using Image Quant software (Tanon, China). The protein levels were presented as the ratio (protein/β-actin) normalized to the WT control group. β-actin was used as an internal control.
Antibodies used for studies were: mouse anti-PPAR-γ (1:2000, Santa Cruz, cat. no. sc-7273); rabbit anti-p-Erk1/2 (1:4000, Cell Signaling, cat. no. 4370); rabbit anti-Erk1/2 (1:4000, Cell Signaling, cat. no. 4695); rabbit anti-β-actin (1:5000, Cell Signaling, cat. no. 4970); HRP-linked anti-Rabbit IgG (1:8000, Cell Signaling, cat. no. 7074); HRP-linked anti-Mouse IgG (1:8000, Cell Signaling, cat. no. 7076).
Nitrite quantification
BV2 microglial cells were plated in 96-well plates (2 × 104 cells/well) and stimulated with LPS (100 ng/ml) for 24 h after pretreatment of 5 µM ISL for 1 h. The production of NO was assessed by measuring the accumulated nitrite released into culture media. Briefly, after treatment as indicated above, the supernatant was collected, and NO production was determined using the Griess reaction40. Griess reagent was composed of 1% sulfonamide and 0.1% N-1-naphthalene ethylenediamine hydrochloride in 5% phosphoric acid (Sigma-Aldrich, USA). 100 µL Griess reagent and 100 µL culture medium was mixed in enzyme label plate. The absorbance of the reaction mixtures was determined at 540 nm on Synergy 2 (BioTek, USA) after incubation for 10 min at room temperature in the dark.
Statistical analyses
All data were analyzed with GraphPad Prism software using one-way ANOVA followed by Bonferroni or Dunnett T3 post hoc tests. Data were expressed as the mean ± S.E.M. A value of p < 0.05 was considered statistically significant, and statistical significance was shown as *p < 0.05 and **p < 0.01.
Results
Identification of differentially expressed genes between normal healthy controls and AD patients
All DEGs were obtained from GSE5281 dataset and 2,535 DEGs were identified by comparison of the gene expression profiles between normal healthy controls and AD patients. The GEO2R tool, with |log2FC| ≥ 1 and adj p-value < 0.01 as the cut-offs, was used to ensure the dataset’s quality is reliable. 1075 upregulated and 1460 downregulated genes were obtained (Fig. 1a). PPI network analysis of top 100 DEGs was performed by Cytoscape software and the node color was set according to their degree values. Our results demonstrated that the network connections of the top 100 DEGs were densely connected (Fig. 1b). The expression changes of these genes were further displayed in heatmap (Fig. 1c).
Functional and pathway enrichment analysis of the ISL against AD
The chemical structure of ISL was illustrated in Fig. 2a. To better explore the potential molecular mechanism by which ISL exerts its anti-AD effect, network pharmacology was performed. Venn analysis showed that 36 overlapping targets were screened by matching 366 targets of ISL with 6073 targets of the AD databases and GSE5281 dataset (Fig. 2b). To further understand the biological activities and pathways associated with the anti-AD effect of ISL, GO and KEGG enrichment analyses of these common targets were analyzed with the Metascape database and visualized by Sangerbox platform. Top 20 enriched items of GO analysis showed that these DEGs were mainly enriched in functions related to positive regulation of phosphorus metabolic process, regulation of MAPK cascade and positive regulation of transferase activity (Fig. 2c). Furthermore, the outcome of KEGG pathway enriching analyses revealed that targeted genes were remarkably enriched in MAPK signaling pathway, central carbon metabolism in cancer and Ras signaling pathway (Fig. 2d,e).
GO analysis and KEGG pathway enrichment analysis. (a) The structure of ISL. (b) Venn diagram between ISL targets and AD targets. (c) Top 20 bubble chart of biological process from GO enrichment analysis. (d) KEGG pathway analysis of ISL-related DEGs in AD. (e) KEGG pathway analysis and related genes.
Network-based clusters identification
Based on the results obtained from the STRING database, 31 relevant target proteins were analyzed using a PPI network (Fig. 3a). The nodes and edges of PPI network represent interacting proteins and interactions41. Further, two PPI clusters were showed by MCODE tool to generate a highly connected sub-network (Fig. 3b,c). Cluster 1 contained 9 nodes and 29 edges with a score of 7.25, and the intersection genes were mainly enriched the regulation of protein kinase B signaling and MAPK cascade (Fig. 3d). Cluster 2 contained 4 nodes and 6 edges with a score of 4, and it mainly related to the monosaccharide metabolic process and glycolytic process (Fig. 3e).
Key targets of ISL in AD
Four different algorithms (BottleNeck, Betweenness, Stress and Radiality) of top 10 DEGs were calculated via topology analyses (Fig. 4a–d). Based on the ranking of top 10 hub nodes of BottleNeck, Betweenness, Stress and Radiality sub-nets, we selected the 7 overlapping genes (ALB, EGFR, SLC2A1, IGF1, MAPK1, PPARA and PPARG) in the PPI network as key targets (Fig. 4e), which may be beneficial as therapeutic targets.
Molecular docking verification
Based on the network pharmacology results, we selected the top 7 target proteins. Molecular docking strategy was used to predict the potential for interaction between ISL and related core targets (Fig. 5a–g). Through docking simulation, we identified the binding affinity. The binding energy values of the therapeutic targets with the ISL acting on them were all less than − 5 kcal/mol (Fig. 5h). It is considered ligands could bind to the target proteins freely when the binding energy is less than 0. And the lower the binding energy, the stronger binding activity42. A score < − 5 kcal/mol represents that the drug molecular ligand binds to the target, forming conformational stability43. The docking results demonstrated ISL had a strong binding affinity to all 7 key therapeutic targets.
Evaluation target genes with ROC
The ROC curve was used to describe the discrimination accuracy of these targets in the diagnosis of AD. The ensemble-based model showed that the AUC of all target genes ranged from 0.65 to 0.85 (Fig. 6a–g). AUC value was greater than 0.5, indicating that all target genes have high sensitivity and specificity for AD44. We also assessed the levels of these genes during the course of AD development and progression. By analyzing the GEO database, levels of IGF-1, ALB, PPARG and MAPK1 were found to be higher in health controls while the rest higher in AD patients (Fig. 6h–n). These target genes might be strongly correlated to the development and progression of AD.
ISL regulated the phosphorylation of ERK1/2 and PPAR-γ in LPS-induced BV2 microglial cells
To further validate the results of network pharmacology, the pharmacological effects of ISL were investigated in LPS-treated BV2 cells. Cells were pre-treated with ISL (5 µM) for 1 h following 100 ng/ml LPS treatment for 5 h. The mRNA levels of PPAR-γ (PPARG), MAPK1 (encoding the extracellular signal-regulated protein kinase 2, ERK2), SLC2A1 which are highly expressed in microglia were measured using qRT-PCR. The results showed that levels of PPAR-γ and MAPK1 were decreased while SLC2A1 was increased compared with the sham group. Treatment of ISL increased the mRNA levels of PPAR-γ and MAPK1 without the difference in SLC2A1 level (Fig. 7a–c). Furthermore, the expression of related proteins PPAR-γ and ERK was examined by WB. Results showed that ISL treatment reversed the ERK1/2 activation and PPAR-γ decrease under LPS stress (Fig. 7d–f). Together, these results indicated that ISL regulated the ERK/PPAR-γ pathway.
ISL altered the morphological changes and proinflammatory responses in LPS-induced BV2 microglial cells
ERK/PPAR-γ pathway has been known to regulate inflammatory responses. Thus, we investigated whether ISL alters LPS-stimulated inflammatory responses and morphology of BV2 microglial cells. The cells in the control group most frequently showed an ovoid shape, while LPS-treated cells exhibited to be a rounded amoeboid morphology with enlargement of cell body and loss of ramifications45. Moreover, ISL pre-treatment prevented the cellular morphological transformation (Fig. 8a). And then proinflammatory cytokine levels were measured. The production of COX-2, iNOS, NO, IL-1β, IL-6 and TNF-α were greatly increased in LPS-treated BV2 microglial cells. Whereas ISL significantly decreased the levels of these proinflammatory cytokines (Fig. 8b–h). These results indicated that pretreatment with ISL inhibited LPS-mediated increase in proinflammatory responses in BV2 microglial cells.
ISL significantly decreased pro-inflammatory mediators in LPS-induced BV2 microglial cells. (a) The morphology of BV2 microglial cells. Scale bar = 5 μm. (b-d) WB detected iNOS and COX-2. (e) Nitric oxide analysis with Griess Assay was used to measure NO concentration. (f-h) IL-1β, IL-6 and TNF-α mRNA expression levels were determined by qRT-PCR. n = 6. **p < 0.01.
Discussion
As the main active pharmacological component in licorice46, ISL has been widely explored in the treatment of AD, but its molecular mechanisms are still unclear. In order to explore the mechanism underlying the anti-AD effects of ISL, network pharmacology was conducted. Results of KEGG enrichment analysis showed that ISL involved in regulation of MAPK signaling pathway. Moreover, based on PPI network analysis and molecular docking technology, we screened 7 potential gene targets of ISL: ALB, EGFR, SLC2A1, IGF1, MAPK1, PPARA and PPARG. Through ROC analysis, we also found that these gene targets had good predictive ability for AD. The above results suggested that ALB, EGFR, SLC2A1, IGF1, MAPK1, PPARA and PPARG might be critical target genes of ISL against AD.
Previous studies have demonstrated that ISL improved cognitive dysfunction in LPS-induced mice models. Meanwhile, ISL has been shown to inhibit the release of inflammatory factors in BV2 cells induced by amyloid-β oligomers (AβOs) or LPS15,47. These findings suggest that ISL could alleviate AD process by inhibiting microglial inflammation. Based on previous studies and our network pharmacology results, LPS-induced BV2 microglial cells were used as an in vitro model to evaluate the anti-inflammatory mechanism of ISL. Related targets highly expressed in microglia were tested with qRT-PCR and WB. Consistent with the network pharmacology results, the experiment results validated that ISL activated PPAR-γ but suppressed ERK1/2 activation, thereby attenuating inflammatory response in LPS-induced BV2 cells.
In health, microglia work to provide neuroprotection and maintain CNS homeostasis by clearing out unwanted chemicals like debris from pathogens or damaged cells. If accumulated debris is not removed timely, it may lead to CNS dysfunction48. Following injury to the CNS, microglia become activated and initiate a neuroinflammatory response. The persistent inflammatory responses in turn induces neurotoxicity. Mounting evidence exists that neuroinflammation occurs early prior to the formation of Aβ plaques, and microglia-mediated inflammation might be a major element in the promotion of cognitive deficits in AD49. Microglia-induced neuroinflammation has emerged as a promising treatment target for neurodegenerative diseases50.
PPAR-γ, one of the members of ligand-inducible nuclear receptor superfamily, plays an effect role in the inflammatory response and production of Aβ in AD51. PPAR-γ is highly expressed in microglia in human brain52. Interestingly, studies have demonstrated a significant reduction in PPAR-γ expression in both AD patients and animal models, while treatment with PPAR-γ Agonist agonists showed positive effects on AD51. PPAR-γ is also known to prevent LPS-induced microglial activation and stimulate changes in polarization from an inflammatory phenotype to an anti-inflammatory phenotype53. Its activation decreases pro-inflammatory cytokines54. The mechanism underlying the activation of PPAR-γ suppressing inflammation may be attributed to the negative regulation of nuclear factor kappa-B (NF-κB) activation55. It is known that ERK1/2 might be an upstream factor that regulates the expression of PPAR-γ56. In activated microglia, the ERK pathway is key to regulating inflammation. Activation of ERK has been shown to result in a decrease of PPAR-γ57. While with the inhibitor of PPAR-γ, the ERK pathway was activated by the increased phosphorylated ERK58. The ERK/PPAR-γ signaling pathway has been shown to regulate microglia polarization and inflammation59. Suppressing ERK phosphorylation was proved to activate of PPAR-γ to exert anti-inflammatory effects in LPS-treated BV2 microglial cells58. The current results illustrated that ISL decreased the expression of iNOS, COX-2, NO, IL-1β and IL-6, TNF-α, inhibited ERK1/2 phosphorylation, and restored PPAR-γ expression. Therefore, we speculated that ISL might regulate inflammatory response in BV2 microglial cells via ERK/PPAR-γ signaling pathway.
Our studies have identified the main targets and crucial pathways through the application of network pharmacology and experimental verification. Based on our findings, ISL appeared to suppress microglial activation and inflammation thereby attenuating AD progression, which may be closely associated with the modulation of the ERK/PPAR-γ signaling pathway. Nevertheless, this study has its limitations. Initially, the in vitro cell model cannot fully simulate translational animal models for AD. Future research will incorporate AD mice to substantiate the effects of ISL on ameliorating learning-memory ability and inhibiting neuroinflammation. Furthermore, although we have preliminarily observed that ISL could inhibit the release of microglial inflammatory factors and regulate the expression of PPAR-γ and ERK1/2 phosphorylation, it is still unclear whether ISL could improve inflammation through ERK/PPAR-γ signaling pathway. The precise mechanism that ISL regulates neuroinflammation through ERK/PPAR-γ signaling pathway warrants further exploration, and we will consider inhibiting PPAR-γ or ERK1/2 to determine if ISL specifically targets the ERK/PPAR-γ signaling pathway. Thus, more detailed work is still required to verify the therapeutic effects of ISL in AD.
Conclusions
In summary, we used network pharmacology and in vitro experimental verification to analyze the molecular mechanism of ISL against AD, and concluded that ISL may exert its effects in treating AD by inducing microglial inflammation through the ERK/PPAR-γ pathway. Our findings revealed that ISL might be a promising candidate for a natural product in AD therapeutics.
Data availability
Data will be made available from the corresponding author on reasonable request.
References
Dos Santos Picanco, L. C. et al. Alzheimer’s disease: A review from the pathophysiology to diagnosis, New perspectives for Pharmacological Treatment. Curr. Med. Chem. 25, 3141–3159. https://doi.org/10.2174/0929867323666161213101126 (2018).
Karran, E. & Hardy, J. A critique of the drug discovery and phase 3 clinical programs targeting the amyloid hypothesis for Alzheimer disease. Ann. Neurol. 76, 185–205. https://doi.org/10.1002/ana.24188 (2014).
Serrano-Pozo, A., Das, S. & Hyman, B. T. APOE and Alzheimer’s disease: Advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 20, 68–80. https://doi.org/10.1016/s1474-4422(20)30412-9 (2021).
Spangenberg, E. E. & Green, K. N. Inflammation in Alzheimer’s disease: Lessons learned from microglia-depletion models. Brain. Behav. Immun. 61, 1–11. https://doi.org/10.1016/j.bbi.2016.07.003 (2017).
Calsolaro, V. & Edison, P. Neuroinflammation in Alzheimer’s disease: Current evidence and future directions. Alzheimers Dement. 12, 719–732. https://doi.org/10.1016/j.jalz.2016.02.010 (2016).
Kinney, J. W. et al. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimers Dement.. 4, 575–590. https://doi.org/10.1016/j.trci.2018.06.014 (2018).
Shen, H., Pei, H., Zhai, L., Guan, Q. & Wang, G. Aurantiamide suppresses the activation of NLRP3 inflammasome to improve the cognitive function and central inflammation in mice with Alzheimer’s disease. CNS Neurosci. Ther. 29, 1075–1085. https://doi.org/10.1111/cns.14082 (2023).
Hayes, A., Thaker, U., Iwatsubo, T., Pickering-Brown, S. M. & Mann, D. M. Pathological relationships between microglial cell activity and tau and amyloid beta protein in patients with Alzheimer’s disease. Neurosci. Lett. 331, 171–174. https://doi.org/10.1016/s0304-3940(02)00888-1 (2002).
Wirths, O. et al. Inflammatory changes are tightly associated with neurodegeneration in the brain and spinal cord of the APP/PS1KI mouse model of Alzheimer’s disease. Neurobiol. Aging. 31, 747–757. https://doi.org/10.1016/j.neurobiolaging.2008.06.011 (2010).
Kempuraj, D. et al. Brain and Peripheral atypical inflammatory mediators Potentiate Neuroinflammation and Neurodegeneration. Front. Cell. Neurosci. 11 https://doi.org/10.3389/fncel.2017.00216 (2017).
vom Berg, J. et al. Inhibition of IL-12/IL-23 signaling reduces Alzheimer’s disease–like pathology and cognitive decline. Nat. Med. 18, 1812–1819. https://doi.org/10.1038/nm.2965 (2012).
Mae, T. et al. A licorice ethanolic extract with peroxisome proliferator-activated receptor-gamma ligand-binding activity affects diabetes in KK-Ay mice, abdominal obesity in diet-induced obese C57BL mice and hypertension in spontaneously hypertensive rats. J. Nutr. 133, 3369–3377. https://doi.org/10.1093/jn/133.11.3369 (2003).
Chen, W. et al. Neuroprotective effect of total flavonoids in stems and leaves of Glycyrrhiza Uralensis Fisch. On oxidative stress in HT-22 cells and Caenorhabditis elegans. Aging. 15, 5290–5303. https://doi.org/10.18632/aging.204627 (2023).
Zulfugarova, P. et al. A mechanistic review of pharmacological activities of homeopathic medicine licorice against neural diseases. Front. Neurosci. 17, 1148258. https://doi.org/10.3389/fnins.2023.1148258 (2023).
Fu, Y. & Jia, J. Isoliquiritigenin confers Neuroprotection and alleviates Amyloid-β42-Induced Neuroinflammation in Microglia by regulating the Nrf2/NF-κB signaling. Front. NeuroSci. 15 https://doi.org/10.3389/fnins.2021.638772 (2021).
Gay, N. H. et al. Butein, isoliquiritigenin, and scopoletin attenuate neurodegeneration via antioxidant enzymes and SIRT1/ADAM10 signaling pathway. RSC Adv. 10, 16593–16606. https://doi.org/10.1039/c9ra06056a (2020).
Fang, J., Wang, C., Zheng, J. & Liu, Y. Network pharmacology study of Yishen capsules in the treatment of diabetic nephropathy. PLoS One. 17, e0273498. https://doi.org/10.1371/journal.pone.0273498 (2022).
Kibble, M. et al. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat. Prod. Rep. 32, 1249–1266. https://doi.org/10.1039/c5np00005j (2015).
Ru, J. et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 6, 13. https://doi.org/10.1186/1758-2946-6-13 (2014).
Liu, Z. et al. BATMAN-TCM: A bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Sci. Rep. 6, 21146. https://doi.org/10.1038/srep21146 (2016).
Liu, X. et al. PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 38, W609–614. https://doi.org/10.1093/nar/gkq300 (2010).
Gfeller, D. et al. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 42, W32–38. https://doi.org/10.1093/nar/gku293 (2014).
UniProt, C. UniProt: The Universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531. https://doi.org/10.1093/nar/gkac1052 (2023).
Amberger, J. S., Bocchini, C. A., Schiettecatte, F., Scott, A. F. & Hamosh, A. OMIM.org: Online mendelian inheritance in man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 43, D789–798. https://doi.org/10.1093/nar/gku1205 (2015).
Barbarino, J. M., Whirl-Carrillo, M., Altman, R. B. & Klein, T. E. PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip Rev. Syst. Biol. Med. 10, e1417. https://doi.org/10.1002/wsbm.1417 (2018).
Wishart, D. S. et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082. https://doi.org/10.1093/nar/gkx1037 (2018).
Chen, X., Ji, Z. L. & Chen, Y. Z. TTD: Therapeutic target database. Nucleic Acids Res. 30, 412–415. https://doi.org/10.1093/nar/30.1.412 (2002).
Fishilevich, S. et al. GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database. https://doi.org/10.1093/database/bax028 (2017). (2017).
Barrett, T. et al. NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991–995. https://doi.org/10.1093/nar/gks1193 (2013).
von Mering, C. et al. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33, D433–437. https://doi.org/10.1093/nar/gki005 (2005).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. https://doi.org/10.1101/gr.1239303 (2003).
Dario Martucci, M., Pinciroli, F. & Masseroli & Gene ontology application to genomic functional annotation, statistical analysis and knowledge mining. Stud. Health Technol. Inf. 102, 108–131 (2004).
Liang, B., Li, C. & Zhao, J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med. Oncol. 33, 111. https://doi.org/10.1007/s12032-016-0829-6 (2016).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523. https://doi.org/10.1038/s41467-019-09234-6 (2019).
Tang, Y., Li, M., Wang, J., Pan, Y. & Wu, F. X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 127, 67–72. https://doi.org/10.1016/j.biosystems.2014.11.005 (2015).
Pinzi, L. & Rastelli, G. Molecular Docking: Shifting paradigms in Drug Discovery. Int. J. Mol. Sci. 20 https://doi.org/10.3390/ijms20184331 (2019).
Bahmani, A., Tanzadehpanah, H., Hosseinpour Moghadam, N. & Saidijam, M. Introducing a pyrazolopyrimidine as a multi-tyrosine kinase inhibitor, using multi-QSAR and docking methods. Mol. Divers. 25, 949–965. https://doi.org/10.1007/s11030-020-10080-8 (2021).
Burley, S. K. et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 49, D437–D451. https://doi.org/10.1093/nar/gkaa1038 (2021).
Kim, S. et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 49, D1388–D1395. https://doi.org/10.1093/nar/gkaa971 (2021).
Yang, G. et al. Guizhi Fuling capsule relieves memory deficits by inhibition of microglial neuroinflammation through blocking JAK2/STAT3 pathway in presenilin1/2 conditional double knockout mice. Front. Immunol. 14, 1185570. https://doi.org/10.3389/fimmu.2023.1185570 (2023).
Zhang, T. et al. Identifying the mechanisms and molecular targets of Yizhiqingxin formula on Alzheimer’s disease: Coupling network pharmacology with GEO database. Pharmgenomics Pers. Med. 13, 487–502. https://doi.org/10.2147/pgpm.S269726 (2020).
Cao, Y. et al. Network pharmacology and experimental validation to explore the molecular mechanisms of Bushen Huoxue for the treatment of premature ovarian insufficiency. Bioengineered 12, 10345–10362. https://doi.org/10.1080/21655979.2021.1996317 (2021).
Du, Y. et al. Revealing the mechanisms of Byu dMar 25 in the treatment of Alzheimer’s Disease through Network Pharmacology, Molecular Docking, and in vivo experiment. ACS Omega. 8, 25066–25080. https://doi.org/10.1021/acsomega.3c01683 (2023).
Liu, Z. et al. Exploring the mechanism of ellagic acid against gastric cancer based on bioinformatics analysis and network pharmacology. J. Cell. Mol. Med. https://doi.org/10.1111/jcmm.17967 (2023).
Yang, X., Xu, S., Qian, Y. & Xiao, Q. Resveratrol regulates microglia M1/M2 polarization via PGC-1α in conditions of neuroinflammatory injury. Brain Behav. Immun. 64, 162–172. https://doi.org/10.1016/j.bbi.2017.03.003 (2017).
Wang, K. L., Yu, Y. C. & Hsia, S. M. Perspectives on the role of Isoliquiritigenin in Cancer. Cancers (Basel). 13 https://doi.org/10.3390/cancers13010115 (2021).
Lee, D. G., Nam, B. R., Huh, J. W. & Lee, D. S. Isoliquiritigenin reduces LPS-Induced inflammation by preventing mitochondrial fission in BV-2 microglial cells. Inflammation 44, 714–724. https://doi.org/10.1007/s10753-020-01370-2 (2021).
Hickman, S., Izzy, S., Sen, P. & Morsett, L. El Khoury, J. Microglia in neurodegeneration. Nat. Neurosci. 21, 1359–1369. https://doi.org/10.1038/s41593-018-0242-x (2018).
Fakhoury, M. Microglia and astrocytes in Alzheimer’s disease: Implications for therapy. Curr. Neuropharmacol. 16, 508–518. https://doi.org/10.2174/1570159x15666170720095240 (2018).
Prinz, M., Jung, S. & Priller, J. Microglia biology: One century of evolving concepts. Cell 179, 292–311. https://doi.org/10.1016/j.cell.2019.08.053 (2019).
Gu, L. et al. Research progress of PPARγ regulation of cholesterol and inflammation in Alzheimer’s disease. Metab. Brain Dis. 38, 839–854. https://doi.org/10.1007/s11011-022-01139-6 (2023).
Krishna, S. et al. PPAR-γ activation enhances myelination and neurological recovery in premature rabbits with intraventricular hemorrhage. Proc. Natl. Acad. Sci. U S A. 118 https://doi.org/10.1073/pnas.2103084118 (2021).
Ji, J. et al. Antagonizing peroxisome proliferator-activated receptor γ facilitates M1-to-M2 shift of microglia by enhancing autophagy via the LKB1-AMPK signaling pathway. Aging Cell. 17, e12774. https://doi.org/10.1111/acel.12774 (2018).
Bernardo, A. & Minghetti, L. PPAR-gamma agonists as regulators of microglial activation and brain inflammation. Curr. Pharm. Des. 12, 93–109. https://doi.org/10.2174/138161206780574579 (2006).
Lee, S. H. et al. Single-cell transcriptomics reveal cellular diversity of aortic valve and the immunomodulation by PPARγ during hyperlipidemia. Nat. Commun. 13, 5461. https://doi.org/10.1038/s41467-022-33202-2 (2022).
Morgan, H. J. et al. CD200 ectodomain shedding into the tumor microenvironment leads to NK cell dysfunction and apoptosis. J. Clin. Invest. 132 https://doi.org/10.1172/jci150750 (2022).
Lu, X. et al. Hypoxia downregulates PPARγ via an ERK1/2-NF-κB-Nox4-dependent mechanism in human pulmonary artery smooth muscle cells. Free Radic Biol. Med. 63, 151–160. https://doi.org/10.1016/j.freeradbiomed.2013.05.013 (2013).
Yao, H., Zhao, J. & Song, X. Protective effects of fraxin on cerebral ischemia-reperfusion injury by mediating neuroinflammation and oxidative stress through PPAR-γ/NF-κB pathway. Brain Res. Bull. 187, 49–62. https://doi.org/10.1016/j.brainresbull.2022.06.010 (2022).
Wang, Z. F., Li, J., Ma, C., Huang, C. & Li, Z. Q. Telmisartan ameliorates Aβ oligomer-induced inflammation via PPARγ/PTEN pathway in BV2 microglial cells. Biochem. Pharmacol. 171, 113674. https://doi.org/10.1016/j.bcp.2019.113674 (2020).
Acknowledgements
This work was supported by the General Program of National Natural Science Foundation of China (82174003, 81773927 to Ying Xu).
Author information
Authors and Affiliations
Contributions
YX and YZ conceived and designed the research. WG, GY, XL, KH, JP, XW performed the research. WG and GY analyzed the data and prepared the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, W., Yang, G., Liu, X. et al. Network pharmacology and experimental verification to investigate the mechanism of isoliquiritigenin for the treatment of Alzheimer’s disease. Sci Rep 15, 4379 (2025). https://doi.org/10.1038/s41598-025-88542-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-88542-y
Keywords
This article is cited by
-
RRBP1 Inhibition Reduces Microglial M1 Polarization and Inflammation-Mediated Neuronal Loss and Oxidative Stress by Regulating ERK Pathway in Alzheimer’s Disease
Molecular Neurobiology (2026)
-
The anionic surfactant Sodium Dodecyl Benzene Sulfonate alleviates inflammatory responses in atopic dermatitis via NFκB/MAPK pathways in a STAT3-dependent manner
Scientific Reports (2025)