Abstract
Despite recent surge of interest in deploying colon capsule endoscopy (CCE) for early diagnosis of colorectal diseases, there remains a large gap between the current state of CCE in clinical practice, and the state of its counterpart optical colonoscopy (OC). This is due to several factors, such as low quality bowel cleansing, logistical challenges around both delivery and collection of the capsule, and most importantly, the tedious manual assessment of images after retrieval. Our study, built on the “Danish CareForColon2015 trial (cfc2015)” is aimed at closing this gap, by focusing on the full integration of AI in CCE’s pathway, where image processing steps linked to the detection, localization and characterisation of important findings are carried out autonomously using various AI algorithms. We developed a family of algorithms based on explainable deep neural networks (DNN) that detect polyps within a sequence of images, feed only those images containing polyps into two parallel independent networks to characterize, and estimate the size of important findings. Our recognition DNN to detect colorectal polyps was trained and validated (\(70\%\)) and tested (\(30\%\)) on an unaugmented database of 1751 images containing colorectal polyps and 1672 images of normal mucosa reached an impressive sensitivity of \(99.9\%\), a specificity of \(99.4\%\), and a negative predictive value (NPV) of \(99.8\%\). The characterisation DNN trained on an unaugmented database of 317 images featuring neoplastic polyps and 162 images of non-neoplastic polyps reached a sensitivity of \(84.3\%\) and a specificity of \(81.5\%\) in classifying polyps. The size estimation DNN trained on an unaugmented database of 280 images reached an accuracy of \(88\%\) in correctly segmenting the polyps. By automatically incorporating important information including size, location and pathology of the findings into CCE’s pathway, we moved a step closer towards the full integration of explainable AI (XAI) in CCE’s routine clinical practice. This translates into a fewer number of unnecessary investigations and resection of diminutive, insignificant colorectal polyps.
Similar content being viewed by others
Introduction
Colon capsule endoscopy (CCE) is a non-invasive procedure with advantages in diagnosing, monitoring, and managing colorectal diseases compared to optical colonoscopy (OC), flexible sigmoidoscopy, and computed tomographic colonography (CTC)1,2,3. Clinical trials show CCE outperforms CTC in detecting polyps larger than 6 mm and is non-inferior for polyps larger than 10 mm4. In cases of incomplete OC, CCE has a higher diagnostic yield than CTC for polyps of any size CCE5,6. Patients also prefer CCE due to its lower complication rate compared to OC, supporting its broader adoption. However, several challenges limit its widespread use, including the dependency on bowel cleansing quality, logistical issues in capsule handling, labor-intensive image review ( 12,000 images per investigation), low-resolution imaging, and low completion rates. To address these, we developed an AI-enhanced wireless capsule featuring real-time image processing, dual-mode imaging (white-light and narrow-band), and bi-directional communication with personal devices for reporting findings7. This novel design significantly improves hardware and software capabilities, enhancing diagnostic accuracy and enabling real-time AI analysis. Despite these advancements, gaps in clinical implementation persist, raising feasibility concerns among professionals8.
The CCE pathway includes bowel preparation, capsule ingestion, manual image analysis, and post-procedure care. This study focuses on fully integrating AI into the image analysis stage, enabling autonomous detection, localization, and characterization of findings using advanced algorithms. Characterization involves analyzing abnormalities’ morphology (e.g., size) and histopathological (HP) properties, such as neoplastic features. Building on previous work in polyp detection and localization5,6,9,10,11,12,13,14,15,16,17, we incorporate size estimation and HP analysis to further optimize CCE’s workflow, advancing toward full AI integration in routine clinical practice.
While AI-based detection and classification of colorectal polyps in optical colonoscopy is well-studied18, few studies (besides our own) have explored similar questions in CCE investigations19,20,21. This is largely due to the lack of publicly available datasets for CCE, unlike optical colonoscopy databases such as Kvasir (A Multi-Class Image Dataset for Computer Aided Gastrointestinal Disease Detection). Additionally, CCE’s reliance on invariant white-light imaging (WLI) results in lower-resolution images, posing significant challenges for AI algorithms to achieve the robustness required for routine clinical practice.
This study builds on the “Danish CareForColon2015 trial (cfc2015),” launched in 2021 as part of the Danish Colorectal Cancer Screening program. As the largest randomized controlled trial on CCE22, its primary aim was to compare detected colorectal cancers and intermediate- or high-risk adenomas between intervention and control groups. Secondary aims included evaluating patient acceptability, complication and completion rates, interval CRC rates, patient-reported outcomes (PRO), long-term cancer incidence, social inequality, CCE applicability, and cost-effectiveness. Of 370, 306 individuals invited to screen, 2015 FIT-positive patients underwent CCE investigations, forming the basis for developing and validating the AI algorithms in this study. Further details can be found at https://clinicaltrials.gov/ct2/show/NCT04049357 or one of our recent publications15.
Contribution
The sketch of the workflow associated with CCE’s pathway automation is presented in Figs. 1 and 2. The steps presented in blue have been published in previous works5,6,11,13,14, while those in orange are under development. This paper presents those steps highlighted in green, which focus on the recognition of important abnormalities, estimating their size and define their histopathology.
This paper is organized as follows: We first introduce a deep neural network capable of detecting abnormalities with high sensitivity and specificity. Detected abnormalities are then processed by two parallel algorithms for size estimation and characterization, with details of their explainability (XAI) criteria provided in the article. Finally, we evaluate the outcomes of these algorithms, highlight their strengths and limitations, and conclude on the feasibility of optimizing CCE image analysis with AI.
Methods and results
Code availability
Codes for recognition, characterization and size estimation algorithms developed in this study are available to interested readers upon request to the corresponding author. Consortium agreements signed with the funding agencies i.e., the European Union and UK Research and Innovation Office (co-funded) prohibits us from sharing the codes in public repositories.
Ethics
The study was approved by the Regional Health Research Ethics committee (journal number S-20190100), was registered with the Regional Data Protection Agency (journal number 19/29858), as well as with ClinicalTrials (identifier NCT04049357). All participants received verbal and written study information prior to participation and signed informed consent was obtained from each individual. The study was conducted in accordance with the declaration of Helsinki.
Recognition
Our prior research on colorectal polyp detection and localization using an enhanced ZF-Net achieved \(98.0\%\) accuracy, \(98.1\%\) sensitivity, and \(96.3\%\) specificity on a dataset of approx. 800 images (400 with polyps, 400 normal mucosa)12. While this remains one of the top-performing networks in the literature, deploying it in the cfc2015 trial risked missing up to four cancers among 2015 FIT-positive patients due to the prevalence of CRC. Additionally, its findings serve as input for size estimation and characterization algorithms, necessitating a new DNN with higher negative predictive value (NPV), sensitivity, and specificity. After evaluating leading architectures like ResNet50 and InceptionV3, we selected NasNetLarge as the backbone for abnormality recognition.
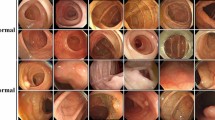
To adapt network’s architecture and to use transfer learning for the purpose of this study, we modified the last 20 learnable layers, and froze the parameters of the remaining layers, accordingly. The learning rate was initially set to \(1\hbox{e}{-}3\), but adaptively reduced after every 2 epochs during the training process, until the validation criteria were met. The epoch size for training process was limited to a maximum of 6, with a mini batch size of 10, and the validation frequency of 798. The dataset containing images of both colorectal polyps (1751) and the normal mucosa (1672) (Fig. 3) was augmented by horizontal and vertical random reflection, random scaling and random translation, along with random rotation, all picked from a continuous uniform distribution, without affecting the contents or the size of the images. This augmentation resulted in a database containing 5838 images of colorectal polyps and 5573 images of normal mucous layer. The dataset was split to \(70\%\) for both training and validation process, while the remaining \(30\%\) was allocated to the test process. NasNetLarge with the aforementioned configuration resulted in a sensitivity of \(99.9\%\), a specificity of \(99.4\%\), and an NPV of \(99.8\%\) on the test set, implying that less than one cancer among the cfc2015 trial subjects will be missed.
To evaluate as whether the network has learnt sufficient features, and knowledge has been transferred, we exposed the recognition network to an additional set of images not previously used for either training or testing. Manual examination by trained CCE readers initially classified three cases as either inflammation or normal tissue, a diagnosis confirmed by specialists. However, during a routine quality check in the cfc2015 trial, medical experts identified these cases as cancers. To evaluate our DNN’s performance, the misdiagnosed images were included in the test set, where the network correctly flagged them as significant findings, demonstrating its generalization capability. Following this incident, the cfc2015 trial experts reexamined all patient images to ensure no critical findings were missed.
Explainable AI (XAI)
To enhance interpretability and trust in our recognition algorithm, three classes of XAI methods for image processing, namely, CartoonX23, Pixel Rate Distortion Explanation (RDE)23 and Class Activation Mapping (CAM) and it’s variations such as GradCam24 were integrated.
Starting off with CAM-based techniques, the explanations from GradCAM, GradCAM++, AblationCAM, and RandomCAM reveal that the proposed recognition model extract most representative features important for the classification task, i.e., the existence of a polyp vs. no-polyp. The parameters, remove and debias nullify the effect of leaking data by a weighted average of its neighbors. Percentile values of [20, 40, and 95] provide the top \(20\%\), \(40\%\), and \(95\%\) of the most important regions identified by various CAM techniques to correlate with the target class. A single score measures the average impact on model confidence by contrasting the removal of least and most important regions of an image. Our overall analysis showed that among the four CAM-based techniques, GradCAM++ explanations map the exact features from the last convolutional layer, which mainly focus on polyps within the image. This is highlighted by the score obtained for each explanation (Fig. 4), hence justifying the decision of the network.
To further confirm the validity of the explanations, we also used CartoonX and pixel rate distortion explanations. Unlike CAM-based models, which rely on output feature maps, CartoonX and pixel RDE explore input features, making them model-independent explainable techniques. To have a faithful comparison between different methods, we used the same image of a polyp for comparing the outputs of CartoonX and pixel RDE with CAM-based techniques. As evident from Fig. 4, both CartoonX and pixel RDE explanations detect the main region of the polyp, being the most critical area of the image. CartoonX provides effective input features due to its ability to extract relevant and piecewise smooth image segments, rather than focusing on sparsely distributed pixel regions. Comparison between CartoonX and pixel RDE explanations reveals that CartoonX explanations show lower distortions for image classification compared to Pixel RDEs, confirming the findings of the original study25. This is since CartoonX provides piecewise smooth explanations, effectively uncovering meaningful patterns that are less apparent with pixel RDE and CAM-based techniques.
Size estimation
The size estimation algorithm builds on our previous work developing AID-U-Net, a novel semantic segmentation network13,14,26. AID-U-Net incorporates direct contracting and expansive paths, along with unique sub-contracting and sub-expansive paths, achieving superior performance (F1 score: \(88.1\%\)) compared to U-Net (\(81.1\%\)) and U-Net++ (\(87.6\%\)), without requiring pre-trained backbones. The optimal architecture for segmenting CCE images was AID-U-Net(2,2), with a depth of two for both the direct path and sub-path. For detailed architecture and performance insights, we refer readers to our earlier work13.
We applied AID-U-Net(2,2) to an augmented dataset of 5,838 colorectal polyp images, achieving correct segmentation in \(81\%\) (4685 images). By comparison, U-Net and U-Net++ achieved \(61\%\) and \(72\%\), respectively. Incorrect segmentation included three scenarios: (1) missing a region of interest (ROI, e.g., a polyp), (2) segmenting the wrong region, or (3) splitting a single ROI into multiple segments. Assuming each image contained one ROI and summing the estimated sizes of all segmented regions improved segmentation accuracy to \(88\%\). Examples of segmented polyps, along with their bounding boxes and fitted ellipses, are shown in Fig. 5.
Examples of the outcome from the segmentation and size estimation algorithm. Purple regions of the images represent pixels associated with detected abnormalities, and the white region is the cropped out version of the same region. Red rectangles represent bounding boxes encompassing the important findings, e.g., polyps.
Precise size estimation requires polyp depth relative to the camera lens, which is unavailable in CCE. Therefore, our algorithm estimates size by calculating the ratio of the largest diameter of the fitted ellipse around the segmented polyp to the total image size, excluding peripheral information (e.g., date and time). Using this approach, we achieved a perfect match with size estimates from the RAPID Reader27 used by trained CCE readers.
Using histopathology reports as the gold standard, the size estimation algorithm mapped polyp sizes from segmented CCE images to pathology outcomes. Currently, 280 polyps are matched between the two datasets (CCE vs. pathology). We observed that CCE generally overestimates polyp sizes compared to pathology reports. The best regression model, using fine Gaussian support vector machines (SVM), achieved a root mean squared error (RMSE) of approximately 6mm. Despite this error, a richer database of matched polyps, currently being expanded through the cfc2015 trial, will improve accuracy.
Characterization
Colorectal polyps are classified as neoplastic or non-neoplastic, with this classification-alongside size, histology, and location (distal vs. proximal colon)-guiding patient management and treatment success. Our characterization algorithm is a binary classifier, taking CCE images flagged as important findings and categorizing them as neoplastic or non-neoplastic.
The dataset includes 479 images of polyps observed during CCE, resected, and matched post-polypectomy. Of these, 317 were neoplastic and 162 non-neoplastic. To address the small sample size and class imbalance (2:1 ratio), we augmented the dataset fourfold using random horizontal/vertical reflections, scaling, translation, and rotation without altering image content or size. Although only polyp segments should inform pathology, we used entire image frames as many featured small polyps occupying minimal space.
Using the same training settings as the recognition DNN, we implemented a VGG16 network as the backbone for the characterization algorithm. The dataset was split into \(70\%\) for training/validation and \(30\%\) for testing, yielding a binary classifier with \(84.3\%\) sensitivity, \(81.5\%\) specificity, and \(82\%\) accuracy.
To enhance the explainability of the VGG16 characterization network, we assigned relevance to the learned patterns using pattern attribution techniques such as Layer-wise relevance propagation (LRP). LRP interprets neural network predictions by attributing relevance scores to individual input features, such as pixels in an image28,29. Starting from the output of the network, relevance is traced backward, layer by layer, down to the input while ensuring that the total relevance remains constant across layers. This process highlights which input features are most influential in the prediction. We implemented the following LRP variants as shown in Fig. 6:
-
Epsilon Rule: incorporating a small positive constant \(\epsilon\) to prevent division by small values when distributing relevance proportionally to the weighted activations of neurons, reducing noise in the relevance attribution. We set \(\epsilon = 0.1\).
-
Alpha-Beta Rule: splitting relevance between positive (\(\alpha\)) and negative (\(\beta\)) contributions, where \(\alpha +\beta =1\), allowing flexible focus on supportive or opposing evidence. We set \(\alpha =1\) and \(\beta =0\) so that only positive contributions are highlighted.
-
Gamma Epsilon Rule: enhancing relevance for positive contributions by applying a factor \(\gamma >0\), to emphasize important features. Further, we set \(\epsilon = 0.1\) to avoid division by small values.
-
Patter net: rather than attributing relevance to individual features based on activations or weights, Patternnet focuses on specific patterns in the input data that are most aligned with the network’s learned features.
-
Pattern attribution: relevance is attributed not just to the individual input features but to the patterns that are learned by the network, highlighting their contribution to the final decision.
In Fig. 6, the regions marked in red have a significant influence on the network’s decision, while the regions marked in blue have a lesser impact. From this figure, we can see that PatternAttribution offers a clearer visualization of the features influencing the polyp characterization network’s predictions. Unlike other methods that focus on pixel-level or activation-level relevance, PatternAttribution captures the relationship between input patterns and the model’s decision-making process, providing a more intuitive and comprehensive understanding of how the network made its decisions.
To enhance performance by incorporating polyp texture, we also applied neural style transfer (NST), a technique effective in image stylization30,31. By leveraging Gram matrices from different convolutional layers, we captured a stationary, multi-scale representation of texture through filter response correlations, independent of global image arrangement. Since neoplastic and non-neoplastic polyps exhibit distinct textures, this texture-based information supplemented the VGG16 classification, utilizing Gram matrix outputs as inner products of vectorized feature maps across layers.
Discussion and conclusions
The Danish National Institute of Health’s Technology Assessment Board recently decided against recommending AI for colonoscopy in diagnosing neoplastic disease. Their decision was based on two main reasons: insufficient evidence, with only a meta-analysis of two trials from the same authors sponsored by the manufacturer, and outdated clinical guidelines that require the removal of all polyps, including many insignificant ones. Current guidelines mandate the removal of all colorectal polyps, which means AI’s ability to detect even insignificant polyps would increase the treatment burden on patients and the healthcare sector. Despite growing interest in AI solutions in radiology, the Danish Treatment Council’s decision highlights the healthcare sector’s unreadiness to adopt AI for gastrointestinal disorder detection. This is due to the lack of robust, generalizable models in gastroenterology, unlike in radiology where extensive public databases exist, and the complexity of AI-based radiology reports that provide detailed diagnostic information.
Clinical trials like CFC2015 and initiatives in NHS Scotland (ScotCap)32 and NHS England33 to implement colon capsule endoscopy in primary care will increase data availability. Additionally, research like this offers AI-based solutions comparable to those in radiology, bridging the gap in AI deployment between gastroenterology and radiology. The goal is to develop algorithms that generate comprehensive patient reports, similar to those in other clinical fields where AI is more established.
To achieve this goal, each algorithm (Fig. 1) must perform reliably. CCE videos are a sequence of images that are sampled at a variable frequency. The main difference between still-image analysis and video analysis is that the temporal information carried by the video, i.e., order in the sequence of images is partially discarded during still-image analysis. While some tasks such as tracking the path of the endoscopic capsule (localization of important findings) based on feature point tracking require temporal information of the video, other tasks such as polyp recognition, characterization or size estimation do not necessarily benefit from the inclusion of the temporal information. We reported these findings in one of our previous studies12 where the performance of the AI algorithms on both still image and video analysis were similar.
The recognition network has shown exceptional sensitivity, specificity, and NPV, making it ready for external validation with over 3000 new CCE videos from the ScotCap trial. For each patient, the network will identify candidate images with polyps and other significant findings. Inclusion of XAI techniques such as heat map, CartoonX, Pixel RDE and GradCam explanations enhances both interpretability and trust in algorithm’s decisions. This is particularly important for misclassified cases, and those such as the one shown in Fig. 4 (second row). Despite correctly classifying the image as one containing polyp, the DNN based its decision partly on regions that feature normal mucosa.
The size estimation algorithm’s first component uses the segmentation results from AID-U-Net(2,2). This network outperformed UNet and UNet++ and matched UNetResNet’s \(84\%\) accuracy, despite having fewer parameters. The second component is a Gaussian SVM-based regression estimator that maps CCE findings to histopathology sizes. Our results showed a systematic overestimation by CCE, with a 6 mm size estimation error (RMSE) compared to histopathology.
Several studies have shown that CCE often overestimates polyp sizes compared to freshly retrieved (OC) and formalin-fixed (histopathology) polyps, especially for polyps smaller than 6 or 10 mm10,17. This discrepancy between CCE and OC is due to differences in morphological assessment, with polyps appearing more “pedunculated” in CCE and more “flat” in OC, likely due to colon inflation during OC34. The 6 mm size estimation error (RMSE) between CCE and histopathology is partly due to a small dataset, which will improve with more data. Switching from a regression-based to a classification-based size estimator, dividing sizes into four classes (\(\le 6\,\hbox{mm}\), \(7\,\hbox{mm} \le \cdots <10\,\hbox{mm}\), \(10\,\hbox{mm} \le \cdots < 20\,\hbox{mm}\), and \(\ge 20\,\hbox{mm}\)) as shown in Fig. 7, can reduce patient classification uncertainty. However, there still remains a gap between the performance of our size estimation algorithm and AI-assisted colonoscopy34.
Knowing the exact location of a polyp is very important for the follow-up colonoscopy. In one of our previous studies11, we developed a novel localization technique using feature point tracking that addressed the issue. However, this algorithm required a considerable amount of time (in order of hours) to run and to reconstruct the path that the capsule has taken through the GI tract. Knowing the precise location of a polyp instantly using CCE videos remains an open challenge, and therefore, we provided an alternative solution. By detecting anatomical landmarks such as flexures, we are capable of reporting the approximate location of polyps, i.e., ascending, transverse or descending colon and guide the colonsocopist to the resection site.
The two strategies for the characterization algorithm-training a VGG16 network and using neural style transfer for texture analysis-have been effective. Inclusion of layer-wise relevance propagation-based XAI to better capture the important regions of the image, by attributing relevance scores to individual input features, has been effective. Despite clear advantage of PatternAttribution over other LRP-based algorithms, it can be observed in Fig. 6 that in one case (top middle), the network highlighted regions that feature normal mucosa. We anticipate that increasing the number of images and including larger polyps will improve the algorithm’s performance. Analyzing texture information of segmented polyps and surrounding tissue will help quantify vascularity, aiding in distinguishing between neoplastic and non-neoplastic classes.
While we are externally validating our algorithms with ScotCap data, improving their performance remains a priority. Annotating data and achieving interobserver and intraobserver agreement in CCE and OC evaluations require hybrid active learning approaches. Strategies like Self Supervised Learning (SSL), Balance Exploration and Exploitation (BEE) and conformal prediction (CP) can reduce the number of samples needed by querying the teacher network for labels. Future work includes completing all algorithms, external validation with ScotCap data, and enhancing algorithm performance. We also plan to adopt radiology workflow solutions, such as PACS and DICOM, and transfer CCE-generated non-DICOM images to cloud-based PACS for real-time analysis, report generation, and sharing with external medical professionals.
Data availability
The data used in this article is part of the cfc2015 trial’s outcome, which belongs to the “Odense University Hospital (OUH)”. The request to access the data, or part of it should be made to Prof. Gunnar Baatrup.
References
Thygesen, M. et al. Screening individuals experiences of colonoscopy and colon capsule endoscopy: A mixed method study. Acta Oncol. 71–76, https://doi.org/10.1080/0284186X.2019.1581372 (2019).
Zammit, S. C. & Sidhu, R. Capsule endoscopy-recent developments and future directions. Expert Rev. Gastroenterol. Hepatol. 15, 127–137. https://doi.org/10.1080/17474124.2021.1840351 (2020).
Gonzalez-Suarez, B. et al. Colon capsule endoscopy versus ct colonography in fit-positive colorectal cancer screening subjects: A prospective randomised trial-the vicoca study. BMC Med. 18, https://doi.org/10.1186/s12916-020-01717-4 (2020).
Atkin, W. et al. Computed tomographic colonography versus colonoscopy for investigation of patients with symptoms suggestive of colorectal cancer (siggar): A multicentre randomised trial. The Lancet 381, 1194–1202. https://doi.org/10.1016/S0140-6736(12)62186-2 (2013).
Buijs, M. et al. Assessment of bowel cleansing quality in colon capsule endoscopy using machine learning: A pilot study. Endosc. Int. Open 6, E1044–E1050. https://doi.org/10.1055/a-0627-7136 (2018).
Schelde-Olesen, B. et al. Interobserver agreement between an artificial intelligence algorithm and colon capsule endoscopy readers on bowel-cleansing quality. iGIE 2, 148–153. https://doi.org/10.1016/j.igie.2023.04.006 (2023).
Sahafi, A. et al. Edge artificial intelligence wireless video capsule endoscopy. Sci. Rep. 12, 13723 (2022).
Koulaouzidis, A. & Baatrup, G. Current status of colon capsule endoscopy in clinical practice. Nat. Rev. Gastroenterol. Hepatol. 1–2 (2023).
Schelde-Olesen, B. et al. Interobserver agreement on landmark and flexure identification in colon capsule endoscopy. Tech. Coloproctol. 1–7 (2023).
Blanes-Vidal, V., Nadimi, E. S., Buijs, M. M. & Baatrup, G. Capsule endoscopy vs. colonoscopy vs. histopathology in colorectal cancer screening: Matched analyses of polyp size, morphology, and location estimates. Int. J. Colorectal Dis. 33, 1309–1312 (2018).
Herp, J. et al. Feature point tracking-based localization of colon capsule endoscope. Diagnostics 11, https://doi.org/10.3390/diagnostics11020193 (2021).
Nadimi, E. S. et al. Application of deep learning for autonomous detection and localization of colorectal polyps in wireless colon capsule endoscopy. Comput. Electric. Eng. 81, 106531. https://doi.org/10.1016/j.compeleceng.2019.106531 (2020).
Tashk, A., Herp, J., Bjørsum-Meyer, T., Koulaouzidis, A. & Nadimi, E. S. Aid-u-net: An innovative deep convolutional architecture for semantic segmentation of biomedical images. Diagnostics 12, https://doi.org/10.3390/diagnostics12122952 (2022).
Tashk, A., Şahin, K. E., Herp, J. & Nadimi, E. S. A cnn architecture for detection and segmentation of colorectal polyps from cce images. In 2022 IEEE 5th International Conference on Image Processing Applications and Systems (IPAS) Vol. 5, 1–6 https://doi.org/10.1109/IPAS55744.2022.10052795 (2022).
Deding, U. et al. Colon capsule endoscopy vs. CT colonography following incomplete colonoscopy: A systematic review with meta-analysis. Cancers 12, https://doi.org/10.3390/cancers12113367 (2020).
Deding, U. et al. Colon capsule endoscopy versus ct colonography after incomplete colonoscopy. application of artificial intelligence algorithms to identify complete colonic investigations. United Eur. Gastroenterol. J. 8, 782–789. https://doi.org/10.1177/2050640620937593 (2020).
Blanes-Vidal, V., Baatrup, G. & Nadimi, E. S. Addressing priority challenges in the detection and assessment of colorectal polyps from capsule endoscopy and colonoscopy in colorectal cancer screening using machine learning. Acta Oncol. 58, S29–S36. https://doi.org/10.1080/0284186X.2019.1584404 (2019).
Lou, S. et al. Artificial intelligence for colorectal neoplasia detection during colonoscopy: A systematic review and meta-analysis of randomized clinical trials. eClinicalMedicine 66, 102341. https://doi.org/10.1016/j.eclinm.2023.102341 (2023).
Moen, S., Vuik, F. E., Kuipers, E. J. & Spaander, M. C. Artificial intelligence in colon capsule endoscopy—a systematic review. Diagnostics 12, 1994 (2022).
Saraiva, M. et al. Artificial intelligence and colon capsule endoscopy: Development of an automated diagnostic system of protruding lesions in colon capsule endoscopy. Tech. Coloproctol. 25, 1243–1248 (2021).
Yamada, A., Niikura, R., Otani, K., Aoki, T. & Koike, K. Automatic detection of colorectal neoplasia in wireless colon capsule endoscopic images using a deep convolutional neural network. Endoscopy 53, 832–836 (2021).
NCT04049357. Care for colon 2015 (cfc2015). https://clinicaltrials.gov/study/NCT04049357 (2021).
Grohs, P. & Kutyniok, G. Mathematical Aspects of Deep Learning (Cambridge University Press, 2022).
Sterkenburg, T. F. & Grunwald, P. D. The no-free-lunch theorems of supervised learning. Synthese 199, 9979–10015 (2021).
Kolek, S., Nguyen, D. A., Levie, R., Bruna, J. & Kutyniok, G. Cartoon explanations of image classifiers. In European Conference on Computer Vision Vol. 13672. https://doi.org/10.1007/978-3-031-19775-8_26 (2022).
Tashk, A., Herp, J. & Nadimi, E. S. Fully automatic polyp detection based on a novel u-net architecture and morphological post-process. In 2019 International Conference on Control, Artificial Intelligence, Robotics & Optimization (ICCAIRO), Vol. 5, 37–41 https://doi.org/10.1109/ICCAIRO47923.2019.00015 (2019).
MedTronic. Rapid reader software. https://www.medtronic.com/covidien/en-us/support/software/gastrointestinal-products/rapid-reader-software-v8-3.html.
Montavon, G., Binder, A., Lapuschkin, S., Samek, W. & Müller, K. R. Layer-wise relevance propagation: An overview. In Explainable AI: Interpreting, Explaining and Visualizing Deep Learning. Lecture Notes in Computer Science Vol. 11700 (eds Samek, W., Montavon, G., Vedaldi, A., Hansen, I., Müller, K. R.) (Springer, Cham, 2019). https://doi.org/10.1007/978-3-030-28954-6_10.
Kindermans, P. J. et al. Learning how to explain neural networks: Patternnet and patternattribution. arXiv:1705.05598 (2017).
Gatys, L. A., Ecker, A. S. & Bethge, M. A neural algorithm of artistic style. Vision 16, 326 (2016).
Li, Y., Wang, N., Liu, J. & Hou, X. Demystifying neural style transfer. In Proceedings of the Twenty-Sixth International Joint Conference on Artificial Intelligence (IJCAI-17) 2230–2236 (2017).
Health, D. & Center, C. I. Scotcap. https://www.dhi-scotland.com/projects/scotcap/.
England, N. NHS England. https://www.england.nhs.uk/2021/03/nhs-rolls-out-capsule-cameras-to-test-for-cancer/.
Park, D. K. et al. A prospective multicenter randomized controlled trial on artificial intelligence assisted colonoscopy for enhanced polyp detection. Sci. Rep. 14, 71–76. https://doi.org/10.1038/s41598-024-77079-1 (2024).
Acknowledgements
This research is part of AICE project (number 101057400) funded by the European Union, and it is part-funded by the United Kingdom government. Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them.
Author information
Authors and Affiliations
Contributions
E.S.N. and G.B. conceptualized the study. E.S.N., J.M.B, S.K., and V.C.G developed the methods. B.S.O. and G.B. conducted the clinical trial and collected, and pre-processed the data. E.S.N., J.M.B., S.K., V.C.G., and V.B.V. analyzed the results. E.S.N. and V.B.V. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nadimi, E.S., Braun, JM., Schelde-Olesen, B. et al. Towards full integration of explainable artificial intelligence in colon capsule endoscopy’s pathway. Sci Rep 15, 5960 (2025). https://doi.org/10.1038/s41598-025-89648-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-89648-z