Abstract
It is well-known that Enterococcus species produce bacteriocins that have antibacterial activity. However, a comprehensive analysis of the bacteriocin distribution among Enterococcus strains has not been conducted. In this study, we identified the bacteriocin genes from 80 Enterococcus faecalis and 38 Enterococcus faecium strains and investigated their antibacterial activity. In the 80 E. faecalis strains, the cytolysin gene (61.3%), enterolysin A gene (27.5%) and BacL1 gene (45.0%) were identified. In the 38 E. faecium strains, the enterocin A gene (97.4%), enterocin B gene (2.6%), enterocin NKR-5-3B gene (21.0%), bacteriocin T8 gene (36.8%) and BacAS9 gene (23.7%) were identified. The antibacterial activity of all strains was tested against E. faecalis and E. faecium. The strains positive for the cytolysin, enterolysin A, BacL1, bacteriocin T8 or BacAS9 genes presented variable antibacterial activity. Several bacteriocin-positive strains showed antibacterial activity against other enterococcal species, but not against Staphylococcus or Escherichia coli. In addition, the enterolysin A-positive strain showed antibacterial activity against vancomycin-resistant E. faecium, and the bacteriocin T8- or BacAS9-positive strains showed activity against vancomycin-resistant E. faecalis and E. faecium. Our findings suggest that E. faecium and E. faecalis strains that carry different bacteriocin genes may affect the composition of the surrounding bacterial community.
Similar content being viewed by others
Introduction
Species of Enterococcusare facultative Gram-positive commensal bacteria that are found mainly in the intestine and oral cavity1. However, Enterococcussometimes causes opportunistic nosocomial infections such as biliary/urinary tract infections, bacteremia and endocarditis2,3,4. Among enterococcal species, Enterococcus faecalis and Enterococcus faeciumare major human pathogens. In recent years, the emergence and spread of drug-resistant Enterococci that exhibited the resistance to β-lactams, aminoglycosides and glycopeptides in health care settings has become a global problem4,5. In particular, vancomycin-resistant Enterococci (VRE) are known to cause serious infections in elderly and immunocompromised individuals6. Therefore, it is expected to discover novel antimicrobial agents to replace conventional antimicrobials.
Bacteriocins are expected to be candidates for new antibacterial agents. Bacteriocins are antibacterial peptides produced by bacteria, mainly Gram-positive bacteria such as lactic acid bacteria7,8. In general, the antibacterial spectra of bacteriocins are narrower than that of currently used antimicrobial agents, and most bacteriocins are effective against bacteria of the same species or genus. Bacteriocins are classified into three main categories: post-translationally modified peptides, including lantibiotics; unmodified bacteriocins, including pediocin-like bacteriocins and two-peptide bacteriocins; and large heat-labile proteins7,8.
Enterococci have been reported to produce multiple bacteriocins7,9. Among enterococcal bacteriocins, cytolysins are well-known lantibiotics10. Cytolysins are composed of two peptides, cytolysin-L and cytolysin-S, which have activities against erythrocytes and Gram-positive bacteria10,11,12. Enterocins are also well-known bacteriocin that has been reported in several Enterococci species, including lantibiotics and non-lantibiotics7,9. In addition, enterolysin A and BacL1(bacteriocin 41), which are peptidoglycan hydrolases, have been reported13,14. Although various studies have been conducted on bacteriocins produced by Enterococci, few comprehensive analyses of bacteriocins among many enterococcal strains have been conducted. Tedim AP et al. (2024) reported the bacteriocin distribution in E. faecium and Enterococcus lactisvia bioinformatic analysis15, but the relationship between bacteriocin genes and their antibacterial activity was not demonstrated. In this study, we comprehensively analyzed the bacteriocin gene status of 38 E. faecium and 80 E. faecalis clinical isolates and their antibacterial activity against enterococcal species, including VRE.
Materials and methods
Bacterial strains used in this study
The bacterial strains used in this study are shown in Table 1. Enterococcus isolates, including E. avium (1 isolate), E. casseliflavus (1 isolate), E. faecalis (80 isolates), E. faecium (38 isolates), E. gallinarum (1 isolate), and E. raffinosus(1 isolate), were previously collected16. Vancomycin-resistant E. faecalis (2 isolates with vanA- positive) and E. faecium (4 isolates with vanA-positive, 2 isolates with vanB-positive) strains were obtained clinically. Escherichia coli XL-II, Acinetobacter baumanniiHN-12517, Staphylococcus epidermidisKSE318, and Staphylococcus aureusMW219 were used. All the strains used in this study were incubated aerobically at 37 °C in trypticase soy broth (Becton, Dickinson and Company).
Identification of bacteriocin genes based on whole-genome sequences
Illumina whole-genome sequencing of 118 clinical enterococcal isolates was performed elsewhere for genomic analysis (DRA015781, PRJDB15342). The bacteriocin genes were searched via the web server BAGEL4 (http://bagel4.molgenrug.nl/). Furthermore, based on the reference7, we obtained the nucleotide sequences of known bacteriocin genes from National center for biotechnology information (NCBI) Genbank (Supl. Table 1), and searched for them using ABRicate (https://github.com/tseemann/abricate). Publicly available complete genome sequences of E. faecalis (273 assemblies) and E. faecium (337 assemblies) were obtained using ncbi-genome-download tool (https://github.com/kblin/ncbi-genome-download) from NCBI Refseq FTP server with an assembly level ‘complete’ in FASTA format on 8 January 2025. The presence of vancomycin resistance genes was analyzed via ABRicate using Resfinder database20.
Phylogenetic tree analysis
Phylogenetic trees of the E. faecalis and E. faeciumisolates were created based on core-genome alignment. The assembled fasta files were annotated by Prokka21, and the obtained gfffiles were aligned to the core genome linked by the Panaroo pipeline22. A maximum-likelihood tree was subsequently generated via RAxML-NG version 1.2.1 with 1000 bootstrap replicates23. The phylogenetic trees were annotated with Interactive Tree of Life (iTOL) software24. Multi-locus sequence typing (MLST) analysis was performed via MLST 2.0 from the Center for Genomic Epidemiology (Lyngby, Denmark). The identities between amino acid sequences were investigated via NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid alignments were obtained using SnapGene (GSL Biotech, Boston, MA, USA). Genetic comparisons were visualized using Easyfig ver. 2.2.525.
Soft-agar overlay assay
To assess the antimicrobial activity of each bacteriocin, the soft-agar overlay method was performed as previously described26, with some modifications. All 118 Enterococcus strains were aerobically precultured in 5 mL of TSB medium at 37 °C overnight, and 2.0 µl of each strain was spotted onto TSA plates. The plates were incubated overnight at 37 °C to form a uniform bacterial growth zone with a diameter of 5 mm.
The indicator strains were precultured overnight aerobically at 37 °C in 5 mL of TSB medium. Fifty microliters of each indicator strain or 5 µL of S. aureus was added to 3.0 mL of prewarmed half-strength TSA agar medium (0.8%). The mixed overlay components were poured onto agar plates containing the previously spotted colonies and incubated aerobically overnight at 37 °C. The diameter of the growth-inhibitory zone surrounding bacteriocin-producing bacteria was measured in three directions, and the mean value was calculated. E. faecalis JARB-HU0683 (no bacteriocin gene), E. faecium JARB-HU0748 (no bacteriocin gene), and eight VRE strains (two E. faecalis strains and six E. faecium strains) were used as indicator bacteria.
Quantification of bacteriocin gene expression
Overnight cultures of the enterococcal isolates were adjusted to an OD660 nm value of 1.0. Then, 10 µl of culture diluted 10-fold using TSB was spotted onto TSA agar medium and incubated at 37 °C for 24 h, after which the colonies were scraped and collected. A FastRNA Pro Blue Kit (MP Biomedicals, Solon, OH, USA) was used to extract total RNA from bacterial cells. After the extraction of RNA, contaminating DNA was removed by using DNase I (Invitrogen, USA). The RNA purity and concentration were measured with a NanoDrop spectrophotometer. One microgram of RNA was converted to cDNA via a Transcriptor first-strand cDNA synthesis kit (Roche, Tokyo, Japan) following the manufacturer’s recommended protocol. Quantitative PCR was performed using FastStart Essential DNA Green Master and a LightCycler 96 instrument (Roche, Tokyo, Japan) with cDNA as the template. The primers used are listed in Supl. Table 2. Two dependent experiments were performed, and the average value was calculated.
Statistical analysis
One-way analysis of variance (ANOVA) for comparisons between groups with Dunnett’s post hoc multiple comparison test was performed using GraphPad Prism version 10.1.0 (GraphPad Software, San Diego, CA, USA).
Results
Identification of the bacteriocin genes among the Enterococcus isolates
Among the 80 E. faecalis and 38 E. faecium strains, we investigated bacteriocin genes that were previously reported (Table 2). In the 80 E. faecalis strains, the cytolysin gene (49 strains [61.3%]), enterolysin A gene (22 strains [27.5%]) and BacL1 gene (36 strains [45.0%]) were identified. Among the 49 cytolysin gene-positive strains, 10 strains, 30 strains and 1 strain were double-positive for both cytolysin/enterolysin A, cytolysin/BacL1, and BacL1/enterolysin A, respectively (Table 3). One strain was positive for the cytolysin, BacL1 and enterolysin A genes. In the 38 E. faecium strains, the enterocin A gene (37 strains [97.4%]), enterocin B gene (1 strain [2.6%]), enterocin NKR-5-3B gene (8 strains [21.0%]), bacteriocin T8 gene (14 strains [36.8%]) and BacAS9 gene (9 strains [23.7%]) were identified (Table 2). Six, 3, 2, and 1 E. faecium bacteriocin double-positive strains possessing enterocin A/bacteriocin T8, enterocin NKR-5-3B/enterocin A, BacAS9/enterocin A and enterocin B/enterocin A were obtained (Table 3). Ten strains were triple-positive, for enterocin NKR-5-3B, bacteriocin T8 and enterocin A (3 strains), enterocin NKR-5-3B, BacAS9 and enterocin A (2 strains) and bacteriocin T8, BacAS9 and enterocin A (5 strains).
Variation in the bacteriocin amino acid sequence
We compared the amino acid sequences of each bacteriocin among the strains (Table 4). The amino acid sequences of enterocin A, enterocin NKR-5-3B and BacAS9 were completely conserved among their respective strains. Among the 49 cytolysin-positive strains, 46 strains presented the same amino acid sequences of cytolysin L (CytL-La) and cytolysin S (CytL-Sa), while one strain presented one amino acid substitution each in cytolysin L (CytL-Lb) and cytolysin S (CytL-Sb), and two strains presented only one cytolysin (CytL-LS-fusion), which showed alignment with the forward portion of cytolysin-L and the latter portion of cytolysin-S (Fig. 1). Among the 22 enterolysin A-positive strains, 18 strains presented the same enterolysin A amino acid sequence (Enterolysin A-a), whereas 4 strains presented one (enterolysin A-b) or 25 amino acid substitutions (enterolysin A-c, d, e) (Suppl. Figure 1). Among the 36 BacL1-positive strains, 33 strains presented the same BacL1 amino acid sequence (BacL1-a), whereas 2 strains presented one (BacL1-c) or two (BacL1-d) amino acid substitutions in BacL1, and the BacL1 gene of one strain was disrupted to generate 2 partial genes (BacL1-b) (Suppl. Figure 2).
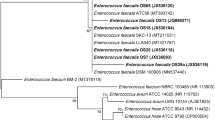
Alignments of the cytolysin-encoding genes of the 3 E. faecalis strains and amino acid alignments of the three cytolysin types. (a) The cytolysin gene cluster (cytL-L1 and cytL-S1, cyt-LS-fusion) and its neighboring region were extracted from 3 strains (JARB-HU0762, HU0680, HU0828) and compared. The grayscale indicates the percent sequence identity. (b) Amino acid alignment of three types of cytolysin, namely, CytL-La + CytL-Sa, CytL-Lb + CytL-Sb and cytolysin-LS-fusion.
Antibacterial activity against E. faecium and E. faecalis
We investigated the antibacterial activity of all the enterococcal strains against E. faecalis JARB-HU0683 (no bacteriocin gene) and E. faecium JARB-HU0748 (no bacteriocin gene) via a soft-agar overlay assay (Fig. 2). Figure 3a and b show the antibacterial activity of each bacteriocin-positive E. faecalis strain (cytolysin, BacL1 and enterolysin A) against E. faecalis JARB-HU0683 and E. faecium JARB-HU0748. Enterolysin A-positive E. faecalis strains, including those double- and triple-positive with bacteriocin genes, presented strong inhibitory activity against E. faecium JARB-HU0748, whereas all but two of the enterolysin A single-positive strains, presented no activity against E. faecalis JARB-HU0683. Compared to the strains with enterolysin A-a, the strains of other types (enterolysin A-b, c, d, or e) presented similar antibacterial activity against E. faecium JARB-HU0748. Cytolysin single-positive strains showed weak activity against E. faecalis JARB-HU0683 and E. faecium JARB-HU0748, with the exception of 1 strain against E. faecalis and 2 strains against E. faecium. The strains with one amino acid substitution (CytL-Lb and CytL-Sb) presented weak activity, whereas one strain with a fused cytolysin (CytL-LS-fusion) showed no activity. All BacL1 single-positive E. faecalis strains except one (BacL1−disrupted) exhibited antibacterial activity against E. faecalis JARB-HU0683 but not E. faecium JARB-HU0748. Compared with either the cytolysin or BacL1 single-positive strains, the cytolysin/BacL1 double-positive strains presented similar or stronger activity against E. faecalis JARB-HU0683. One strain with BacL1 and enterolysin A showed activity against E. faecalis JARB-HU0683 and E. faecium JARB-HU0748, although its activity against E. faecium JARB-HU0748 was weak compared with that of the enterolysin A single-positive strains. Cytolysin and enterolysin A double-positive strains showed activity against both strains, but the activity against E. faecium was weaker than that of the enterolysin A single-positive strains. One strain with cytolysin/BacL1/enterolysin A showed weaker activity against E. faecium JARB-HU0748 than that of the enterolysin A single-positive strains, while its activity against E. faecalis JARB-HU0683 was similar to that of the BacL1 single-positive strains.
Evaluation of the antibacterial activity of E. faecalis and E. faecium strains harboring bacteriocin genes via a soft-agar overlay assay. A soft-agar overlay assay was performed to evaluate the antibacterial activity against E. faecalis JARB-HU0683 and E. faecium JARB-HU0748. Overnight cultures of enterococcal strains (2.0 µl) were spotted onto TSA plates and incubated for 24 h at 37 °C. The indicator strain was added to 3.0 mL of prewarmed soft TSA agar medium, and the mixed medium was poured onto agar plates containing the previously spotted colonies and incubated aerobically overnight at 37 °C. The diameter of the growth-inhibitory zone surrounding the bacteriocin-producing bacteria was measured.
Antibacterial activity of the E. faecalis and E. faecium strains that carry bacteriocin genes against E. faecalis and E. faecium. A soft-agar overlay assay was performed using E. faecalis JARB-HU0683 (a, c) and E. faecium JARB-HU0748 (b, d). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 (one-way ANOVA for comparisons between groups by Dunnett’s post hoc multiple comparison test).
Figure 3c and d show the antibacterial activity of each bacteriocin-positive E. faecium strain (enterocin A, enterocin B, enterocin NKR-5-3B, bacteriocin T8 and BacAS9) against E. faecalis JARB-HU0683 and E. faecium JARB-HU0748. All but two enterocin A- or enterocin B-single-positive E. faecium strains presented no activity against E. faecium JARB-HU0748, and 4 of the 5 enterocin NKR-5-3B strains also presented no activity, whereas the bacteriocin T8- or BacAS9-positive strains presented strong activity against E. faecium JARB-HU0748. Bacteriocin T8- or BacAS9-positive strains presented antibacterial activity against E. faecalis JARB-HU0683, whereas the other bacteriocin-positive strains presented no activity against E. faecalis JARB-HU0683, with a few exceptions.
Expression of the bacteriocin genes among the isolates
We investigated the expression of each bacteriocin gene in several representative strains (Fig. 4). We first evaluated the expression of strains single-positive for BacL1, cytolysin, enterolysin A, and enterocin A. However, since the bacteriocin T8- or BacAS9-positive E. faecium strains also expressed enterocin A, we investigated the expression of both together. BacL1, cytolysin and enterolysin A were expressed in each bacteriocin single-positive strain that showed antibacterial activity, although the expression of cytolysin (ratio to gyrA) was lower than that of the other bacteriocins. Expression in the strain with the cytolysin-LS fusion (JARB-HU0828) was undetectable. Among 15 strains single-positive for enterocin A, we investigated enterocin A expression in two of the strains with antibacterial activity against E. faecalis and two strains that showed no activity and detected the expression in all 4 strains. Next, we investigated the expression of bacteriocin genes in the double- and triple-positive strains. Strains with both the enterolysin A and cytolysin genes presented lower enterolysin A gene expression compared with that of the enterolysin A single-positive strains. This result was consistent with the antibacterial activity against E. faecalis or E. faecium. In the bacteriocin T8 or BacAS9/enterocin A double-positive strains, which exhibited antibacterial activity against E. faecalis and E. faecium, the expression of both genes was observed. In the strain with the BacL1 and cytolysin genes, which showed antibacterial activity against E. faecalis, expression of the cytolysin and BacL1 genes was observed, although the expression of the BacL1 gene was lower than that of the BacL1 single-positive strain. In the cytolysin, enterolysin A, and BacL1 triple-positive strain, the expression of BacL1 and cytolysin was similar to that in the single-positive strains, but enterolysin A expression was lower. In bacteriocin T8, BacAS9, and enterocin A triple-positive strain, the expression of three genes was observed.
Expression of bacteriocin genes and antibacterial activity of Enterococcus strains with or without bacteriocin genes against several bacterial species. Antibacterial activity and bacteriocin gene expression of representative enterococcal strains. RNA extraction and quantitative PCR were performed on representative isolates of each bacteriocin type to observe the correlation between the zones of inhibition and gene expression. * JARB-HU notation is abbreviated to HU.
Antibacterial activity against other enterococcal species and other bacteria
We investigated the antibacterial activity of several strains with one of the 6 bacteriocins (cytolysin, enterolysin A, BacL1, enterocin A, BacAS9 and bacteriocin T8) (Fig. 4). Strains with one, two or three bacteriocin genes were selected for evaluation of their antibacterial activity against E. avium, E. raffinosus, E. casseliflavus, E. gallinarum, E. coli, A. baumannii, S. epidermidis, and S. aureus. The cytolysin-single positive isolate (JARB-HU0762) where expression was observed showed antibacterial activity against other Enterococci (E. avium, E. raffinosus, E. casseliflavus, and E. gallinarum) but not E. coli, A. baumannii, S. epidermidis or S. aureus, while the cytolysin-single positive isolate (JARB-HU0813) where expression was 1ower that that of JARB-HU0762 only showed activity against E. faecalis JARB-HU0683. The enterolysin A single-positive isolate showed antibacterial activity against other Enterococci but not other species. The BacL1 single-positive isolate (JARB-HU0783) showed antibacterial activity against E. avium, E. raffinosus, and E. casseliflavus. The bacteriocin T8 and BacAS9-positive isolates showed antibacterial activity against E. avium, E. raffinosus and E. casseliflavus. Two enterocin A single-positive strains that showed activity against E. faecalis also exhibited antibacterial activity against other Enterococci, whereas two enterocin A single-positive strains that did not show activity against E. faecalis also exhibited no activity against other Enterococci. The strain expressing enterocin NKR-5-3B showed no antibacterial activity against any of the strains tested.
Phylogenetic tree analysis
Among the E. faecalis 34 sequence types (STs), ST179 (25 strains) and ST16 (11 strains, 13.8%) were present in high proportions (Fig. 5a). The ST16 isolates had the cytolysin L and S genes (except for one isolate) and the enterolysin A gene and exhibited antibacterial activity against E. faecalis JARB-HU0683 and E. faecium JARB-HU0748. The ST179 isolates also presented cytolysin genes together with BacL1 and showed strong activity against E. faecalis JARB-HU0683 but not E. faecium JARB-HU0748.
Among the E. faecium 11 STs, ST17 and ST547 were present in high proportions (Fig. 5b). All the E. faecium isolates except 1 had an enterocin A gene. However, there was no clear trend of STs with respect to the bacteriocin genes, although the strains with ST17 genes presented a high proportion of bacteriocin T8.
Antibacterial activity against VRE
We investigated the effects of 5 bacteriocins (cytolysin, bacteriocin T8, BacAS9, enterolysin A and BacL1) against 8 VRE strains, including 2 E. faecalis strains and 6 E. faecium strains (Fig. 6). Before investigating antibacterial activity, we first determined the presence of bacteriocin genes in each VRE strain. Two VR-E. faecalis strains (VREFs1, 2) had the vanA, cytolysin and BacL1 genes, while 4 VR-E. faecium strains (VREFm1, 2, 4, 6) were vanA- and bacteriocin T8-positive, and 2 VR-E. faecium strains (VREFm3, 5) were vanB- and bacteriocin T8-positive.
Antibacterial activity of bacteriocin-producing strains against VRE. A soft-agar overlay assay was performed using 3 E. faecalis strains, including 2 vancomycin-resistant strains, and 7 E. faecium strains, including 6 vancomycin-resistant strains. VS: vancomycin susceptible, VR: vancomycin resistant, EFs: E. faecalis, EFm: E. faecium. VSEFs1: JARB-HU0683 (no bacteriocin gene), VSEFm1: JARB-HU0748 (no bacteriocin gene), VREFs1, 2: vanA, cytL-L, cytL-S and bacL1 positive, VREFm1-4: vanA, enterocin A and bacteriocin T8 positive, VREFm5, 6: vanB, enterocin A and bacteriocin T8 positive.
The cytolysin gene-positive strain showed weak antibacterial activity against E. faecalis without the cytolysin gene (VSEFs1) and E. faecium strains, but this strain did not show activity against the VREFs and VSEFm strains. The BacL1-gene-positive strain also showed antibacterial activity against only VSEFs1. The enterolysin A-positive strain showed antibacterial activity against only the E. faecium strains, including the VR and VSEFm strains, whereas the bacteriocin T8- or BacAS9-positive strains showed antibacterial activity against all the VS and VR strains.
Discussion
In this study, we comprehensively analyzed bacteriocins reported previously in E. faecium and E. faecalis. To date, many enterococcal bacteriocins, including lantibiotics, nonlantibiotics, cyclic peptides and large proteins, have been identified7,27. We identified 3 bacteriocin genes from E. faecalis and 5 genes from E. faeciumamong our collections of enterococcal strains previously isolated16 (Table 2). Compared the distribution of bacteriocin genes with previous reports, 62.4% of BacL128 among 327 E. faecalis clinical isolates and 47.4% of Cytolysin-L among 97 E. faecalisisolates29 have been reported. It was also reported the distribution of entA (81.3%), entB (13.7%), bac43 (T8) (20.7%), bacAS9 (10.1%) based on 883 E. faeciumgenomes15. In addition, we also investigated the distribution of bacteriocin genes using 273 E. faecalis genomes including 15 VRE and 337 E. faecium genomes including 202 VRE. Regarding bacteriocin genes found in this study, we found 32.6% of cytolycin, 38% of enterolysin A and 17% of BacL1 in total E. faecalis genomes, while 84.0% of enterocin A, 11.3% of enterocin B, 4.7% of enterocin P, 30.3% of NKR-5-3B, 19.3% of bacteriocin T8 and 8.0% of bacAS9 in total E. faecium genomes (Suppl. Table 3). However, the percentage of each bacteriocin retained tended to be higher in VRE than in vancomycin sensitive Enterococci (VSE). The strains examined in this study except one strain were VSEs, and compared to previous reports and analysis from genomic data, they showed mostly the same trend in the percentage of each bacteriocin retained.
It has been reported that cytolysins, which are composed of two peptides containing lanthionine, cytolysin-L and cytolysin-S, are active against eukaryotic cells (erythrocytes) and Gram-positive bacteria7. However, the detailed antibacterial activity of cytolysins among bacterial species has not been well analyzed. In this study, we identified only cytolysin-positive strains and investigated their antibacterial activity against several bacterial species. Cytolysin single-positive strains had antibacterial activity against E. faecalis, E. faecium (very weak activity), E. avium, E. raffinosus, and E. casseliflavus but had no activity against S. epidermidis, S. aureus, E. coli or A. baumannii. Therefore, cytolysins are mostly effective in treating enterococcal species. In addition, we found that two E. faecalis strains had only a single cytolysin derived from cytolysin-L or cytolysin-S (Fig. 1). Two cytolysins are bound and inserted into the bacterial membrane, resulting in pore formation. However, one strain that has only one bacteriocin of a single cytolysin (CytL-LS-fusion) showed no activity against E. faecalis or E. faecium because its gene was not expressed. Therefore, it remains unknown whether the CytL-LS fusion has activity against E. faecalis. Among 49 cytolysin-positive E. faecalis strains, enterolysin A (11 strains) or BacL1(29 strains), which are peptidoglycan hydrolases14,30, were also identified. The mode of action of BacL1 is cleavage between D-isoglutamine (D-iGln) and L-lysine (L-Lys) in E. faecalispeptidoglycan with recognition of the L-alanine (L-Ala)-L-Ala structure31. Enterolysin A cleaves the interpeptide links between L-Ala and D-glutamine (D-Glu) and between L-lysine (L-Lys) and D-aspartic acid (D-Asp) in Lactococcus lactis ssp. cremorispeptidoglycan32. Since E. faecalis stem peptide (L-Ala-D-iGln-L-Lys-D-Ala-D-Ala) is same with E. faecium33,34 (Suppl. Figure 3) and is different with that of Lactococcus lactis ssp. Cremoris with D-Glutamine instead of D-iGln, it is considered enterolysin A cleaves the interpeptide links between L-Ala and D-iGln in E. faecium. Although the cleavage site of BacL1 (D-iGln-L-Lys) or enterolysin A (L-Ala and D-iGln) is found in stem peptide of E. faecalis and E. faeciumpeptidoglycan35, enterolysin A is only active in E. faecium, and BacL1 is only active in E. faecalis. In addition, BacL1-positive strains showed no antibacterial activity against S. aureus and S. epidermidis which also have the cleavage site of enterolysin A and BacL136. Since the cross-linked structure of peptidoglycan differs among E. faecalis (L-Ala-L-Ala), E. faecium (L-Asx: aspartic acid or asparagine), S. aureus (pentaglycine) and S. epidermidis (tetra-glycine and one serine), the peptidoglycan structure adjacent to the cleavage site is required for their enzymatic activity. In addition, BacA was reported to be required for the BacL1bacteriolytic activity37, and identified 3 subtypes, type I, IIa and IIb38. We investigated BacA among 36 BacL1 positive strains, and found 33 strains (91.7%) with type I, one strain with one amino acid substitution of type I and one strain with 3 amino acid substitution of type IIa (Suppl. Figure 4). However, the BacL1-positive strains with the mutated type I and type IIa showed same antibacterial activity with those with type I, although the strain with the truncated BacA showed weak antibacterial activity. Therefore, it is considered that BacL1-positive strains in this study showed antibacterial activity against E. faecalis (BacL1 negative) by coordinately functioning BacL1 and BacA.
Compared with BacL1- or cytolysin single-positive strains, cytolysin and BacL1 double-positive strains presented stronger antibacterial activity against E. faecalis, while most enterolysin A single-positive strains presented stronger activity than both positive strains. Therefore, we investigated the expression of the enterolysin A gene and observed decreased enterolysin A expression in both positive strains (Fig. 4). Enterolysin A single-positive E. faecalis (JARB-HU0760), which showed higher enterolysin A gene expression than the double-positive strains, exhibited stronger antibacterial activity against E. faecium (JARB-HU0748). Only one E. faecalis strain (JARB-HU0734) was triple-positive for bacteriocin genes (cytolysin, enterolysin A and BacL1) and showed high activity against E. faecalis and moderate activity against E. faecium, and we found that the expression of enterolysin A was suppressed compared to enterolysin A single positive E. faecalis(JARB-HU0760). Therefore, we believe that some bacteriocin genes in strains with two or three bacteriocin genes are suppressed for unknown reasons. Cytolysin-encoded genes39,40 and BacL1-encoded genes27 have been reported to be located on the plasmid, although these genes were not found on the same plasmid, whereas the enterolysin A-encoded gene is located on the chromosome according to the NCBI database (CP028724, CP113832). In this study, 30 of the 80 E. faecalis strains possessed cytolysin and BacL1, so these strains have two different plasmids. In the MLST analysis, we observed some correlations between the STs and bacteriocins. All the ST179 strains contained cytolysin and BacL1, and 10 of the 11 ST16 strains contained cytolysin and enterolysin A. Among the various STs found in Japan, ST16 and ST179, which belong to CC16, are major STs41,42. Since we demonstrated the coexistence of cytolysin genes and enterolysin A/BacL1 in ST179 and ST16, these two bacteriocins may be associated with infection at human sites such as the intestine and pharynx with individual microflora.
We also investigated E. faecalis bacteriocins (cytolysin, enterolysin A and BacL1) against vancomycin-resistant E. faecalis (VREFs) and E. faecium (VREFm). Although BacL1 had strong activity against E. faecalis, BacL1 had no activity against VR-E. faecalis. The two VR-E. faecalis strains used in this study are BacL1 positive, so the immunity factor, bacI1bac/I2 is located downstream of BacL1and BacA gene38. Therefore, two VR-E. faecalis showed resistance to BacL1. In contrast, enterolysin A was effective against all E. faecium strains (4 vanA-positive strains and 2 vanB-positive strains) because the cleavage site of enterolysin A and the cross-linked structureVR-E. faecium are not changed. Since two VR-E. faecalisstrains showed no activity against cytolysins, we investigated the presence of cytolysin genes and identified these genes. Cyl-I has been demonstrated to function in immunity, although its precise function remains unknown43. Therefore, these two strains were resistant to cytolysins. Bacrteriocin T8/enterocin A and BacAS9/enterocin A-positive strains showed antibacterial activity against all VRE strains. In this study, we did not find the antibacterial activity of enterocin A against E. faecalis and E. faecium strains (Fig. 3), so it is speculated that bacteriocin T8 is effective to VRE strains. However, 6 VR-E. faeciumstrains possessed bacteriocin T8 gene. Downstream of bacteriocin T8 gene, the gene coding the predicted immunity factor is located44. Therefore, bacteriocin T8 still shows the antibacterial activity against even bacteriocin T8-positive E. faecium strains although the susceptibility of VR-E. faecium strains were lower than that of vancomycin sensitive E. faecium without T8 gene.
Among the E. faecium strains used in this study, we identified 5 bacteriocin genes, namely, enterocin A, enterocin B, enterocin NKR-5-3B, bacteriocin T8 and BacAS9, but only bacteriocin T8 and BacAS9 exhibited antibacterial activity against E. faecalis and E. faeciumstrains. Bacteriocin T8 has been reported to be a class IIa sec-dependent bacteriocin44 produced by E. faeciumand to have antibacterial activity against several bacterial strains, whereas the BacAS9 gene has only been identified genetically15. BacAS9 showed similarity with bacteriocin T8, Sakacin P from Lactobacillus sake45 and leucocin A from Leuconostoc gelidum46 (Fig. 7). Therefore, this is the first report demonstrating the antibacterial activity of BacAS9. Amino acid alignment of T8, Leucocin A, Sakacin P and BacAS9 revealed a conserved YGNGX2CX4CXV motif, which is typically found in class IIa bacteriocins (Fig. 7). These two peptides showed antibacterial activity against E. faecium, including VR strains; E. faecalis, including VR strains; E. avium; and E. raffinosus. All VR E. faeciumstrains used in this study were positive for the bacteriocin T8 gene. Although the immunity factor against bacteriocin T8 is located downstream of the bacteriocin T8 gene44, these VR E. faeciumstrains were susceptible to the bacteriocin T8 gene-positive strain. This result implies that immune factors do not provide full resistance against high concentrations of bacteriocin T8. Mills EM et al. (2024) reported the potential for the clinical use of bacteriocin T8 for treating enterococcal infections47. Therefore, bacteriocin T8 and BacAS9 are thought to be candidates for the prevention of VRE infections. In this study, we found no activity for 4 bacteriocins against E. faecalis or E. faecium strains. In particular, enterocin A was identified from E. faecium48 as having strong antibacterial activity against Lactobacillus, Pediococcus and Listeria species, while its activity against E. faecalis and E. faecium varied among strains. Enterocin A gene-positive E. faecium strains possess an enterocin A synthesis operon including the immunity factor, EntI. In this study, we also found that 37 of the 38 E. faecium strains were enterocin A positive. Furthermore, we found that the single enterocin A-positive E. faecium strain, which did not possess other bacteriocin genes, did not exhibit an inhibitory effect on entA-negative E. faecium strains. We also investigated the expression of the entA gene in several strains via quantitative PCR and found no relationship between entA expression and antibacterial activity. Based on these results, we concluded that enterocin A has no antibacterial activity against E. faecium or E. faecalis. Since some entA single-positive strains exhibited antibacterial activity against E. faecalis (JARB-HU0683) (Fig. 4), other unconfirmed bacteriocin genes are likely involved. Among the 12 STs from the E. faecium strains, we found no typical correlations between the STs and the bacteriocin genes.
A limitation of this study is that the distribution of the bacteriocin genes among the E. faecalis and E. faeciumstrains may not reflect a general tendency because the strains used in this study were collected from one university hospital16. However, we previously reported no obvious transmission within the hospital. Additionally, the STs of E. faecium and E. faecalis used in this study were varied. Therefore, our collections are considered not to be significantly biased.
In conclusion, our study clarified the distribution of bacteriocin genes among E. faecalis and E. faecium strains based on genomic analysis and the antibacterial activity profile. Some strains carrying multiple bacteriocin genes have been identified, and different antibacterial patterns of each bacteriocin type have been demonstrated. Additionally, we evaluated the antibacterial activity against several VRE strains. Our findings suggest that the different bacteriocin-carrying E. faecalis or E. faecium strains may affect the composition of the bacterial flora. Furthermore, some Enterococcus bacteriocins are possible candidates for antibacterial agents against VRE infection.
Data availability
The genome data have been deposited into the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA) accession number DRA015781 under BioProject accession no. PRJDB15342.
References
Krawczyk, B., Wityk, P., Gałęcka, M. & Michalik, M. The many faces of Enterococcus spp.-commensal, probiotic and opportunistic pathogen. Microorganisms 9(9), 1900. https://doi.org/10.3390/microorganisms9091900 (2021).
Neelakanta, A. et al. Impact of changes in the NHSN catheter-associated urinary tract infection (CAUTI) surveillance criteria on the frequency and epidemiology of CAUTI in intensive care units (ICUs). Infect. Control Hosp. Epidemiol. 36(3), 346–349. https://doi.org/10.1017/ice.2014.67 (2015).
Fernández-Guerrero, M. L. et al. Nosocomial enterococcal endocarditis: A serious hazard for hospitalized patients with enterococcal bacteraemia. J. Intern. Med. 252(6), 510–515. https://doi.org/10.1046/j.1365-2796.2002.01061.x (2002).
Guzman Prieto, A. M. et al. Global emergence and dissemination of Enterococci as nosocomial pathogens: Attack of the clones? Front. Microbiol. 7, 788. https://doi.org/10.3389/fmicb.2016.00788 (2016).
Hollenbeck, B. L. & Rice, L. B. Intrinsic and acquired resistance mechanisms in Enterococcus. Virulence 3(5), 421–433. https://doi.org/10.4161/viru.21282 (2012).
Faron, M. L., Ledeboer, N. A. & Buchan, B. W. Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant Enterococcus in the health care setting. J. Clin. Microbiol. 54(10), 2436–2447. https://doi.org/10.1128/jcm.00211-16 (2016).
Franz, C. M., van Belkum, M. J., Holzapfel, W. H., Abriouel, H. & Gálvez, A. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol. Rev. 31(3), 293–310. https://doi.org/10.1111/j.1574-6976.2007.00064.x (2007).
Solis-Balandra, M. A. & Sanchez-Salas, J. L. Classification and multi-functional use of bacteriocins in health, biotechnology, and food industry. Antibiotics (Basel). 13(7), 666. https://doi.org/10.3390/antibiotics13070666 (2024).
Wu, Y., Pang, X., Wu, Y., Liu, X. & Zhang, X. Enterocins: Classification, synthesis, antibacterial mechanisms and food applications. Molecules 27(7), 2258. https://doi.org/10.3390/molecules27072258 (2022).
Booth, M. C. et al. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol. Microbiol. 21(6), 1175–1184. https://doi.org/10.1046/j.1365-2958.1996.831449.x (1996).
Van Tyne, D., Martin, M. J. & Gilmore, M. S. Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins (Basel). 5(5), 895–911. https://doi.org/10.3390/toxins5050895 (2013).
Cox, C. R., Coburn, P. S. & Gilmore, M. S. Enterococcal cytolysin: A novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr. Protein Pept. Sci. 6(1), 77–84. https://doi.org/10.2174/1389203053027557 (2005).
Zheng, B., Tomita, H., Inoue, T. & Ike, Y. Isolation of VanB-type Enterococcus faecalis strains from nosocomial infections: First report of the isolation and identification of the pheromone-responsive plasmids pMG2200, encoding VanB-type Vancomycin resistance and a Bac41-type bacteriocin, and pMG2201, encoding erythromycin resistance and cytolysin (Hly/Bac). Antimicrob. Agents Chemother. 53(2), 735–747. https://doi.org/10.1128/aac.00754-08 (2009).
Nilsen, T., Nes, I. F., Holo, H. & Enterolysin, A. A cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 69(5), 2975–2984. https://doi.org/10.1128/AEM.69.5.2975-2984.2003 (2003).
Tedim, A. P., from the ESCMID Study Group on Food- and Water-borne Infections (EFWISG) et al. Bacteriocin distribution patterns in Enterococcus faecium and Enterococcus lactis: Bioinformatic analysis using a tailored genomics framework. Appl. Environ. Microbiol. 90(10), e0137624. https://doi.org/10.1128/aem.01376-24 (2024).
Fujii, A. et al. Antibiotic susceptibility and genome analysis of Enterococcus species isolated from inpatients in one hospital with no apparent outbreak of Vancomycin- resistant Enterococcus in Japan. Microbiol. Immunol. 68(8), 254–266. https://doi.org/10.1111/1348-0421.13155 (2024).
Kawayanagi, T. et al. The oral cavity is a potential reservoir of gram-negative antimicrobial-resistant bacteria, which are correlated with ageing and the number of teeth. Heliyon 10(21), e39827 (2024).
Nakazono, K. et al. Complete sequences of epidermin and nukacin encoding plasmids from oral-derived Staphylococcus epidermidis and their antibacterial activity. PLoS One 17(1), e0258283. https://doi.org/10.1371/journal.pone.0258283 (2022).
Centers for Disease Control and Prevention (CDC). Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus — Minnesota and North Dakota, 1997–1999. MMWR Morb Mortal. Wkly. Rep.(1999).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67(11), 2640–2644. https://doi.org/10.1093/jac/dks261 (2012).
Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30(14), 2068–2069. https://doi.org/10.1093/bioinformatics/btu153 (2014).
Tonkin-Hill, G. et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 21(1), 180. https://doi.org/10.1186/s13059-020-02090-4 (2020).
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35(21), 4453–4455. https://doi.org/10.1093/bioinformatics/btz305 (2019).
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M. & Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9(8), e104984. https://doi.org/10.1371/journal.pone.0104984 (2014).
Sullivan, M. J., Petty, N. K. & Beatson, S. A. Easyfig: A genome comparison visualizer. Bioinformatics 27(7), 1009–1010. https://doi.org/10.1093/bioinformatics/btr039 (2011).
Kawada-Matsuo, M. et al. Three distinct two-component systems are involved in resistance to the class I bacteriocins, Nukacin ISK-1 and Nisin A, in Staphylococcus aureus. PLoS One 8(7), e69455. https://doi.org/10.1371/journal.pone.0069455 (2013).
Ness, I. F., Diep, D. B. & Ike, Y. Enterococcal bacteriocins and antimicrobial proteins that contribute to niche control. Enterococci: Commensals Lead. Causes Drug Resistant Infect(2014).
Kurushima, J., Ike, Y. & Tomita, H. Partial diversity generates effector immunity specificity of the Bac41-Like bacteriocins of Enterococcus faecalis clinical strains. J. Bacteriol. 198(17), 2379–2390. https://doi.org/10.1128/jb.00348-16 (2016).
Anderson, A. C. et al. Enterococcus faecalis from clinicalispecimenscimensoraldsites sprevalencealence of virulence factors in association with biformationmation. Front. Microbiol. 6, 1534. https://doi.org/10.3389/fmicb.2015.01534 (2015).
Tomita, H., Kamei, E. & Ike, Y. Cloning and genetic analyses of the bacteriocin 41 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI14: A novel bacteriocin complemented by two extracellular components (lysin and activator). J. Bacteriol. 190(6), 2075–2085. https://doi.org/10.1128/jb.01056-07 (2008).
Kurushima, J., Nakane, D., Nishizaka, T. & Tomita, H. Bacteriocin protein BacL1 of Enterococcus faecalis targets cell division loci and specifically recognizes L-Ala2-cross-bridged peptidoglycan. J. Bacteriol. 197(2), 286–295. https://doi.org/10.1128/JB.02203-14 (2015).
Khan, H., Flint, S. H. & Yu, P. L. Determination of the mode of action of enterolysin A, produced by Enterococcus faecalis B9510. J. Appl. Microbiol. 115(2), 484–494. https://doi.org/10.1111/jam.12240 (2013).
Yang, H., Singh, M., Kim, S. J. & Schaefer, J. Characterization of the tertiary structure of the peptidoglycan of Enterococcus faecalis. Biochim. Biophys. Acta Biomembr. 1859(11), 2171–2180. https://doi.org/10.1016/j.bbamem.2017.08.003 (2017).
Patti, G. J., Kim, S. J. & Schaefer, J. Characterization of the peptidoglycan of Vancomycin-susceptible Enterococcus faecium. Biochemistry 47(32), 8378–8385. https://doi.org/10.1021/bi8008032 (2008).
Arbeloa, A. et al. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J. Biol. Chem. 279(40), 41546–41556. https://doi.org/10.1074/jbc.M407149200 (2004).
Schleifer, K. H. & Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36(4), 407–477. https://doi.org/10.1128/br.36.4.407-477.1972 (1972).
Kurushima, J., Hayashi, I., Sugai, M. & Tomita, H. Bacteriocin protein BacL₁of Enterococcus faecalis is a peptidoglycan D-isoglutamyl-L-lysine endopeptidase. 288(52), 36915–36925. (2013). https://doi.org/10.1074/jbc.M113.506618
Kurushima, J., Ike, Y. & Tomita, H. Partial diversity generates effector immunity specificity of the Bac41-Like bacteriocins of Enterococcus faecalis clinical strains. J. Bacteriol. 198(17), 2379–2390. https://doi.org/10.1128/jb.00348-16 (2016).
Clewell, D. B. Properties of Enterococcus faecalis plasmid pAD1, a member of a widely disseminated family of pheromone-responding, conjugative, virulence elements encoding cytolysin. Plasmid 58(3), 205–227. https://doi.org/10.1016/j.plasmid.2007.05.001 (2007).
Singh, N. V. et al. Colocalization of linezolid resistance (cfr) and virulence factors Cytolysin and Hemolysin (cln and hln) on a plasmid in Enterococcus faecalis. Antimicrob. Agents Chemother. 67(6), e0025923. https://doi.org/10.1128/aac.00259-23 (2023).
Aung, M. S. et al. Antimicrobial resistance, virulence factors, and genotypes of Enterococcus faecalis and Enterococcus faecium clinical isolates in Northern Japan: Identification of optrA in ST480 E. faecalis. Antibiot. (Basel). 12(1), 108. https://doi.org/10.3390/antibiotics12010108 (2023).
Harada, S. et al. Prevalence of high-level Aminoglycoside Resistance and genes encoding aminoglycoside-modifying enzymes in Enterococcus faecalis and Enterococcus faecium isolated in a University Hospital in Tokyo. Jpn J. Infect. Dis. 73(6), 476–480. https://doi.org/10.7883/yoken.JJID.2019.416 (2020).
Coburn, P. S., Hancock, L. E., Booth, M. C. & Gilmore, M. S. A novel means of self-protection, unrelated to toxin activation, confers immunity to the bactericidal effects of the Enterococcus faecalis cytolysin. Infect. Immun. 67(7), 3339–3347. https://doi.org/10.1128/iai.67.7.3339-3347.1999 (1999).
De Kwaadsteniet, M., Fraser, T., Van Reenen, C. A. & Dicks, L. M. Bacteriocin T8, a novel class IIa sec-dependent bacteriocin produced by Enterococcus faecium T8, isolated from vaginal secretions of children infected with human immunodeficiency virus. Appl. Environ. Microbiol. 72(7), 4761–4766. https://doi.org/10.1128/AEM.00436-06 (2006).
Tichaczek, P. S., Vogel, R. F. & Hammes, W. P. Cloning and sequencing of sakP encoding sakacin P, the bacteriocin produced by Lactobacillus sake LTH 673. Microbiol. (Reading). 140(Pt 2), 361–367. https://doi.org/10.1099/13500872-140-2-361 (1994).
Hastings, J. W. et al. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J. Bacteriol. 173(23), 7491–7500. https://doi.org/10.1128/jb.173.23.7491-7500.1991 (1991).
Mills, E. G. et al. Bacteriocin production facilitates nosocomial emergence of Vancomycin-resistant Enterococcus faecium. medRxiv [Preprint]. https://doi.org/10.1101/2024.08.01.24311290 (2024).
Aymerich, T. et al. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl. Environ. Microbiol. 62(5), 1676–1682. https://doi.org/10.1128/aem.62.5.1676-1682.1996 (1996).
Author information
Authors and Affiliations
Contributions
M.K.-M, H.S., T.A., and H.K. contributed to designing and conceptualizing the study. A.F., M.K.-M, M.L., Y.S., and S.N. contributed to the majority of the experiments and participated in interpreting the data. A.F., M.K.-M., Y.S., S.N., and H.K. contributed to writing the manuscript, and M.L., H.S., and T.A. contributed to revising the manuscript. All the authors read and approved the final version of the manuscript as submitted and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fujii, A., Kawada-Matsuo, M., Le, M.NT. et al. Comprehensive analysis of bacteriocins produced by clinical enterococcal isolates and their antibacterial activity against Enterococci including VRE. Sci Rep 15, 4846 (2025). https://doi.org/10.1038/s41598-025-89518-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-89518-8
This article is cited by
-
Expression of enterocin E-760 in the fusion form with SUMO-3 in Pichia pastoris X33 and its antimicrobial characterization
Vegetos (2026)
-
Unveiling the probiotic potential of Enterococcus spp.: Mechanisms and roles in animal and human health, A comprehensive review
World Journal of Microbiology and Biotechnology (2025)
-
Current Status and Future Challenges in the Use of Bacteriocins to Modulate Ruminant Gut Microbiota
Probiotics and Antimicrobial Proteins (2025)
-
Genomic surveillance of vancomycin-resistant Enterococcus faecium: a study on Resistome, Plasmidome, and mobilome profiling
Current Genetics (2025)