Abstract
A hyperinflammatory state with highly elevated concentrations of inflammatory biomarkers such as C-reactive protein (CRP) is a characteristic feature of severe coronavirus disease 2019 (COVID-19). To examine a potential role of common genetic factors that may influence COVID-19 outcomes, we investigated whether individuals with a polygenic predisposition for a pro-inflammatory response (in the form of Polygenic Scores) are more likely to develop severe COVID-19. The innovative approach of polygenic scores to investigate genetic factors in COVID-19 severity should provide a comprehensive approach beyond single-gene studies. In our cohort of 156 patients of European ancestry, two overlapping Polygenic Scores (PGS) predicting a genetic predisposition to basal CRP concentrations were significantly different between non-severe and severe COVID-19 cases and were associated with less severe COVID-19 outcomes. Furthermore, specific single nucleotide polymorphisms (SNPs) that contribute to either of the two Polygenic Scores predicting basal CRP levels are associated with different traits that represent risk factors for COVID-19 disease initiation (ACE2 receptor, viral replication) and progression (CRP). We suggest that genetically determined enforced CRP formation may contribute to strengthening of innate immune responses and better initial pathogen control thereby reducing the risk of subsequent hyperinflammation and adverse course of COVID-19.
Similar content being viewed by others
Introduction
Coronavirus disease 2019 (COVID-19) caused by Severe Acute Respiratory Syndrome Coronavirus Type 2 (SARS-CoV-2) has been a major health issue over the past few years. A severe disease course associated with hospitalization and critical care support is now rare due to vaccination, COVID-19 recovery, and possibly less pathogenic virus variants, but was frequent in the early stages of this pandemic1,2,3,4. One of the factors associated with severe COVID-19 and a poor prognosis is an overwhelming host immune reaction, termed hyperinflammation5,6. Initial uncontrolled viral replication can be associated with an insufficient innate immune response, as e.g. reflected by a poorer outcome in subjects with defects in type I interferon signaling or the presence of anti-interferon-antibodies7,8, but also defects in adaptive immune responses leading to a dysregulated, excessive, aberrant and non-effective immune response in severely ill COVID-19 patients7,9,10,11. This hyperinflammatory cytokine storm triggers at least part of acute respiratory distress syndrome which can lead to respiratory failure and death in severely affected COVID-19 patients12,13. Severe COVID-19 is associated with considerably higher concentrations of C-reactive protein (CRP) at hospital admission compared to patients with a milder course14,15, and circulating CRP has been suggested as a biomarker to predict COVID-19 severity. However, CRP serum concentrations are not only determined by infection severity but also by other factors including age, sex or individual genetic characteristics16. Serum CRP concentrations are higher in adults than in children, with the highest median concentration observed in the elderly17. In this regard, Ying et al. (2021) explored the genetic relationship between aging and COVID-19 risk, identifying genetic factors that link lifespan with susceptibility to severe disease. This underscores the importance of considering age as a modifier of disease outcomes in our study.
Several studies have investigated genetic risk factors for severe COVID-19. A locus on chromosome 3 constituting a core “Neanderthal” haplotype of 13 single nucleotide polymorphisms (SNPs) showed a significant association with severe COVID-19 on a genome-wide level18. Other studies have also emphasized the genetic risk of severe COVID-19 and pointed out the potential involvement of SNPs in severity of COVID-1919. However, few studies utilized polygenic scores (PGS) for the identification of COVID-19-associated genetic risk. Polygenic scores are individual predictors of specific traits calculated as the sum of the allele dosages multiplied by their corresponding effect sizes obtained from relevant genome-wide association studies (GWAS). They allow a weighted summation for a large number of genetic variants throughout the whole genome associated to a certain trait. A strength and novelty of our PGS-based approach is its quality as a vector for multigene expression, not limited to single genes but encompassing a wide range of variants in relevant pathways contributing to a functional multi-gene network. Using this concept, we may also be able to identify relevant single gene variants driving differences between patient cohorts that may be missed by other approaches only screening for known genomic regions. Considering the importance of inflammatory processes and associated biomarkers for COVID-19 outcomes, we therefore examined whether individuals with a predicted genetic predisposition for a pro-inflammatory response, based on Polygenic Score predictions, are more likely to develop severe COVID-19.
Results
Severe COVID-19 is more frequent and associated with higher CRP and IL-6 concentrations in males
Baseline parameters of 156 patients who tested positive for SARS-CoV-2 are shown in Table 1. Only patients with predicted European ancestry – as determined by Principal Component Analysis (PCA) based SNP array data – were included in the study. 119 patients had non-severe COVID-19 (outpatient treatment or hospitalization), while 37 patients suffered from severe (critical care) or fatal disease (Supplementary Table S1). The median age was not significantly different between groups. Patients in the severe disease group were more frequently male (62.8%; t-test, p-value: 0.004) and had a minimally, but not significantly higher body mass index (non-severe: BMI 26.2, severe: BMI 27.1; t-test, p-value: 0.348). Patients with severe COVID-19 showed higher median maximal CRP serum concentrations (non-severe: 6.3 mg/dl, severe: 29.3 mg/dl; t-test, p-value: 2.2 × 10−11) as well as higher maximal IL-6 concentrations (non-severe: 5.2 ng/l, severe: 408 ng/l; t-test, p-value: 4.5 × 10−5). Furthermore, the number of patients with diabetes mellitus was higher in the severe disease group (non-severe: 12.6% (15/119 cases), severe: 29.7% (11/37 cases); t-test, p-value: 0.042). Other comorbidities were also evaluated; however, no significant differences between patient groups were found (Table 1).
Polygenic score for CRP differs between non-severe and severe cases
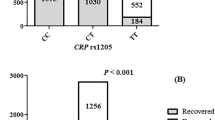
We investigated 24 published polygenic scores that we regarded as potentially relevant multi-locus genetic predisposition surrogates linked to COVID-19 severity, as described in Supplementary Table 2. Only one of the examined scores – PGS00031420 for CRP – showed a significant difference between non-severe and severe COVID-19 patients (t-test; p-value: 0.001, corr. p-value: 0.031; Table 2). Severely ill patients had a lower PGS predicting a genetic predisposition to lower basal CRP concentrations, compared to patients in the non-severe group showing a higher PGS (Fig. 1A).
(A) Polygenic Score for CRP (PGS000314) in non-severe versus severe COVID-19 cases. Boxplot of distribution of calculated weights for the initially applied Polygenic Score PGS000314 [20] predicting genetic predisposition for basal CRP concentrations in the patient cohort (n = 156) stratified by COVID-19-severity. Data shows significant differences in mean PGS weight between non-severe (n = 119) and severe (n = 37) COVID-19-cases, showing a lower mean PGS value in severe COVID-19 cases; * indicating statistical significance (t-test; p- value: 0.001, corr. p-value (Benjamini-Hochberg): 0.031); box with line indicates median and 25% and 75% quantile, whiskers indicate 95% CI, points outside of whiskers indicate outliers. (B) Polygenic Score for CRP (PGSHua2021) in non-severe versus severe COVID-19 cases. Boxplot of distribution of calculated weights for the second overlapping Polygenic Score PGSHua2021 [20] predicting genetic predisposition for basal CRP concentrations in the patient cohort (n = 156) stratified by COVID-19-severity. Data shows significant differences in mean PGS weight between non-severe (n = 119) und severe (n = 37) COVID-19-cases, showing also a lower mean PGS value in severe COVID-19 cases; * indicating statistical significance (t-test; p- value: 0.006); box with line indicates median and 25% and 75% quantile, whiskers indicate 95% CI, points outside of whiskers indicate outliers.
To substantiate a possible link between genetic determinants of CRP concentrations with COVID-19 severity, we calculated a second marginally independent CRP-related score, PGSHua202121. The two scores PGS000314 and PGSHua2021 comprise 77 and 51 SNPs respectively. However, the remaining non-identical SNPs located at the same genomic position between both scores are still proxy-SNPs providing virtually the same signals and effects. All SNPs for both PGS000314 and PGSHua2021 including the overlapping SNPs are documented in Supplementary Table S3. Furthermore, all the individual SNPs were substantially correlated between the two PGS (R² = 0.68, Supplementary Figure S2) and similar weights for the same SNPs in PGS00314 and PGSHua2021 were observed (Supplementary Figure S3). Nevertheless, PGSHua2021 also showed a lower score in severely ill COVID patients (Fig. 1B), with significant differences between the two groups (t-test; p-value: 0.006; Table 3).
Binary logistic regression analysis corrected for sex and age of both scores - PGS000314 and PGSHua2021 - revealed that patients with a lower CRP PGS were more likely to be in the group of severe COVID cases (PGS000314: odds ratio [OR], 6.35; 95% CI: 1.79–22.57; p = 0.004; PGSHua2021: odds ratio [OR], 7.58; 95% CI: 1.57–36.60; p = 0.012). However, no significant correlation could be found between measured CRPmax levels and a PGS-score or one of the most relevant SNPs (Table 4).
Three SNPs may explain the group differences in COVID-19 severity
For the identification of genetic variants with the highest relevance for COVID-19 severity, we performed multi-step recalculations for both PGS after successive removal of the highest-impact SNPs. Significance was lost for PGS000314 after removal of three SNPs, rs7310409, rs3091244 and rs141729353 (new name: rs12734169). For PGSHua2021 significance was lost after removal of rs7310409, indicating that these SNPS may be linked to genetic loci associated with COVID-19 severity in our cohort (Table 5).
Variants rs3091244 (NC_000001.11:g.159714875G > A) and rs12734169 (NC_000001.11:g.159734040 C > T) are intergenic SNPs located on chromosome 1q23.2 between the DUSP23 (dual specificity phosphatase 23; important for dephosphorylation) and the CRP (C-reactive protein) gene. Previously published associations for rs3091244 include CRP concentrations22, a higher susceptibility to dengue-chikungunya co-infection23and abdominal aortic aneurysms24, whereas little information is available on rs12734169. It is not in linkage disequilibrium with variant rs3091244, although they are located at a similar genomic position, and it is also not associated with any trait yet. Variant rs7310409 is located in intron 1 of the HNF1A (Liver Specific Transcription Factor gene) linked to diabetes and dyslipidemia25 (NC_000012.12:g.120987058 A > G). It has been reported to be associated with CRP concentrations by various studies in an east Asian population (p-value: 3.0 × 10−8, beta coefficient: 0.07)26, two European populations (p-value: 7.0 × 10−17, beta coefficient: 0.015)27 (p-value: 3.0 × 10−269, beta coefficient: 0.147)22, and a mixed population (p-value: 3.0 × 10−44, beta coefficient: 0.11)28 (Table 6).
Discussion
In this study of 156 patients of European ancestry who tested positive for SARS-CoV-2, our main interest was to determine whether multi-locus polygenic SNP scores (PGS) can be identified as risk factors of adverse COVID-19 outcomes. Among 24 PGS studied, two overlapping PGS – which predict a genetic predisposition to higher CRP concentrations – showed a significant difference between non-severe and severe COVID-19 cases after correction for multiple testing. This observation suggests a possible link between genetic variants determining basal CRP concentrations and COVID-19 outcomes. CRP is an acute-phase protein synthesized by the liver in response to IL-6 secretion by macrophages and other immune cells. As such it is involved in early innate immune activation, ameliorated pathogen recognition and elimination mainly by phagocytic cells29. High CRP concentrations upon patient admission to hospitals have been associated with an unfavorable course of COVID-195,30. On the other hand, the results of our study may point to a protective effect of genetic variants in patients with COVID-19 disease, predicting increased basal CRP concentrations in the non-severe cohort. Genetic factors and infection severity, as well as viral load and bacterial load are independent determinants of CRP concentrations in the individual case and may directly have differing effects on COVID-19 prognosis. It is interesting to note that in a study on colorectal cancer, an increase of the CRP PGS was associated with reduced lethality21. There are few studies on the association of genetic factors that influence CRP concentrations with infectious disease-related outcomes or severity. In a previous study on a possible association of COVID-19 outcomes with genetic predictors for CRP and venous thromboembolism, a higher PGS for CRP showed a marginal protective effect on death due to COVID-19 but not in regard to any other severe outcome such as hospitalization, critical care or need of mechanical ventilation support31,32. Large-scale GWAS studies, such as the Severe Covid-19 GWAS Group (2020), identified significant loci associated with severe respiratory failure in COVID-19. Our findings align with those of Pairo-Castineira et al. (2021), who identified genetic mechanisms underlying critical illness in COVID-19, reinforcing the polygenic and multifactorial nature of severe COVID-19 outcomes51. However, many confounders, including age, sex, and pre-existing chronic conditions such as obesity, cardiovascular diseases, hypertension, chronic lung, liver, or kidney conditions, and cancer, have been shown to influence COVID-19 disease outcomes47,48,51,52and may also skew the results of our study. However, apart from sex and diabetes mellitus, the occurrence of above mentioned confounding factors was not significantly different between our patient groups. Lima-Martínez et al. (2021) reported that COVID-19 disease left diabetics more often hospitalized, with severe pneumonia, and a higher mortality. Diabetics show a low-grade chronic systemic inflammatory state favoring an exaggerated inflammatory response probably worsening the effects of the viral infection. On the contrary, COVID-19 is capable of directly damaging the pancreas worsening symptoms of the Diabetes53. Of course, Diabetes was also linked to other confounding factors like hypertension and obesity49,54, which were not significantly differing between our patient cohorts, but could also drive disease severity indirectly.
In our study, a possible protective effect concerning COVID-19 severity could be driven by three SNPs, rs7310409, rs3091244 and rs12734169. Variant rs3091244 is a CRP gene promoter polymorphism that is frequent in European and Asian populations and has been formally validated as a functional regulator of CRP expression in cohorts of cancer patients and patients with atrial fibrillation. Compared to the common G allele, heterozygous or homozygous presence of an A allele at SNP rs3091244 causes higher baseline serum CRP concentrations due to an effect on transcription factor binding and altered transcriptional activity in the CRP gene promoter33,34. The higher frequency of the A allele (effect allele) – and a higher PGS score – in non-severely ill COVID-19 patients predicts higher baseline CRP concentrations. In theory, genetically influenced higher baseline CRP levels could assist in pathogen clearance in the early course of a viral disease but also positively impact elimination of bacteria upon bacterial superinfection. In mouse models, CRP has been shown to provide protection against certain bacteria, by binding to the cell wall and activation of the complement pathway, since treatment with CRP increased survival in these mice35. CRP during inflammation is, among other factors, responsible for opsonization of bacteria via the complement pathway36.
On the other hand, a lower Polygenic Score – as seen in severely ill COVID-19 patients - does not exclude high CRP concentrations due to various disease-related factors. However, CRP-related SNPs respectively a CRP-PGS may be a better indicator of basal physiology than CRP measurements from patient plasma. Our data may give a hint that a genetically determined predisposition to higher CRP-concentrations may be a protective factor against a severe COVID-19 disease course. This must be differentiated from reactively elevated CRP-concentrations due to failure of initial (viral) and subsequent (bacterial) pathogen control, a higher viral and/or bacterial load and consecutive induction of hyperinflammation in severely ill patients. Hence most likely, correlation analysis between SNPs and available CRP max values of our patients did not show any significant relations. However, there are no clinical data available in our study to support this concept as neither data on baseline CRP concentrations before SARS-CoV-2 infection nor on the percentage of bacterial superinfections in severely ill COVID-19 patients were documented in our cohort of COVID-19 patients. Furthermore, sex-specific differences of basal CRP concentrations as described by e.g. Khera et al.37 could not be taken into consideration due to cohort size.
Of the three highest-impact SNPs, the only overlapping variant in both PGS is rs7310409. HNF1A genetic variant rs7310409 is in high linkage disequilibrium with rs7139079 (TopLD: EUR: r2 = 0.84, D’=0.97)38. It is one of three loci reported to be associated with higher plasma ACE2 receptor concentrations at genome-wide significance in men, which explain 4.91% of the variation in plasma ACE2 concentration39. The exact functional connection between rs7139079 and higher plasma ACE2 concentrations (sACE2) remains to be clarified. Our data may also hint at genetically determined predispositions to higher ACE2-concentrations being a protective factor against a severe COVID-19 disease course, besides CRP. This is further supported by Yang et al. (2022), who analyzed the genetic landscape of the ACE2 receptor and identified key loci influencing plasma ACE2 concentrations, including one locus, which is a coding variant in the HNF1A gene, the same gene rs7139079 is located in. Their findings of these loci possibly influencing virus entry into the host cell via regulating SARS-CoV-2 spike protein glycosylation, provides additional context for the relevance of rs7310409 and related variants in modulating disease severity in COVID-1949. However, many other factors have been shown to elevate plasma ACE2 concentrations. For example, diabetes mellitus, which is also a risk factor for severe COVID-19 disease, elevated angiotensin II and SARS-CoV-2 infection itself can initiate shedding of membrane bound ACE2, which can lead to higher plasma ACE2 concentrations40,41,42. Plasma ACE2 is able to bind SARS-CoV-2. However, since plasma ACE2 bound SARS-CoV-2 is not an antibody-antigen complex marked for classical immune clearance, it remains to be clarified whether higher basal plasma ACE2 concentrations are able to impact viral entry or the efficacy of immune response against the virus and thus influence the disease course.
Our study indicates that PGS analyses could represent a promising and applicable method for the identification of specific genetic factors in multifactorial diseases. Applying this approach for COVID-19, we observed genetic variants potentially associated with protective traits, such as an increased propensity for higher CRP concentrations and lower ACE2 plasma concentrations. While these findings suggest possible implications for patient management, particularly for identifying individuals at increased genetic risk for severe COVID-19 who might benefit from intensified early medical intervention, further studies are needed to validate these observations. Additionally, PGS analysis may hold potential as a future screening tool for other viral infections, to identify genetic risk factors influencing clinical outcomes, but this concept requires further exploration.
Our study has several limitations. The first limitation is the rather small number of patients of European ancestry that we were able to include in our cohort, due to time point of collection, availability and quality of samples. Unfortunately, due to the unique circumstances of the COVID-19 pandemic, it was not possible to establish a larger cohort. This study was conducted with patient samples from the onset of the pandemic, which included only individuals not previously exposed to SARS-CoV-2 and without vaccinations. There are no more individuals without previous exposure to SARS-CoV-2, thus the situation is unique and cannot be replicated by inclusion of additional patients reasoning our rather small cohort. However, this limits the statistical power of the study and findings may not be generalizable. This especially may influence the effects seen in our single SNP studies. Also, sex-specific differences of basal CRP concentrations, as described by e.g. Khera et al.37, could not be taken into consideration due to our cohort size. Although we only included patients with European ancestry, as confirmed by Principal component analysis, definition of patient groups and regional differences within the same ancestry may further influence our effects seen on single SNPs and in clinical outcomes of single patients. However, this study being a single center study should have reduced missing more subtle regional influences.
A second limitation of our study is that neither data on initial baseline CRP concentrations before SARS-CoV-2 infection nor viral load, bacterial load nor the percentage of bacterial superinfections in severely ill COVID-19 patients were available in our cohort of COVID-19 patients to determine a functional context or correlate our genetic findings. These factors and clinical parameters could only be determined on admission to hospital when patients usually had been ill for some time. Viral load, bacterial load and initial CRP levels in patients may be also be associated with patient’s outcome and would need to be further investigated.
As a third limitation, data on sACE2 and genetic determinants thereof in COVID-19 patients is lacking. Studies on baseline ACE2 concentrations without infection and their association with genotype data would be required for further exploration of a functional association. Other unvaccinated cohorts collected during the COVID-19 pandemic may provide suitable opportunities to confirm our observations, and it would be of interest to extend the analyses to other infections. Determining genetic factors, especially PGS, in the blood of individuals could therefore aid in the stratification of patients at risk for severe COVID-19.
However, the use of PGS as a screening tool may be applicable in other viral infections to search for multi-gene affected associated clinical factors. In prevalent diseases, like cancers, which are possibly associated with a polygenic background, if there is no monogenic cause, PGS are already investigated to serve as a predictive tool of patient prognosis in the future. It remains to be seen whether determination of Polygenic Scores in the individual may be clinically useful for the prediction of personal risk of an adverse disease course and for choosing effective prevention strategies.
Methods
Patient cohort and baseline characteristics
A total of 162 patients, tested positive for SARS-CoV-2 confirmed by polymerase chain reaction (PCR), as per definition provided by the Austrian Federal Ministry of Social Affairs, Health, Care and Consumer Protection, were initially recruited at the Medical University of Innsbruck between March and November 2020. Although at this time no virus genotyping was routinely performed, mainly wild-type and eventually alpha variant cases were present. Further inclusion criteria comprise of patients of both genders aged 18 years or older. 43 patients were included during hospitalization due to COVID-19 (median time to blood withdrawal 23 days; range 0–57 days) and 119 patients, who were either treated as outpatients or also being hospitalized during the acute phase, were included upon follow-up evaluation performed 31–119 days (median: 56 days) after initial diagnosis of SARS-CoV-2 infection. 156 patients remained in the analyzed cohort after exclusion of patients with non-European ancestry estimated via Principal Component Analysis (PCA) conducted with LASER43 and collection of relevant clinical data (Supplementary Figure S1). Further exclusion criteria included pregnancy, known HIV or Hepatitis B/C infection.
The patient cohort was divided into groups by severity of COVID-19 (Fig. 1). Non-severe COVID-19 outcomes were defined as: (1) SARS-CoV-2 positive PCR test and (2) outpatient treatment or (3) hospitalization with and without respiratory support or additional oxygen supply. Severe COVID-19 outcomes were defined as: (1) SARS-CoV-2 positive PCR test, (2) hospitalization, (3) need for critical care (intensive care unit), (4) need for critical respiratory support or 4) death (Supplementary Table S1).
Patient parameters, like the need for critical care as well as respiratory support were identified using the hospital inpatient admissions data. Common laboratory analyses and total blood counts stated in the baseline characteristics were assessed by standard methods as part of patient care at the hospital’s central laboratory.
The study and the trial protocols (EK-Nr: 1091/2020 and 1103/2020) were approved by the ethic institutional review board at Innsbruck Medical University and conducted in accordance with the tenets of the Declaration of Helsinki principles. All study subjects provided a signed and dated declaration of consent in accordance with ICH-GCP Guidelines and participated voluntarily.
Sample Preparation
Genomic DNA was obtained from frozen peripheral blood EDTA samples using QIAamp DNA Blood Midi Kit (Qiagen, Hilden, DE) for manual preparation. DNA concentrations were obtained by Qubit dsDNA BR Assay Kit (Life Technologies, Carlsbad, CA) on a Qubit Fluorometer (Life Technologies, Carlsbad, CA).
Global screening array
To interrogate variants across the entire genome to obtain the most comprehensive view of genomic variation, the Infinium Global Screening Array 24 v3 kit with Multi Disease Content (Illumina, San Diego, CA) for genome-wide genotyping was utilized. The array was performed according to the manufacturer’s recommendations. Data from the array was acquired via iScan (Illumina, San Diego, CA). For data processing, Illumina Genome Studio 2.0 (Illumina, San Diego, CA) was used. After careful evaluation of array and sample quality, raw data was processed according to Technical note “Infinium Genotyping Data Analysis” (Pub.No 970.2007.0050) and Guo et al.44. SNP clusters were optimized by custom re-clustering and poorly performing SNPs (Cluster separation rate < 0.3, Call frequency < 0,97 and a mean of normalized R-values for the AB genotypes > 0.2) were excluded from the data set. Data was exported in PLINK format for further analyses.
Quality control and Genotype Imputation
For imputation of Global Screening array data to accurately assign genotypes at untyped markers improving genome coverage, Michigan Imputation Server was utilized45. Samples were excluded if the call rate was < 0.9. On variant level, if invalid alleles other than A, C, T or G occur, if there are duplicates, indels, monomorphic sites, allele mismatches between reference panel and study, if a SNP call rate below 90% is detected or if it deviated from the Hard-Weinberg equilibrium (p-value < 10−5), the variant was excluded. Genotypes were imputed with the Michigan Imputation Server using Minimac4, Eagle2 (Reference-based phasing using the Haplotype Reference Consortium panel) and the 1000 genomes (Phase 3 v5) reference panel46.
Calculation of polygenic scores
From the imputed genomic data, 24 scores per patient were calculated to estimate predisposition to certain traits. PGS were calculated using pgs-calc (Available: http://github.com/lukfor/pgs-calc). 23 of these calculated PGS were derived from the PGS Catalog (www.PGSCatalog.org, access date 16.12.2020), an open resource for already calculated PGS (Lambert et al. 2021; PMID: 33692568). PGSs were calculated for the following traits: Body Mass Index (BMI) (PGS000027 and PGS000320), Interleukin-6 serum levels (IL-6) (PGS000252), Venous thromboembolism (VTE) (PGS000043), Type 2 diabetes mellitus (T2DM)(PGS000330), Hemoglobin A1c (HbA1c) (PGS000127), leukocyte count (Leuko) (PGS000191), lymphocyte count (Lympho) (PGS000172), neutrophil count (Neutro) (PGS000182), platelet count (Plate) (PGS000186), hemoglobin measurement (HemConc) (PGS000168), Lung Function (FEV1/FVC Ratio) (LungFunct) (PGS000210), Macrophage colony-stimulating factor 1 serum levels (CSF-1) (PGS000225), Interleukin-18 serum levels (IL-18) (PGS000249), Interleukin-6 receptor subunit alpha serum levels (IL-6RA) (PGS000253), Growth/differentiation factor 15 serum levels (GDF-15) (PGS000243), N-terminal prohormone brain natriuretic peptide serum levels (NTproBNP) (PGS000270), low density lipoprotein cholesterol (LDL) (PGS000115), High density lipoprotein cholesterol (HDL) (PGS000064), C-reactive protein (CRP) (PGS000314), Coronary heart disease (CHD) (PGS000329), Thrombomodulin serum levels (TM) (PGS000286) and Total cholesterol (TotalChol) (PGS000311). Details for each PGS are shown in Supplementary Table S2.
For confirmation of findings in one trait, a second PGS for the same trait (CRP), originally established by Hua et al.21 (PGSHua2021) was calculated. A comparison of both scores and their overlapping SNPs is shown in Supplementary Table S3 as well as Supplementary Figure S2 and S3.
For power analysis of single SNPs included into the PGS000314 and PGSHua2021, we calculated 128 different scores including only n-1 SNPs. For further analysis of allele dosage in the cohort, we calculated one score per each individual significant SNP from PGS000314.
Statistical analysis
Statistical Analysis was performed using SPSS (IBM Corp. Released 2019. IBM SPSS Statistics for Windows, Version 26.0. Armonk, NY: IBM Corp) and RStudio (R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/, R version 4.1.2 (2021-11-01)). To explore differences in traits (PGS, serum concentrations, haplotypes) between COVID outcome groups and controls, we performed Kolmogorov-Smirnov Test for normal distribution. Independent samples 2-sided T-test including Benjamini-Hochberg correction for multiple comparisons was performed for normally distributed samples. To assess correlations between PGS and laboratory measures, correlation coefficient using Spearman rho rank-order correlation was calculated. To explore the association of specific PGS with COVID outcomes, we performed binary logistic regression analysis, adjusting for age and sex. Statistical significance was defined as α < 0.05.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Williamson, E. A. O. X. et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 584 (7821), 430–436 (2020).
McCoy, K., Peterson, A., Tian, Y. & Sang, Y. A. O. Immunogenetic Association underlying severe COVID-19. Vaccines (Basel). 8 (4), 700 (2020).
Burkert, F. R., Lanser, L., Bellmann-Weiler, R. & Weiss, G. Coronavirus disease 2019: Clinics, treatment, and prevention. Front. Microbiol. 12, 761887 (2021).
Ulloa, A. C., Buchan, S. A., Daneman, N. & Brown, K. A. Estimates of SARS-CoV-2 Omicron variant severity in Ontario, Canada. JAMA 327 (13), 1286–1288 (2022).
Manson, J. J. et al. COVID-19-associated hyperinflammation and escalation of patient care: a retrospective longitudinal cohort study. Lancet Rheumatol. 2 (10), e594–e602 (2020).
McKechnie, J. L. & Blish, C. A. The Innate Immune System: fighting on the Front lines or fanning the flames of COVID-19? Cell. Host Microbe. 27 (6), 863–869 (2020).
Giamarellos-Bourboulis, E. J. et al. Complex Immune Dysregulation in COVID-19 patients with severe respiratory failure. Cell. Host Microbe. 27 (6), 992–1000 (2020).
Mazzeffi, M. A., Chow, J. H. & Tanaka, K. COVID-19 Associated Hypercoagulability: manifestations, mechanisms, and management. Shock 55 (4), 465–471 (2021).
Mehta, P. et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395 (10229), 1033–1034 (2020).
Bastard, P. et al. Vaccine breakthrough hypoxemic COVID-19 pneumonia in patients with auto-abs neutralizing type I IFNs. Sci. Immunol. 14, eabp8966 (2022).
Zhang, B. et al. Clinical characteristics of 82 cases of death from COVID-19. PLoS One. 15 (7), e0235458 (2020).
Hasan, S. A. O. et al. Mortality in COVID-19 patients with acute respiratory distress syndrome and corticosteroids use: a systematic review and meta-analysis. Expert Rev. Respir Med. 14 (11), 1149–1163 (2020).
Gustine, J. N. & Jones, D. Immunopathology of Hyperinflammation in COVID-19. Am. J. Pathol. 191 (1), 4–17 (2021).
Yitbarek, G. A. O. et al. The role of C-reactive protein in predicting the severity of COVID-19 disease: a systematic review. SAGE Open. Med. 9, 20503121211050755 (2021).
Bellmann-Weiler, R. A. O. X. et al. Neopterin predicts Disease Severity in hospitalized patients with COVID-19. Open. Forum Infect. Dis. 8 (1), ofaa521 (2020).
Kathiresan, S. et al. Contribution of clinical correlates and 13 C-reactive protein gene polymorphisms to interindividual variability in serum C-reactive protein level. Circulation 113 (11), 1415–1423 (2006).
Schlenz, H. et al. C-reactive protein reference percentiles among pre-adolescent children in Europe based on the IDEFICS study population. Int J. Obes. (Lond). 38 (Suppl 2), S26–31 (2014).
Zeberg, H. A. O. & Pääbo, S. A. O. The major genetic risk factor for severe COVID-19 is inherited from neanderthals. Nature 587 (7835), 610–612 (2020).
Kaser, A. Genetic risk of severe Covid-19. N Engl. J. Med. 383 (16), 1590–1591 (2020).
Xie, T. et al. Genetic risk scores for Complex Disease traits in Youth. Circ. Genom Precis Med. 13 (4), e002775 (2020).
Hua, X. A. O. et al. Genetically predicted circulating C-Reactive protein concentration and colorectal Cancer survival: a mendelian randomization Consortium Study. Cancer Epidemiol. Biomarkers Prev. 30 (7), 1349–1358 (2021).
Ligthart, S. et al. Genome analyses of > 200,000 individuals identify 58 Loci for Chronic Inflammation and highlight pathways that Link inflammation and Complex disorders. Am. J. Hum. Genet. 103 (5), 691–706 (2018).
Sengupta, S., Bhattacharya, N. & Tripathi, A. Association of C-reactive protein polymorphisms with serum-CRP concentration and viral load among dengue-chikungunya mono/co-infected patients. Antiviral Res. 197, 105225 (2022).
Saratzis, A. et al. C-reactive protein polymorphism rs3091244 is associated with abdominal aortic aneurysm. J. Vasc Surg. 60 (5), 1332–1339 (2014).
Li, L. M., Jiang, B. G. & Sun, L. L. HNF1A: from monogenic diabetes to type 2 diabetes and gestational diabetes mellitus. Front Endocrinol. (Lausanne). 13, 829565 (2022).
Okada, Y. et al. Genome-wide association study for C-reactive protein levels identified pleiotropic associations in the IL6 locus. Hum. Mol. Genet. 20 (6), 1224–1231 (2011).
Ridker, P. M. et al. Loci related to metabolic-syndrome pathways including LEPR, HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the women’s Genome Health Study. Am. J. Hum. Genet. 82 (5), 1185–1192 (2008).
Kilpeläinen, T. O. et al. Multi-ancestry study of blood lipid levels identifies four loci interacting with physical activity. Nat. Commun. 10 (1), 376 (2019).
Rhodes, B., Fürnrohr, B. G. & Vyse, T. J. C-reactive protein in rheumatology: biology and genetics. Nat. Rev. Rheumatol. 7 (5), 282–289 (2011).
Bellmann-Weiler, R. et al. Prevalence and Predictive Value of Anemia and Dysregulated Iron Homeostasis in patients with COVID-19 infection. J. Clin. Med. 9 (8), 2429 (2020).
Guo, B. et al. Association between Genetic Predictors for C-Reactive Protein and venous thromboembolism with severe adverse coronavirus Disease 2019 outcomes. Crit. Care Explor. 3 (12), e0602 (2021).
Guo, Y. et al. Genome-wide association study of hippocampal atrophy rate in non-demented elders. Aging (Albany NY). 11 (22), 10468–10484 (2019).
Szalai, A. J. et al. Single-nucleotide polymorphisms in the C-reactive protein (CRP) gene promoter that affect transcription factor binding, alter transcriptional activity, and associate with differences in baseline serum CRP level. J. Mol. Med. (Berl). 83 (6), 440–447 (2005).
Carlson, C. S. et al. Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels. Am. J. Hum. Genet. 77 (1), 64–77 (2005).
Mold, C., Nakayama, S., Holzer, T. J., Gewurz, H. & Du Clos, T. W. C-reactive protein is protective against Streptococcus pneumoniae infection in mice. J. Exp. Med. 154 (5), 1703–1708 (1981).
Sproston, N. R. & Ashworth, J. J. Role of C-Reactive protein at sites of inflammation and infection. Front Immunol. 9, 754 (2018).
Khera, A. et al. Race and gender differences in C-reactive protein levels. J. Am. Coll. Cardiol. 46 (3), 464–469 (2005).
Huang, L. et al. TOP-LD: a tool to explore linkage disequilibrium with TOPMed whole-genome sequence data. Am. J. Hum. Genet. 109 (6), 1175–1181 (2022).
Nelson, C. P. et al. Genetic associations with plasma angiotensin converting enzyme 2 concentration: potential relevance to COVID-19 risk. Circulation 142 (11), 1117–1119 (2020).
Wallentin, L. et al. Angiotensin-converting enzyme 2 (ACE2) levels in relation to risk factors for COVID-19 in two large cohorts of patients with atrial fibrillation. Eur. Heart J. 41 (41), 4037–4046 (2020).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181 (2), 271–280e8 (2020).
Sama, I. E., Voors, A. A. & van Veldhuisen, D. J. New data on soluble ACE2 in patients with atrial fibrillation reveal potential value for treatment of patients with COVID-19 and cardiovascular disease. Eur. Heart J. 41 (41), 4047–4049 (2020).
Taliun, D. et al. LASER server: ancestry tracing with genotypes or sequence reads. Bioinformatics 33 (13), 2056–2058 (2017).
Zhao, S. et al. Strategies for processing and quality control of Illumina genotyping arrays. Brief. Bioinform. 19 (5), 765–775 (2018).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48 (10), 1284–1287 (2016).
Loh, P. R. et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 48 (11), 1443–1448 (2016).
Jalaleddine, N. et al. ACE2 polymorphisms impact COVID-19 severity in obese patients. Sci. Rep. 12(1), 21491 (2022).
Ejaz, H. et al. COVID-19 and comorbidities: deleterious impact on infected patients. J. Infect. Public. Health. 13 (12), 1833–1839 (2020).
Yang, Z. et al. Genetic Landscape of the ACE2 coronavirus receptor. Circulation 145 (18), 1398–1411 (2022).
Ying, K. et al. Genetic and phenotypic analysis of the causal relationship between aging and COVID-19. Commun. Med. (Lond). 1, 35 (2021).
Pairo-Castineira, E. et al. Genetic mechanisms of critical illness in COVID-19. Nature 591 (7848), 92–98 (2021).
Severe Covid-19 GWAS Group et al. Genomewide Association Study of Severe Covid-19 with respiratory failure. N Engl. J. Med. 383 (16), 1522–1534 (2020).
Lima-Martínez, M. M., Carrera Boada, C., Madera-Silva, M. D., Marín, W. & Contreras, M. COVID-19 and diabetes: a bidirectional relationship. Clin. Investig Arterioscler. 33 (3), 151-157 (2021).
Li, C. et al. Associations of diabetes, hypertension and obesity with COVID-19 mortality: a systematic review and meta-analysis. BMJ Glob Health. 8 (12), e012581 (2023).
Author information
Authors and Affiliations
Contributions
I.T., T.S., J.L.-R., V.P., D.H., E.P.-S. and M.G. recruited the cohort, V.V., D.H., L.F., S.S., S.C., F.K., J.Z., E.J. and G.W. conceived the experiments, V.V. and L.F. conducted the experiments, V.V., D.H., L.H., S.S. J.Z., E.J. and G.W. analyzed the results. V.V. and D.H. wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Vogi, V., Haschka, D., Forer, L. et al. Severe COVID-19 disease is associated with genetic factors affecting plasma ACE2 receptor and CRP concentrations. Sci Rep 15, 4708 (2025). https://doi.org/10.1038/s41598-025-89306-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-89306-4