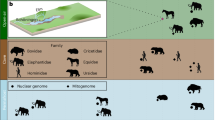
Abstract
Mitochondria are semi-autonomous organelles that play a crucial role in the energy budget of animal cells and are closely related to the locomotor abilities of animals. Equidae is renowned for including two domesticated species with distinct purposes: the endurance-oriented donkey and the power-driven horse, making it an ideal system for studying the relationship between mitochondria and locomotor abilities. In this study, to cover the genetic diversity of donkeys, we sequenced and assembled six new mitochondrial genomes from China. Meanwhile, we downloaded the published mitochondrial genomes of all species within Equus and conducted a comprehensive pan-mitochondrial genome analysis. We found that the mitochondrial genomes of Equus are highly conserved, each encoding 37 genes, including 13 protein-coding genes (PCGs). Phylogenetic analysis based on mitochondrial genomes supports previous research, indicating that the extant species in Equus are divided into three main branches: horses, donkeys, and zebras. Specifically, 761 genetic variants were identified between donkeys and horses, 68 of which were non-synonymous mutations in PCGs, potentially linked to their different locomotor abilities. Structural protein modeling indicated that despite genetic differences, the overall protein structures between donkeys and horses remain similar. This study revealed the mitochondrial genome variation patterns of domesticated animals, offering novelty perspectives on domestication imprints. Additionally, it provides reliable candidate molecular markers for the identification of donkeys and horses.
Similar content being viewed by others
Introduction
The mitochondria, as the only organelle in animal cells besides the nucleus that possesses unique genetic material, originated from endosymbiosis1,2. Over billions of years, the number of genes encoded within the mitochondrial genome (mitogenome) has decreased from thousands in ancestral protobacteria to only dozens in typical animal mitochondria, which encoded 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and 2 ribosome RNA (rRNA) genes3,4,5. These genes are closely related to the electron transport chain of cellular respiration in animals, providing over 90% of the cell’s energy budget6,7. Previous studies have confirmed that the mitogenome of flightless birds (e.g., Pavo, Gallus, and Numida) is under significantly lower selection pressure compared to flying birds, suggesting that flightless birds require less energy to sustain their movement. Even if minor mutations affecting mitochondrial gene functions occur in the mitogenome of flightless birds, the energy efficiency decreases without posing significant harm to the survival and reproduction of individuals8. Similarly, the flight ability of insects is closely related to the evolutionary rate of their mitogenomes9. This hypothesis was also supported in the analysis of 214 mammalian mitogenomes, indicating that the mitogenomes of weakly locomotive groups experience significantly relaxed selection pressure compared to strongly locomotive groups8,10. Despite previous analyses of selective pressures on the whole mitogenome, there is still an urgent need for more in-depth research to identify which specific genes and variants have profound effects on mitochondrial metabolism and animal locomotor abilities. Additionally, animal mitogenomes are typically single circular DNA molecules, ranging from 15 to 20 kb (kilobase pairs) in size11,12 They are primarily maternally inherited and exhibit rare recombination13. Notably, the evolutionary rate of animal mitogenomes is approximately 5–10 times faster than that of nuclear genomes14. Due to these advantages, mitogenomes are widely used in reconstructing phylogenetic relationships across various taxonomic levels in animals15,16,17.
Equidae contains only one extant genus, Equus, including eight living species. These species are grouped into horses (Equus caballus, Equus przewalskii), donkeys (Equus asinus, Equus homionus, and Equus kiang), and zebras (Equus zebra, Equus quagga, and Equus grevyi)18,19,20,21. Among those species, horses and donkeys are the most renowned domesticated animals. In traditional agricultural societies, both of them were served as vital means of transportation, extensively used for riding and carrying goods22,23,24. Horses, known for their speed, are ideal for rapid movement and long-distance travel20. Donkeys, while slower, excel in heavy load-bearing and short-distance transport25. Additionally, donkeys exhibit remarkable adaptability to harsh environments, especially in mountainous and arid regions, where they are frequently employed for labor. Compared to horses, donkeys are less costly to maintain, making them ideal for resource-limited environments25,26,27,28. Given that both horses and donkeys belong to the Equus and have been domesticated to suit different purposes, there are a large amount of publicly available data. This makes them ideal subjects for studying the mitochondrial imprints of domestication on locomotion abilities. Furthermore, donkey meat is characterized by its fine texture and palatable taste, being rich in protein and low in fat. In traditional Chinese medicine, it is reputed for its blood-nourishing, energy-boosting, and muscle-strengthening properties29,30. Due to limited supply and high demand, donkey meat is typically more expensive31. Conversely, horse meat is more widely available and therefore tends to be more affordable. Consequently, there is an urgent need to develop molecular markers for the accurate identification of these two species.
Over the past century, the origin, diversification, and domestication history of Equus have been extensively studied, establishing it as a model organism for the study of macroevolution. However, previous phylogenetic studies of this genus primarily relied on morphological characters, which were often subject to the judgments of researchers32,33,34,35,36. Recently, a study reconstructed the phylogenetic relationships of extant and middle-late Pleistocene equids using the whole mitogenomes and 17 nuclear gene fragments, resulting in a robust phylogenetic tree. This research demonstrates that mitogenomes are suitable for reconstructing the phylogenetic relationships of Equus. Although the sample size was relatively small, the study produced a robust phylogenetic tree. However, the study lacked sampling of the Equus przewalskii, one of the eight extant species22. Therefore, studying the phylogenetic relationships and adaptive evolution of extant species within Equus, more comprehensive sampling across the genus is still needed.
In this study, in order to cover the diversity of donkeys as much as possible, we assembled six mitogenomes of donkeys distributed in China using second-generation sequencing technology and downloaded the published mitogenomes of all species in Equus. In particular, we conducted pan-genomic analyses of Equus mitogenomes to identify independent domestication signatures in donkeys and horses and to develop reliable candidate molecular markers between donkeys and horses.
Materials and methods
Sample collection, DNA extraction and sequencing
The samples of six donkeys were collected from Dezhou (Shandong, China) in July 2023. A total of five mL blood was obtained from each accession and all samples were stored at -20 ℃. Then, the genomic DNA was extracted by the DNeasy Tissue Kit (QIAGEN, Beijing, China) and following the manufacturer’s instructions. The quality of the DNA was assessed by 1% agarose gel electrophoresis and NanoDrop 2000 (Thermo Fisher). Subsequently, 1 μg of genomic DNA was fragmented to ~ 500 bp using the Covaris M220 system. The fragmented DNA was then used to construct short-insert libraries according to the manufacturer’s instructions (TruSeq Nano DNA Sample Prep Kit, Illumina). Sequencing was performed on an Illumina NovaSeq 6000 platform (BIOZERON Co., Ltd., Shanghai, China) with a 150 bp paired-end read length.
All samples used in this study were obtained from breeding farms. Approval was granted by the Ethics Committee of Institute of Forensic Science, Ministry of Public Security (MPS) (protocol code 2023-002 and approved on March 16, 2023). The study was conducted in accordance with the local legislation and institutional requirements. All experiments were performed in accordance with ARRIVE guidelines.
Mitogenome assembly and annotation
First, we performed quality control of the raw data using FastQC v0.12.0 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Trimmomatic v0.3937, removing bases with quality scores below 20, adapter sequences, and reads shorter than 75 bp. Next, we used GetOrganelle v1.7.5 to perform the initial assembly of the clean data38. Candidate mitochondrial scaffolds were identified using BLAST v2.8.1 against the NT database, and these scaffolds were connected based on overlaps, ultimately completing the assembly of the six donkey mitogenomes39,40.
Subsequently, we used MITOS to predict protein-coding, tRNA, and rRNA genes within these six mitogenomes, selecting codon Table transl_table=2 (The Vertebrate Mitochondrial Code)41. We then manually corrected the positions of the start and stop codons using BioEdit v7.7.142. Finally, we utilized Proksee43 to create a circular visualization of the newly assembled mitogenomes and submitted these mitogenomes to NCBI (https://www.ncbi.nlm.nih.gov/) (Accession Number: PP049795-PP049800). We also used our annotation pipeline to re-annotate previously published, unannotated mitogenomes.
Evolutionary analyses
To reconstruct the phylogenetic relationships within Equus, we integrated six newly sequenced mitogenomes with 46 previously published ones, using Tapirus bairdii (NC_063943) and Tapirus terrestris (NC_053962) as outgroups (Table S1)44. We reconstructed phylogenetic relationships of Equus based on all 13 PCGs. The specific workflow was as follows: (1) The whole mitogenomes and the 13 PCGs were aligned using MAFFT v7.045, and then hypervariable regions were removed by Gblocks v. 0.9146 to obtain reliable alignment matrices. (2) Maximum likelihood (ML) trees based on the whole mitogenome and the concatenated matrices of 13 PCGs were constructed by IQ-TREE v1.6.10 with 1,000 bootstrapped replicates (BP), respectively47. (3) The phylogenetic trees were visualized and refined using the tvBOT online website48.
We employed DnaSP v6.12.03 to calculate the Ka, Ks, and Ka/Ks for the reliable alignment matrices of the 13 PCGs obtained in the previous step49. Additionally, we selected PP049795 as the representative of the six newly assembled mitogenomes and analyzed the relative synonymous codon usage (RSCU) of the 13 PCGs by PhyloSuite v1.2.350.
Mitochondrial variants between horses and donkeys identification
To comprehensively cover the genetic diversity of donkeys and horses, we downloaded and analyzed all published mitogenomes from NCBI (April, 2024), totaling 161 donkey and 560 horse mitogenomes (Table S1). First, we checked and modified the annotation information of all donkey and horse mitogenomes using our pipeline in section "Mitogenome assembly and annotation". Next, the annotated mitogenomes were imported into PhyloSuite v1.2.3 to extract the PCGs. Then, each PCG was aligned by MAFFT v7.0 and manual corrected by BioEdit v7.7.1 to obtain reliable alignment matrices. Finally, specific variants between donkey and horse mitogenomes were manually identified by BioEdit v7.7.1, and the variant types, including synonymous and nonsynonymous mutations, were determined.
Protein structure prediction
The protein structures were predicted using the online prediction tool available on the SWISS-MODEL website (https://swissmodel.expasy.org)51. The parameters included finding suitable templates first, building the best model, and selecting the model with the highest GMQE value. The models were exported in PDB format to PyMol v3.0.0 (https://pymol.org/) for analysis and further refined using UCSF ChimeraX v1.7.152.
Results
General features of Equus mitogenomes
To cover the genetic diversity of donkeys, we newly sequenced, assembled, and annotated six donkey mitogenomes. These mitogenomes are typical circular molecules, ranging from 16,536 to 16,696 bp in size, and each contains 37 genes, including 13 PCGs (atp6, atp8, cox1, cox2, cox3, cytb, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6), 22 tRNA genes, and 2 rRNA genes (Fig. 1, Table S2). Atp8 is the shortest PCG among the 13 PCGs, which is 204 bp in length, whereas nad5 is the longest PCG, which is 1821 bp in length. The lengths of all 22 tRNA genes range from 66 to 75 bp. The length of the 12S rRNA gene is 975 bp, while the length of the 16S rRNA gene is 1582 bp. The overlapping regions are generally small, with the largest being only 10 bp between nad5 and nad6. In addition, the highest frequency of start codon ATG and stop codon TAA. The nucleotide composition of the E. asinus mitogenome is as follows (Table S3): A = 32.4%, T = 25.6%, G = 13.2%, and C = 28.9%. The entire GC-skew and AT-skew of the E. asinus mitogenome were − 0.374 and 0.117, respectively. The AT skew for the E. asinus mitogenome was slightly positive. This suggests that A nucleotides are more abundant than T nucleotides. The GC-skew for the E. asinus mitogenome was scarcely negative, with C nucleotides outnumbering G nucleotides.
Starting with atp6, we mapped the mitochondrial gene sequences of the Equus species and compared them with our samples (Fig. 2A). We found that, except for E. a. somalicus and E. hydruntinus, all other species possess 37 mitochondrial genes. Specifically, the transcription directions of tRNA-P and tRNA-Y in E. a. africanus and tRNA-C in E. kiang, differ from those in other species. E. a. somalicus has lost tRNA-F, while E. hydruntinus has only one copy of nad5. In addition to this, we analyzed the base content of the genomes of all existing species in the entire Equus, the base content of different codon sites, codon usage, etc. (Table S4). In the relative synonymous codon usage (RSCU) of our samples, codons CUA (L), CGA (R), UCA (S), and UGA (W) are used more frequently, and codon GCG (A) is less used (Fig. 2B). Taking the genus Equus on average, the analysis of amino acid content showed that the contents of Leu, Ile, Thr, Ser, Ala, and Phe were higher than 6%; especially, Leu was the highest over 16% (Table S5, Fig. S1), and the contents of Cys and Arg were lower at 0.6% and 1.69%, respectively. E. asinus had a higher rate of Ile (9.3%) than the other species (9.1%). Among the mitochondrial protein-encoding genes of Equus species, the codons CUA (L), CGA (R), UCA (S), UGA (W), AAA (K), ACA (T), GCC (A), and GUA (V) are used more frequently, while the codon GCG (A) is used less frequently, particularly in our sample. The start codon of Equus mitochondrial genes cox1, cox2, cox3, nad1, atp6, atp8, and nad6 is ATG, and nad3 is ATA.
(A) Part of complete mitogenome alignment map of Equus; rectangles for PCGs, lozenges for rRNA, circles for tRNA, the arrows are their transcriptional direction. Unshown species are same with our samples. (B) The relative synonymous codon usage (RSCU) in the mitogenome of E. asinus (PP049795). (C) Comparative analysis of selective pressures exerted on PCGs for all Equus. Ka/Ks ratios for all 13 PCGs of all Equus; red for Ka value, bule for Ks value, and yellow line graph refers to Ka/Ks value. The left vertical axis represents the Ka or Ks, while the right vertical axis represents the Ka/Ks ratio.
Many sites are missing in E. asinus somalicus, presumably due to inaccurate sequencing or annotation errors. There are two species (E. caballus, E. przewalskii) whose initiation codon of the nad5 gene is ATG, and the initiation codon of the other species is ATA. Only E. grevyi has an ATG start codon for nad3, while the other species have an ATA. Only E. hydruntinus has the nad5 copy2 gene with the start code ACC. Each animal has three or four incomplete stop codons, which are completed to TAA by Poly-A addition at the mRNA 3′ end. Most Equus animals’ atp8 genes stop with TAA, except E .caballus, which has a TAG stop codon. The nad2 genes in E. asinus, E. asinus somalicus, and E. caballus, the atp8 genes in E. caballus, and the nad1 gene in E. caballus all feature TAG stop codons (Table S6).
The analysis showed that the Ka/Ks values of the 13 PCGs were all significantly less than 1 (Fig. 2C), with atp8 having the maximum value of 0.158 and cox1 having the minimum value of 0.002. The mutations resulted in synonymous substitutions, and Equus species were subject to purifying selection during the evolutionary process.
Phylogenomics of Equus based on mitogenomes
We reconstructed the phylogenetic tree of Equus using the complete mitogenomes and the 13 PCGs. This analysis included all extant species as well as extinct species with published mitogenomes. Branches with bootstrap support values below 50 were collapsed. The topologies of both trees were nearly consistent (Fig. 3). The eight extant species in Equus clustered into three main clades with large bootstrap values: Subgenus Equus, Subgenus Asinus, and Subgenus Zebra, consistent with the previous studies of phylogeny of Equus22,53,54. The first diverged clade was Subgenus Equus, which includes the E. przewalskii and the domestic horse (E. caballus). E. przewalskii was nested within the domestic horse. Next, the Subgenus Asinus and Subgenus Zebra lineages diverged. Subgenus Zebra consists of three monophyletic branches: E. grevyi, E. quagga, and E. zebra. E. zebra was the first diverged species, followed by the sister species of E. grevyi and E. quagga. The last clade, Subgenus Asinus, is composed of E. asinus, E. hemionus, and E. kiang. The six newly sequenced domestic donkey individuals in this study were distributed across different positions within the domestic donkey clade, indicating that our sampling was highly representative. Notably, the ten accessions of domestic horses (including Przewalski’s horse) and 21 accessions of domestic donkeys (including African wild donkeys) each clustered into distinct branches with very high support. This provides an excellent opportunity to explore the domestication signatures in domestic horses and donkeys.
The ML tree of Equus inferred from the 13 PCGs. The terminal branch numbers represent GenBank accession numbers. The left color blocks designate different species. The right narrow color blocks designate the different subgenus. Gray for extinct (E. h. hemippus, E. h. hydruntinus, E. q. quagga, E. ovodovi). The black asterisk are our samples. Support values and tree scale are shown.
Mitochondrial variations between donkeys and horses
We downloaded the complete mitogenome data of donkeys and horses available on NCBI as of February 2024 and annotated some unannotated data by our pipeline. For interspecies analyses, we excluded unsuitable data, such as low-quality sequences (N > 1%)55, to ensure accuracy. Since the products encoded by PCGs are closely related to the electron transport chain in respiration, we focused our analysis primarily on the PCGs.
There are 761 species-specific variants between domestic donkeys and horses, including 693 synonymous variants and 68 nonsynonymous variants within the 13 PCGs (Table 1 and Fig. 4A). The percentage of site changes ranged from 5.5 to 7.9%, with an overall rate of 6.7%. The number of species-specific sites ranges from 15 in atp8 to 141 in nad5. For nonsynonymous species-specific sites, nad5 has the most with 16 sites, while cox1, cox2, cox3, and nad3 each have only one variant. The atp8 gene had the highest percentage of amino acid differences at 9.0%. The percentage of changes ranged from 0.20 to 9.0%, with an overall rate of 2.1%. Among the 13 PCGs, the rate of site changes was relatively stable and most of the changes were synonymous. The rate of protein changes, on the other hand, was more variable among the 13 PCGs. (Table S7).
(A) Nucleotide substitutions and amino acid changes in different genes of donkey and horse. Horizontal coordinates are the different genes, sorted in order of nucleotide change. Left horizontal coordinates are the number of substitutions, light blue for nucleotides and purple for amino acids. Right vertical coordinates are the proportion of changes, red for nucleotide and yellow for amino acids. (B) Proportional distribution of amino acid use in donkeys and horses. Vertical coordinates are amino acids and horizontal coordinates are proportions. Donkeys are shown in blue and horses in red. The higher the probability of use, the larger the circle. (C) Nucleotide substitutions in PCG between donkey and horse species. Horizontal coordinates are the nucleotide types of donkeys at the corresponding loci, vertical coordinates are the number of substitutions of different nucleotides at the corresponding loci in horses, T in red, G in yellow, C in blue and A in green. (D) Distribution of codon sites for donkey and horse changes. Vertical coordinates are the number of substitutions and horizontal coordinates are the different genes. Red for first coden, green for second coden, blue for third coden, and gray is the total number.
Comparative analyses of amino acid usage rates between domestic donkeys and horses (Fig. 4B) showed that the most significant difference was for Leu, with a difference of 0.47%, followed by Val with a difference of 0.24%. Five amino acids had identical usage rates: Cys, Glu, Lys, Pro, and Trp. Additionally, we analyzed the changes between the four types of nucleotides (Fig. 4C). Among the 761 substitutions in PCGs, the most frequent was between C and T at 481, followed by A and G at 222, with almost no substitutions between other bases. This indicates that transitions significantly outnumber transversions56, which is consistent with known conclusions. Codon position substitutions are also consistent with the known conclusions (Fig. 4D), with the majority of substitutions occurring in the third codon position, with 644 substitutions (84.63%). The first position has 96 substitutions (12.61%), and the second position has only 21 substitutions (2.76%).
Three-dimensional structural predictions of the proteins show that the mitochondrial DNA (mtDNA) of donkeys and horses encodes proteins with essentially similar structures but with minor differences (Fig. 5, Fig. S2). In the atp8 gene, which has the highest percentage of amino acid changes, 6 out of 67 amino acids differ, RMSD (Root Mean Square Deviation) is 0.003 Å. However, the overall structure of the proteins did not change.
Predicted three-dimensional structure of the protein encoded by the atp8 gene in donkey and horse. Blue is donkey, red is horse, and green are the changed amino acids. This image was generated by ChimeraX v1.7.1 (https://www.rbvi.ucsf.edu/chimerax).
Discussion
Relatively conserved mitogenomes of Equus
Studies of mitochondrial DNA in Equus have a long history57,58,59. Fundamental research forms the basis for understanding the genetic dynamics and evolutionary history of Equus species60, offering a framework for conservation efforts and further studies on their adaptation and diversity16,61. In this study, we sequenced and de novo assembled the mitogenomes of six donkey samples. We found that the mitogenomes of all six donkey samples were circular, encoding 37 genes, including 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes. Our samples showed consistency with previous studies in both coding and non-coding regions16,62,63,64. Comparative analyses with donkeys worldwide revealed minimal mitogenome variation among donkey breeds, likely due to the similar roles donkeys play globally32. We compared these mitochondrial gene sequences with those of other Equus species and found that, except for E. a. somalicus and E. hydruntinus, all species possess 37 mitochondrial genes and have a high degree of similarity. We cannot rule out whether this is due to lack of data or sequencing issues, and, selection pressure analyses indicated that all PCGs were under purifying selection.
A well resolved phylogeny of Equus based on mitogenomes
Interest in the origin and evolution of Equus dates back to over a century, but there is still no consensus on the definition of the genus or its phylogenetic position19. Recent studies focusing on the phylogenetic relationships within the genus Equus have leveraged mitochondrial DNA to elucidate evolutionary linkages and genetic diversity among species. For instance, Steiner et al. enhanced the resolution of Equus phylogeny by analyzing 22 partial mitochondrial and nuclear genes, revealing the complex relationship between African wild asses and Asiatic asses and indicating that ancestral polymorphisms contribute to the unresolved nature of this phylogeny65. And, research on mtDNA variations among horse populations in Egypt and Ethiopia has revealed low genetic diversity, further highlighting the distinct maternal lineages present within the genus66,67. Additionally, Sun et al. characterized the complete mitogenome of the Dezhou donkey, supporting the hypothesis that Chinese domestic donkeys may have originated from the Somali wild ass rather than the Asian wild ass25. Similarly, the phylogenetic analysis of ancient and modern Equids using complete mitogenomes highlights the monophyletic nature of zebras and the close relationship between the extinct quagga and plains zebras, suggesting rapid radiation within non-caballine lineages53. In this study, we performed a complete phylogenetic reconstruction of the entire Equus, including extant and extinct species, using 13 PCGs of mitochondrial DNA. This has allowed the evolutionary relationships between some of the extinct species and all of the extant species to be further clarified. Our research indicates that Przewalski’s horse has nested with domestic horses, despite the divergence of domestic horses and Przewalski’s horses occurring long before domestication20. Domestic donkeys form a single clade, offering a valuable opportunity to investigate domestication signatures in both domestic horses and donkeys. These findings collectively underscore the importance of mitogenome analysis in understanding the evolutionary history and conservation of Equus species.
Independent domestication imprints and reliable molecular makers in donkeys and horses mitogenomes
Mitochondria play a crucial role in the adaptability of horses and donkeys, primarily through their involvement in energy metabolism and their capacity to adjust to various environmental conditions16,68,69. Thanks to the wide global distribution of domestic donkeys and horses and the large amount of accessible data, our study was made possible. In both horses and donkeys, differences in the evolutionary pressures on the mitogenome and gene variants may influence their locomotor abilities and environmental adaptability68,70,71. Horses, known for their speed, are well-suited for rapid movement and long-distance travel72,73. This suggests that the mitogenome in horses may be under stronger selection pressure to optimize energy metabolism18,70,74. This need for efficient ATP production supports sustained and quick movement, similar to the evolution seen in flying birds, where high-energy demands drive the evolution of more efficient mitochondrial function8. Donkeys, on the other hand, are renowned for their remarkable adaptability to harsh environments, particularly in mountainous and arid regions20,21,74. The mitogenome in donkeys may have evolved under different selection pressures that allow them to function effectively even in conditions where energy demands are lower14,61,63,64. Given the distinct survival strategies of horses and donkeys in different environments, their mitogenomes likely reflect these differences18. Donkey mitochondria might prioritize endurance and adaptability, whereas horse mitochondria emphasize rapid and efficient energy utilization. These genomic differences may provide adaptive advantages in their respective ecological niches. Thus, studying the mitogenomes of horses and donkeys not only enhances our understanding of how these species have evolved to meet different survival demands but also offers valuable molecular markers and genetic resources for their conservation and utilization25,75,76,77. Our study identified variations in protein-coding genes (PCGs) and amino acid sequences within the mitogenomes of donkeys and horses. Similarly, by comparing the predicted three-dimensional structures of the proteins, we also found differences between the two domesticated species. These differences may contribute to the distinct adaptations of donkeys to semi-arid environments and the enhanced long-distance endurance observed in horses, indicating evolutionary adaptations to their respective environments and functional roles.
However, the lack of comprehensive data and the extinction of many species limit our ability to conduct thorough analyses, potentially overlooking critical evolutionary information.
Conclusions
This study investigates the mitochondrial genomes of horses and donkeys to assess the impact of domestication on their energy production and locomotion capabilities. Mitochondrial DNA provides insights into the evolutionary adaptations of species over time, including phylogenetic relationships. We analyzed the mitochondrial DNA of various species within the Equus. We reconstruct phylogenetic relationships for all extant and partially extinct species, uncovering significant genetic differences linked to energy metabolism and locomotor adaptations. Notably, horses exhibit adaptations for speed and long-distance travel, while donkeys are optimized for carrying heavy loads in challenging environments. By identifying these genetic distinctions, the study seeks to develop molecular markers for species identification and enhance our understanding of the evolutionary history of these animals. Our study has potential societal benefits, including informing conservation strategies and improving breeding programs for donkeys and horses, particularly in resource-limited regions where these animals are vital for transportation and agriculture.
Data availability
Data have been submitted to the GenBank database with accession number PP049795-PP049800 and will be available upon request the corresponding author (wzhang@sdu.edu.cn).
References
Martijn, J., Vosseberg, J., Guy, L., Offre, P. & Ettema, T. J. G. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557, 101–105 (2018).
Roger, A. J., Muñoz-Gómez, S. A. & Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192 (2017).
Chinnery, P. F. Mitochondrial disease in adults: What’s old and what’s new?. Embo Mol. Med. 7, 1503–1512 (2015).
Baker, Z. N., Forny, P. & Pagliarini, D. J. Mitochondrial proteome research: The road ahead. Nat. Rev. Mol. Cell Biol. 25, 65–82 (2024).
Hu, Z. et al. A novel protein CYTB-187AA encoded by the mitochondrial gene CYTB modulates mammalian early development. Cell Metab. 36, 1586-1597.e7 (2024).
Rackham, O. & Filipovska, A. Organization and expression of the mammalian mitochondrial genome. Nat. Rev. Genet. 23, 606–623 (2022).
Nunnari, J. & Suomalainen, A. Mitochondria: In sickness and in health. Cell 148, 1145–1159 (2012).
Shen, Y.-Y., Shi, P., Sun, Y.-B. & Zhang, Y.-P. Relaxation of selective constraints on avian mitochondrial DNA following the degeneration of flight ability. Genome Res. 19, 1760–1765 (2009).
Chang, H. et al. Evolutionary rates of and selective constraints on the mitochondrial genomes of Orthoptera insects with different wing types. Mol. Phylogenet. Evol. 145, 106734 (2020).
James, J. E., Piganeau, G. & Eyre-Walker, A. The rate of adaptive evolution in animal mitochondria. Mol. Ecol. 25, 67–78 (2016).
Boore, J. L. Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767–1780 (1999).
Wang, J. et al. Plant organellar genomes: much done, much more to do. Trends Plant Sci. 29, 754–769 (2024).
Barr, C. M., Neiman, M. & Taylor, D. R. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168, 39–50 (2005).
Brown, W. M., George, M. & Wilson, A. C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 76, 1967–1971 (1979).
Yoshizawa, K. et al. Mitochondrial phylogenomics and genome rearrangements in the barklice (Insecta: Psocodea). Mol. Phylogenet. Evol. 119, 118–127 (2018).
Li, M. et al. Mitochondrial phylogenomics provides insights into the phylogeny and evolution of spiders (Arthropoda: Araneae). Zool. Res. 43, 566–584 (2022).
Bernt, M. et al. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol. Phylogenet. Evol. 69, 352–364 (2013).
Librado, P. & Orlando, L. Genomics and the evolutionary history of equids. Annu. Rev. Anim. Biosci. 9, 81–101 (2021).
Barrón-Ortiz, C. I. et al. What Is Equus? Reconciling taxonomy and phylogenetic analyses. Front. Ecol. Evol. 7 (2019).
Orlando, L. et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78 (2013).
Todd, E. T. et al. The genomic history and global expansion of domestic donkeys. Science 377, 1172–1180 (2022).
Heintzman, P. D. et al. A new genus of horse from Pleistocene North America. Elife 6, e29944 (2017).
Jónsson, H. et al. Speciation with gene flow in equids despite extensive chromosomal plasticity. Proc. Natl. Acad. Sci. USA 111, 18655–18660 (2014).
Rossel, S. et al. Domestication of the donkey: Timing, processes, and indicators. Proc. Natl. Acad. Sci. USA 105, 3715–3720 (2008).
Sun, Y. et al. Characterization of complete mitochondrial genome of Dezhou donkey (Equus asinus) and evolutionary analysis. Curr. Genet. 62, 383–390 (2016).
Liu, S. et al. Current genetic conservation of Chinese indigenous horses revealed with Y-chromosomal and mitochondrial DNA polymorphisms. G3 Genes Genomes Genetics 11, jkab008 (2021).
Steiner, C. C. & Ryder, O. A. Molecular phylogeny and evolution of the Perissodactyla. Zool. J. Linn. Soc. 163, 1289–1303 (2011).
Vilà, C. et al. Widespread origins of domestic horse lineages. Science 291, 474–477 (2001).
Zhang, L. et al. Efficacy and Safety of Ejiao (Asini Corii Colla) in Women With Blood Deficient Symptoms: A Randomized, Double-Blind, and Placebo-Controlled Clinical Trial. Front. Pharmacol. 12, 718154 (2021).
Wang, D. et al. Chemical constituents and bioactivities of Colla corii asini. Drug Discov. Ther. 8, 201–207 (2014).
Liu, T. et al. Protective effect of Colla corii asini against lung injuries induced by intratracheal instillation of artificial fine particles in rats. Int. J. Mol. Sci. 20, 55 (2019).
Bennett, D. K. Stripes do not a zebra make, Part I: A cladistic analysis of Equus. Syst. Zool. 29, 272–287 (1980).
Hulbert, R. C. Taxonomic evolution in North American Neogene Horses (Subfamily Equinae): The rise and fall of and adaptive radiation. Paleobiology 19, 216–234 (1993).
Prado, J. L. & Alberdi, M. R. T. A CLADISTIC ANALYSIS OF THE HORSES OF THE TRIBE EQUINI.
Kelly, T. S. & Kelly, T. S. New Middle Miocene equid crania from California and their implications for the phylogeny of the Equini. Contrib. Sci. 473, 1–43 (1998).
Cirilli, O. et al. Evolution of the Family Equidae, Subfamily Equinae, in North, Central and South America, Eurasia and Africa during the Plio-Pleistocene. Biology (Basel) 11, 1258 (2022).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Jin, J.-J. et al. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241 (2020).
Camacho, C. et al. BLAST+: Architecture and applications. BMC Bioinform. 10, 421 (2009).
Wheeler, D. L. et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 36, D13–D21 (2008).
Bernt, M. et al. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319 (2013).
Dagona, A. G. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series (1999).
Grant, J. R. et al. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51, W484–W492 (2023).
Gutiérrez, E. G., Ortega, J., Savoie, A. & Baeza, J. A. The mitochondrial genome of the mountain wooly tapir, Tapirus pinchaque and a formal test of the effect of altitude on the adaptive evolution of mitochondrial protein coding genes in odd-toed ungulates. Bmc Genom. 24, 527 (2023).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Minh, B. Q. et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Xie, J. et al. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 51, W587–W592 (2023).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302 (2017).
Zhang, D. et al. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355 (2020).
Waterhouse, A. et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Meng, E. C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Vilstrup, J. T. et al. Mitochondrial phylogenomics of modern and ancient equids. PLoS One 8, e55950 (2013).
Oakenfull, E. A., Lim, H. N. & Ryder, O. A. A survey of equid mitochondrial DNA: Implications for the evolution, genetic diversity and conservation of Equus. Conserv. Genet. 1, 341–355 (2000).
Shi, H., Li, W. & Xu, X. Learning the Base Sequence Quality and Content of Bioinformatics Analysis Method 563–566 (Atlantis Press, 2016). https://doi.org/10.2991/meici-16.2016.118.
Guo, C. et al. Transversions have larger regulatory effects than transitions. Bmc Genom. 18, 394 (2017).
George, M. & Ryder, O. A. Mitochondrial DNA Evolution in the Genus Equus’.
Xu, X., Janke, A. & Arnason, U. The complete mitochondrial DNA sequence of the greater Indian rhinoceros, Rhinoceros unicornis, and the Phylogenetic relationship among Carnivora, Perissodactyla, and Artiodactyla (+ Cetacea). Mol. Biol. Evol. 13, 1167–1173 (1996).
Xu, X., Gullberg, A. & Arnason, U. The complete mitochondrial DNA (mtDNA) of the donkey and mtDNA comparisons among four closely related mammalian species-pairs. J. Mol. Evol. 43, 438–446 (1996).
Beja-Pereira, A. et al. African origins of the domestic donkey. Science 304, 1781 (2004).
Xia, J. et al. Next-generation sequencing of the complete huaibei grey donkey mitogenome and mitogenomic phylogeny of the Equidae Family. Animals-basel 13, 531 (2023).
Han, H. et al. Genetic diversity and paternal origin of domestic donkeys. Anim. Genet. 48, 708–711 (2017).
Huang, B., Khan, M. Z., Chai, W., Ullah, Q. & Wang, C. Exploring genetic markers: Mitochondrial DNA and genomic screening for biodiversity and production traits in donkeys. Animals-basel 13, 2725 (2023).
Zeng, L. et al. Genomes reveal selective sweeps in kiang and donkey for high-altitude adaptation. Zool. Res. 42, 450–460 (2021).
Steiner, C. C., Mitelberg, A., Tursi, R. & Ryder, O. A. Molecular phylogeny of extant equids and effects of ancestral polymorphism in resolving species-level phylogenies. Mol. Phylogenet. Evol. 65, 573–581 (2012).
Othman, O. E., Mahrous, K. F. & Shafey, H. I. Mitochondrial DNA genetic variations among four horse populations in Egypt. J. Genet. Eng. Biotechnol. 15, 469–474 (2017).
Effa, K., Rosenbom, S., Han, J., Dessie, T. & Beja-Pereira, A. Genetic Diversities and Historical Dynamics of Native Ethiopian Horse Populations (Equus caballus) Inferred from Mitochondrial DNA Polymorphisms. Genes-basel 12, 155 (2021).
Wang, Y., Hua, X., Shi, X. & Wang, C. Origin, evolution, and research development of donkeys. Genes-basel 13, 1945 (2022).
Xi-Yao, M. et al. Potential dual expansion of domesticated donkeys revealed by worldwide analysis on mitochondrial sequences. Zool. Res. 41, 51–60 (2020).
Ning, T., Xiao, H., Li, J., Hua, S. & Zhang, Y. P. Adaptive evolution of the mitochondrial ND6 gene in the domestic horse. Genet. Mol. Res. 9, 144–150 (2010).
Lippold, S., Matzke, N. J., Reissmann, M. & Hofreiter, M. Whole mitochondrial genome sequencing of domestic horses reveals incorporation of extensive wild horse diversity during domestication. BMC Evol. Biol. 11, 328 (2011).
Dagg, A. I. Giraffe: Biology, Behaviour and Conservation (Cambridge University Press, 2014).
Cao, Q. L., Pukazhenthi, B. S., Bapodra, P., Lowe, S. & Bhatnagar, Y. V. Equid adaptations to cold environments. In The Equids: A Suite of Splendid Species (eds Prins, H. H. T. & Gordon, I. J.) 209–246 (Springer International Publishing, 2023). https://doi.org/10.1007/978-3-031-27144-1_8.
Librado, P. et al. Ancient genomic changes associated with domestication of the horse. Science 356, 442–445 (2017).
Ning, T., Xiao, H., Li, J., Hua, S. & Zhang, Y. P. Adaptive evolution of the mitochondrial ND6 gene in the domestic horse. Genet. Mol. Res. (2010).
Janis, C. M. & Bernor, R. L. The evolution of equid monodactyly: A review including a new hypothesis. Front. Ecol. Evol. 7 (2019).
Linklater, W. L. Adaptive explanation in socio-ecology: lessons from the Equidae. Biol. Rev. 75, 1–20 (2000).
Acknowledgements
We thank Dr. Zhaozhao Xue and miss Changxin Wang for the assistance in sample collection and bioinformatics analysis.
Funding
This research was funded by the National Key Research and Development Program (2022YFC3341002-2) and Shandong Provincial Natural Science Foundation (ZR2023QC278).
Author information
Authors and Affiliations
Contributions
Conceptualization, W.Z. and Q.S.; methodology, W.D. and S.K.; software, S.K.; validation, S.H., W.Z. and Q.S.; formal analysis, W.D. and S.K.; investigation, W.Z and P.Y.; resources, Q.S. and P.Y.; data curation, Q.S. and W.Z.; writing—original draft preparation, W.D.; writing—review and editing, W.Z. and Q.S.; visualization, W.D.; supervision, W.Z. and Q.S.; project administration, W.Z. and Q.S.; funding acquisition, W.Z., P.Y., and Q.S. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The animal study protocol was approved by the Institutional Review Board of Institute of Forensic Science, Ministry of Public Security (MPS) (protocol code 2023-002 and approved on 16 Mar. 2023).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Du, W., Sun, Q., Hu, S. et al. Equus mitochondrial pangenome reveals independent domestication imprints in donkeys and horses. Sci Rep 15, 6803 (2025). https://doi.org/10.1038/s41598-025-91564-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-91564-1