Abstract
Inter-continental study systems are crucial for testing ecological hypotheses, such as the widely cited Enemy Release Hypothesis (ERH), which seeks to explain the superior performance of plant species when they are introduced to new regions. Pinus contorta (lodgepole pine), native to North America, has been extensively introduced to Europe and the Southern Hemisphere, making it an ideal tree species for studying invasion hypotheses from a biogeographical perspective. We compared foliar fungal communities, especially pathogens, of P. contorta across two native–introduced region pairs (NIRPs): a northern NIRP (from Canada to Sweden) and a southern NIRP (from the USA to Patagonia), while also examining the differences between source plantations and invasion fronts within Patagonia. P. contorta underwent significant fungal community shifts and experienced pathogen release during its large-scale introduction from North America to Sweden and Patagonia. The fungal richness and relative abundance changes were more pronounced for the southern NIRP pair, where no closely related tree species to P. contorta are present in Patagonia. In Sweden, the presence of the phylogenetically related P. sylvestris and its associated local fungal community appears to play a role in influencing the foliar fungal communities associated with introduced P. contorta. In Patagonia, the incomplete co-invasion of fungal taxa from the USA emerges as a principal driver of the observed variability in fungal community composition and pathogen release following the introduction of P. contorta. In Patagonia, fungal community composition differences between source plantations and invasion fronts provided insufficient evidence that pathogen release occurs at this local scale. Integrating both biogeographical and phylogenetic perspectives, our study suggests that priority effects of local fungi appear to be a dominant community assembly process when introduction is done in a phylogenetically similar community; whereas, co-invasion of fungal communities is the dominant process in phylogenetically distant communities.
Similar content being viewed by others
Introduction
Over the past century, fast-growing non-native tree species have been extensively introduced to new environments across the globe, where they often demonstrate superior performance, such as increased growth rates and larger individual sizes compared to those in their native habitats1,2. The foliar biological communities associated with introduced tree species play a pivotal role in establishing interactions with local ecosystems, contributing to pest and disease dynamics, facilitating resistance and adaptability, contributing to decomposition after senescence3,4, and ultimately influencing the successful establishment of these species within new ecological contexts5,6. For example, Otsing, et al.3 demonstrated that the community structure of all fungi, saprotrophs, and plant pathogens in foliage shapes leaf litter decomposition in mixed forest ecosystems. Moreover, interactions between pathogens and other fungal functional groups can also shape disease dynamics and, consequently, the successful introduction of tree species to new regions. For example, fungal saprotrophs associated with American beech (Fagus grandifolia) have been shown to influence the progression of beech bark disease7. Given the known importance of foliar fungal communities for tree performance, investigating how the tree introduction and subsequent potential invasion change the composition of foliar fungal communities can deepen our understanding of tree success following their introduction to new environments.
Numerous hypotheses have been proposed to explain the success of introduced plant species, with the Enemy Release Hypothesis (ERH) being particularly influential8. According to ERH, introduced plants can escape the specialist herbivores and pathogens in their native range9,10, and this reduced biotic stress allows resources that would typically be allocated to defense and recovery to be redirected toward biomass accumulation, resource acquisition, competition, and reproduction in the introduced range11,12. However, ecological factors, such as the environmental adaptability of introduced plant species and their phylogenetic relatedness to native species can significantly shape the relevance of ERH. For instance, during the introduction phase, species that share close phylogenetic relationships with native species may remain vulnerable to specialist pathogens due to similar host traits, whereas species that are phylogenetically distant from the local community may experience a greater release from natural enemies, as they are less likely to host specialist pathogens13,14. Moreover, even within the same region, pathogen transmission can exhibit spatial heterogeneity, with pathogens sometimes lagging behind their hosts, such as from source plantations to invasion fronts15. Given these complex influences, biogeographical studies comparing the communities of enemies in a species’ introduced versus native regions have emerged as one of the most compelling approaches to addressing invasion hypotheses such as ERH16,17,18. Further, on a local scale, a comprehensive investigation of exotic species’ fungal communities, especially pathogens, between source plantations and adjacent invasion fronts is also essential for evaluating the spatial and temporal scales over which ERH occurs19. Pinus contorta (lodgepole pine), native to western North America, stands out as a very successful introduced species in both Europe (e.g., Sweden, Scotland, Finland, and Ireland) and the Southern Hemisphere (e.g., Chile, Argentina, and New Zealand)20,21,22,23. In these introduced regions, P. contorta typically exhibits faster growth, earlier maturity, and greater reproductive output than in its native regions17,24. These traits facilitate its successful establishment and growth in the introduced ranges25,26, and in the Southern Hemisphere, they contribute to its success as an aggressive invader27. Given its extensively documented history of introduction, P. contorta represents an ideal “model” study system for investigating ecological hypotheses through a biogeographical perspective28. A key advantage of this large-scale study system is that the taxonomic composition of the communities into which P. contorta has been introduced varies substantially. In Sweden, P. contorta was introduced into environments dominated by native conifers, including P. sylvestris, a phylogenetically similar species18,29. In contrast, in the Southern Hemisphere, such as in Patagonia, no native Pinus species are present in the regions where P. contorta has been introduced18,21. Consequently, the P. contorta introduction study system represents an excellent opportunity for investigating the ERH across a diverse array of regions with varying levels of phylogenetic relatedness.
Since the introduction of P. contorta to Europe and the Southern Hemisphere in the 1970s, research has focused on the trees’ adaptability30,31, productivity21,32, invasiveness25,26,33, impact on local diversity34,35,36, and plant-soil feedback mechanisms18,37. However, no research has yet evaluated how introduction and invasions correspond with shifts in the foliar fungal communities, including pathogens. Given the diverse range of community contexts that P. contorta has been introduced to, investigating its foliar fungal communities has the potential to advance our understanding of key hypotheses that seek to explain exotic species’ success (e.g., ERH), and other microbial community assembly processes, and also to shed new light on the role these foliar microbial communities may play in explaining the higher productivity of P. contorta in its introduced compared to native environments17,21.
Our study aimed to investigate the foliar fungal community differences in the globally introduced P. contorta across its native (i.e., Canada and the USA) and introduced regions (i.e., Sweden and Patagonia), with a particular emphasis on assessing the occurrence of pathogen release during its introduction. Additionally, for the highly invasive Patagonia population, we further explored how fungal communities and associated pathogens shifted as P. contorta expanded from source plantations to invasion fronts over kilometer spatial scales. We employed P. contorta populations from two distinct native-introduced region pairs (NIRPs). In the northern NIRP, P. contorta was introduced from northern British Columbia, Canada, to Sweden, while in the southern NIRP, the species was introduced from the Pacific Northwest, USA, to Patagonia (Fig. 1)18,38. We tested the following hypotheses: After a large-scale introduction to new regions,
(H1a): P. contorta will exhibit shifts in composition and reduced richness of the whole foliar fungal community, as well as pathogens specifically (i.e., ERH), and
(H1b): This change will be more pronounced in the southern compared to northern NIRP, due to P. contorta’s greater phylogenetic difference from the dominant native community in Patagonia, which we anticipated would limit the pool of suitable local taxa.
(H2): In Patagonia, individuals at the invasion fronts will demonstrate a shifted foliar fungal community, reduced fungal richness, and experience pathogen release compared to the introduced plantations. We expected this because previous research has shown that invading P. contorta trees have a markedly lower root endophytic and ectomycorrhizal community diversity, suggesting that fungal co-invasion lags tree dispersal18,39,40.
Materials and methods
Study sites
For our study, we selected 40 sampling sites: 10 in Northern British Columbia, Canada; 10 in Sweden; 10 in the Pacific Northwest, USA; and 10 in the Patagonia region (i.e., Argentina and Chile). The distances between sampling plantation sites were 0.6–321.9 km in Canada, 1.7–57.7 km in Sweden, 0.6–321.7 km in the USA, and 0.8–179.4 km in Patagonia. Specifically, within Patagonia, plantation distances ranged from 0.8 to 5.7 km in Argentina and 1.9–39.9 km in Chile. Further, for the 10 sites in Patagonia, we identified adjacent invasions extending from plantations (referred to as “plantations” or “invasion fronts”), with the distances between invasion fronts and their paired adjacent plantations ranging from 0.1 to 0.9 km in Argentina and from 0.05 to 0.6 km in Chile (Fig. 1). In contrast, the 30 sites in North America and Sweden included only a single stand type: natural-origin stands in North America and introduced plantations in Sweden. The establishment of the study system began by identifying P. contorta plantations between 30 and 55 years of age in Sweden and the Southern Hemisphere18. The sites also represent native and introduced populations of two distinct subspecies, where P. contorta subsp. latifolia from Fort Nelson and Fort St. John British Columbia, Canada38 was introduced to Sweden and P. contorta subsp. murrayana from Central Oregon, USA was introduced to Patagonia. We refer to these as two distinct native-introduced region pairs (NIRPs, as mentioned above): a northern and a southern pair (Fig. 1). For introduced ranges, commercial plantations of P. contorta were extensively established in the 1970s. In Sweden, the 10 P. contorta subsp. latifolia stands were initiated as field trials in 1970 38. The precise seed origin of Patagonia plantations is not known, but trait and anecdotal analysis have indicated the P. contorta subsp. murrayana population in central Oregon, USA as the origin41, where we distributed our native range sampling (Fig. 1).
A map depicting two Pinus contorta native-introduced region pairs (NIRPs, panel a). The northern NIRP is represented in blue, indicating native range locations in northern British Columbia, Canada (b), and corresponding introduced populations in Sweden (c). The southern NIRP is depicted in red, indicating native range populations in the Pacific Northwest, USA (d), and corresponding introduced populations in Patagonia (e). In Patagonia, two paired stand types (i.e., plantations and invasion fronts) were sampled and represented in red and pink, respectively.
Needle sampling
During the 2019 growing season (July in the Northern Hemisphere and January in Patagonia), we randomly selected and sampled needles from eight trees in each stand, ensuring that the trees were evenly spaced. From each tree, we sampled sun-exposed branches within reach using long-handled pruning shears and subsequently collected needles, including old diseased, old healthy, young diseased, and young healthy needles (with “old” referring to 2-year-old needles and “young” to current-year growth). Needles were combined into one sample per tree, resulting in 397 needle samples in total. Upon collection, the needles were immediately placed in paper bags and frozen. Samples in Canada, the USA, and Patagonia were freeze-dried before they were shipped to the Forest Vegetation Ecology laboratory in Umeå, Sweden for further processing. In the laboratory, the needles were cleaned using deionized water and 0.5% Tween 20 to remove surface adherents before DNA extraction. For details about DNA extraction, PCR, and PacBio sequencing see Supplementary Methods.
Bioinformatics analysis
After sequencing, data were processed using the nf-core collection of workflows [nf-core/ampliseq version 2.7.142,43], which relies on reproducible software environments provided by the Bioconda44 and Biocontainers45 projects. Data quality was evaluated with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and summarized with MultiQC46. Primers and adapters were trimmed using Cutadapt47, and reads without matching primers or adapters were discarded. The adapter- and primer-free sequences were then processed on a per-sample basis with DADA248, which corrected sequencing errors, removed chimeric sequences, and inferred true biological sequences, resulting in the generation of high-resolution amplicon sequence variants (ASVs). Finally, taxonomic annotation was performed and ASVs were assigned to corresponding taxa using the UNITE fungal database (Version 9.0)49.
After filtering, all downstream analyses, including plotting, were conducted using the statistical software R 4.4.050. Firstly, each sample was rarified to 1000 sequence reads using the “rrarefy” function in the “vegan” package51. Then ITS ASVs were assigned functional guild annotations using FUNGuild52 through the “FUNGuildR” package, accepting all “probable” or higher-level annotations. We adopted the classifications of “plant pathogen”, “endophyte”, “epiphyte”, and “saprotroph”. Unidentified ASVs were consistently categorized as “unknown ecology”, and groups that fell outside the scope of our research focus (e.g., lichen, parasites, animal pathogens, etc.) were collectively classified as “others”. When an ASV was assigned to multiple functional groups, it was counted separately within each relevant group. For example, if an ASV was identified as “endophyte- leaf saprotroph- plant pathogen”, it was assigned to each of the corresponding categories: “endophyte”, “saprotroph”, and “plant pathogen”. The relative abundance of each functional group was defined as the total fractional abundance of all ASVs in that functional group, and richness was defined as the number of ASVs identified per sample per 1000 reads.
Statistical analysis
For each of our hypotheses, forest stands served as the units of replication, and all univariate data were assessed for adherence to parametric assumptions (i.e., normality and homoscedasticity). All permutational multivariate analyses of variance (PERMANOVA) and tests for homogeneity of multivariate dispersion were performed using the “adonis2” and “betadisper” functions, respectively, from the “vegan” package. When significant interaction effects were detected (α = 0.05), post-hoc pairwise comparisons were made. For our first hypothesis, two-way PERMANOVA with “introduction status” (IS, i.e., P. contorta growing in its native or introduced range) and “native-introduced region pair” (NIRP, i.e., Swedish P. contorta subsp. latifolia originating from Canada or Patagonian P. contorta subsp. murrayana originating from the USA) as two fixed factors were used to assess variations in community composition. Due to the non-normality and heteroscedasticity of the richness and relative abundance data across P. contorta in native and introduced regions, Kruskal-Wallis tests were conducted with the aforementioned fixed factors. Similarly, for our second hypothesis, one-factor PERMANOVAs were employed to compare the community composition differences between P. contorta subsp. murrayana plantations and their paired invasion fronts in Patagonia, and homogeneity of multivariate dispersion tests were conducted to assess within-group uniformity. A Wilcoxon rank sum test was employed to compare the richness of the “others” guild between the plantations and invasion fronts in Patagonia, given the non-normality and heteroscedasticity of the data. The richness of all categories, excluding the “others” guild, and the relative abundance of all guilds were examined using Welch’s t-tests. Principal coordinate analysis based on Bray-Curtis distance (PCoA, “cmdscale” function in “vegan” package) was employed to visually represent multivariate comparisons. For the plant pathogens, we conducted multivariate comparisons based on Bray-Curtis and Jaccard distances, which rely on species relative abundance and presence/absence, respectively. All distance matrices were constructed using the “vegdist” function in the “vegan” package, and species-relative abundance data were used after normalization. Kruskal-Wallis tests were conducted using the “scheirerRayHare” function from the “rcompanion” package. In cases where significant interaction effects were detected (α = 0.05), we followed up with pairwise Wilcoxon rank sum tests, employing Holm-Bonferroni adjusted p-values for multiple comparisons. Further, we conducted a “contribution of variables to similarity (SIMPER)” analysis using the “simper” function within the “vegan” package. This analysis aimed to identify the fungal species that primarily contributed to the similarity within groups as well as the dissimilarity among groups.
Results
After adapter removal and quality control, a total of 6,259 unique ASVs across all sites and stand types were obtained. The number of ASVs in each category for each region and tree type is reported in Table S1. The ASVs that contributed the most to the similarity within each of these groups are reported in Table S2.
Pinus contorta fungal communities across different regions
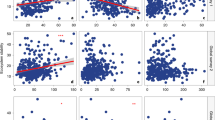
Community composition of all fungal categories associated with P. contorta was responsive to the main effects of introduction status (IS; P. contorta in its native vs. introduced range), native-introduced region pairs (NIRPs; northern NIRP, Canada—Sweden; or southern NIRP, USA—Patagonia), and their interactions (Table 1; Fig. 2). The differences in fungal community dissimilarity among the northern and the southern NIRP regions led to interaction effects between IS and NIRP. For all fungi, plant pathogens, saprotrophs, and “others”, the interaction effects among IS and NIRP arose because communities across the southern NIRP were more similar than the northern NIRP (Fig. 2a,b,c,f,g). For endophytes, epiphytes, and unknown ecology, the interaction effects occurred because the communities across the northern NIRP were more similar than the southern NIRP (Fig. 2d,e,h). Additionally, the multivariate dispersion of each fungal category across native and introduced regions was significantly different, with the average distance to the median ranging from 0.45 to 0.68. Notably, in the northern NIRP, the dispersion of all fungal categories, except for endophytes, was significantly higher in Canada as compared to Sweden, whereas the pattern in the southern NIRP was less pronounced (Fig. S1).
For the northern NIRP, ASVs assigned to Lophodermium baculiferum, Phaeotheca fissurella, Lophodermella concolor, Perusta inaequalis, and Cladosporium basiinflatum were the dominant plant pathogens contributing the most to the dissimilarity between P. contorta subsp. latifolia in Sweden and Canada (Table S3). The relative abundance of these pathogens was generally higher in Canada than in Sweden, except P. inaequalis, which exhibited a marginally higher abundance in Sweden (Table S3). For the southern NIRP, similarly, ASVs assigned to P. fissurella, L. baculiferum, as well as Lophodermium pinastri, Meristemomyces arctostaphyli, and Neophaeomoniella constricta were the predominant plant pathogens that contributed the most to the dissimilarity between P. contorta subsp. murrayana in Patagonia and the USA (Table S4). Among these, P. fissurella, L. baculiferum, and M. arctostaphyli had higher relative abundance in the USA, whereas L. pinastri and N. constricta were more abundant in Patagonia (Table S4).
For richness and relative abundance variables, introduction status (IS) significantly affected the richness of all fungi, plant pathogens, endophytes, epiphytes, and saprotrophs, as well as the relative abundance of plant pathogens, endophytes, epiphytes, saprotrophs, and unknown ecology (Table 1). The native-introduced region pair (NIRP) significantly affected the richness of total fungi, plant pathogens, endophytes, epiphytes, saprotrophs, and unknown ecology, as well as the relative abundance of endophytes, saprotrophs, “others”, and unknown ecology (Table 1). Further, the richness of all fungi, plant pathogens, epiphytes, saprotrophs, “others”, and unknown ecology, as well as the relative abundance of endophytes, epiphytes, saprotrophs, “others”, and unknown ecology were affected by the interaction of IS and NIRP (Table 1). Within the northern NIRP, the richness of all fungi, “others”, and unknown ecology was higher in the introduced relative to native range (Fig. 3a,f,g), whereas the opposite was observed in endophytes and epiphytes (Fig. 3c,d). However, no significant differences in the richness of plant pathogens and saprotrophs were detected between the two regions (Fig. 3b,e). In the southern NIRP, a lower richness of all categories except epiphytes was detected in the introduced relative to the native range (Fig. 3).
For the relative abundance data, plant pathogens and endophytes of P. contorta plantations comprised a greater proportion in the native as compared to the introduced regions, while the opposite occurred for the unknown ecology group (Fig. 3h). For saprotrophs, they had a greater relative abundance of the fungal community in the native than in the introduced regions in the northern NIRP (i.e., higher in Canada than Sweden) while the opposite was observed in the southern NIRP (i.e., higher in Patagonia than the USA) (Fig. 3h).
A principal coordinate analysis (PCoA) showing the results of a PERMANOVA test evaluating differences in community composition of (a) all fungi, (b, c) plant pathogens, (d) endophytes, (e) epiphytes, (f) saprotrophs, (g) “others” and (h) fungi with unknown ecology associated with Pinus contorta needles in four distinct regions (Canada, USA, Sweden, and Patagonia). Results of the PERMANOVA are reported in Table 1. The R2 of post-hoc pairwise comparisons are shown on each panel, number with *, and ** indicates a significant difference at α = 0.05 and α = 0.01 respectively.
Amplicon sequence variants (ASVs) richness (a–g) and relative abundance (h) of fungal communities associated with Pinus contorta needles in two native range populations (Canada and USA) and two introduced populations (Sweden and Patagonia), respectively. Richness is presented as the average rarified richness for each sample. Results from the corresponding two-way Kruskal-Wallis test for all variables are reported in Table 1. Different letters above boxplots (a–g) or across bar segments with the same shade (h) indicate significant pairwise differences (α = 0.05) determined using nonparametric post hoc comparisons.
Fungal communities of introduced Pinus contorta subsp. murrayana plantations vs. invasion fronts in Patagonia
All fungal community composition variables, except epiphytes and “others”, significantly differed between Patagonia plantations and adjacent invasion fronts (Table S5; Fig. 4). Differences in plant pathogen communities were observed irrespective of whether Bray-Curtis or Jaccard distance metrics were applied (Fig. 4b,c). Additionally, a significantly greater multivariate dispersion of unknown ecology was detected within invasion fronts compared to plantations, with average distances to the median of 0.56 and 0.49, respectively (W = 16,537,411, p = 0.004, Wilcoxon rank sum test). The total dissimilarity between P. contorta subsp. murrayana plantations and invasion fronts in Patagonia was 78.2%. Among plant pathogens, P. fissurella, L. pinastri, and N. constricta contributed the most to the dissimilarity (Table S6). Specifically, the abundance of P. fissurella was greater in the invasion front compared to the plantations, whereas the abundance of L. pinastri and N. constricta was similar in both the plantations and the invasion fronts (Table S6).
The richness of endophytes and saprotrophs was significantly greater in invasion fronts as compared to plantations (Table S5; Fig. 5c,e). For the relative abundance data, significant differences were observed across all fungi and other categories, except endophytes (Table S5). The relative abundance of plant pathogens and saprotrophs was higher in P. contorta subsp. murrayana invasion fronts compared to plantations, whereas fungi with unknown ecology exhibited a lower relative abundance in the invasion fronts relative to plantations (Fig. 5h).
A principal coordinate analysis (PCoA) showing the results of a PERMANOVA test evaluating differences in community composition of (a) all fungi, (b, c) plant pathogens, (d) endophytes, (e) epiphytes, (f) saprotrophs, (g) “others” and (h) fungi with unknown ecology associated with Pinus contorta needles in introduced plantations and invasion fronts growing from plantations in Patagonia. Results of the PERMANOVA are reported in Table S5, with α = 0.05 indicating statistically significant differences.
Amplicon sequence variants (ASVs) richness (a–g) and relative abundance (h) of fungal communities associated with Pinus contorta subsp. murrayana needles in introduced plantations and invasion fronts growing from plantations in Patagonia, respectively. Richness is presented as the average rarified richness for each sample. Results from the corresponding Welch’s t or Wilcoxon rank sum tests are reported in Table S5. Different letters above boxplots (a–g) or across bar segments with the same shade (h) indicate significant differences at α = 0.05.
Discussion
We observed significant differences in foliar fungal composition, species richness, and relative abundance following the large-scale introduction of P. contorta from North America to both Sweden and Patagonia. Additionally, we identified substantial variation in the foliar fungal community between invasion fronts and source plantations within Patagonia. We now discuss these patterns in relation to our initial hypotheses.
Foliar fungal communities of Pinus contorta in its native versus introduced ranges
Exotic species interact with local ecosystems through mechanisms such as microbial co-evolution and adaptation, leading to shifts in the community composition of both introduced and native species within new environments53. In the context of P. contorta introduction from North America to Sweden and Patagonia, we predicted that P. contorta would be associated with distinct and less species-rich foliar fungal communities in its introduced compared to native range (H1a). In support of our first hypothesis, we observed a significant alteration of the foliar fungal community composition across all native and introduced populations (Fig. 2). Additionally, the relative abundance of plant pathogens on average was significantly reduced in the introduced compared to native regions (Fig. 3h), and we found a significantly decreased richness of pathogens, endophytes, epiphytes, and saprotrophs in the introduced regions, as compared to native regions (Fig. S2). These findings indicate that in general P. contorta has undergone foliar fungal community composition shifts, including pathogen release following large-scale introductions, which could be one of several factors contributing to its enhanced growth or successful invasion in its introduced areas21,26.
Contrasting Northern versus Southern native-introduced region pairs
The presence or absence of phylogenetically related native species in areas of introduction has the potential to influence various ecological processes that shape associated fungal community composition54. For instance, within our study system, Gundale, et al.18 observed that soil fungal community composition was less similar between P. contorta plantations in the United States and the Southern Hemisphere, where phylogenetically related tree species are absent, compared to populations in Canada and Europe. Given that foliage is more susceptible to airborne fungal spores from the surrounding environment and considering the presence of the phylogenetically related P. sylvestris in Sweden, we predicted that differences in the P. contorta foliar fungal community between native and introduced ranges would be more pronounced in the southern compared to the northern NIRP (H1b). We found that the composition, richness, and relative abundance of nearly all fungal categories were significantly affected by native-introduced region pair (NIRP; Table 1; Figs. 2 and 3).
In the northern NIRP, the greater influx of local, wind-dispersed fungal spores from the phylogenetically close P. sylvestris into the P. contorta plantations in Sweden14,34 may explain the distinctly different foliar fungal communities between Sweden and Canada (Figs. 2 and 3). In support of this, several generalist species such as Phaeotheca sp.55, Lophodermium conigenum56, and Hyphodiscus sp.57, sequenced from Swedish P. contorta needles are species well known to occur in European P. sylvestris forests, but were absent from the Canadian sites examined in our study (Tables S2 and S3). These local fungal taxa, as well as widely distributed generalists at Swedish sites, could also exert priority effects that impede the establishment of the common Canadian taxa20,21.
For plant pathogens, although the overall richness did not differ significantly between Canada and Sweden (Fig. 3b), we found a significantly lower relative abundance of plant pathogens in Swedish plantations compared to Canadian stands (Fig. 3h). Further, several prominent plant pathogens were present in Canadian sites but absent in Swedish sites, including L. baculiferum, L. concolor, and L. resinosum (Tables S2 andS3). These taxa are primarily distributed or originate in North America58,59, and their absence at our Swedish sites offers robust support for ERH in the northern NIRP pair60,61. Furthermore, the generalist plant pathogen Neophaeomoniella constricta was recently identified as a new taxon62, and in our study, this species existed both in Sweden and Canada but had a higher abundance in Swedish P. contorta sites compared to Canadian counterparts (Table S3). Taken together, P. contorta underwent significant foliar fungal community changes, including release from some specific pathogens, during its introduction across the northern NIRP, with the literature indicating that communities commonly associated with phylogenetically close P. sylvestris in Sweden influenced the fungal communities of P. contorta.
In the southern NIRP, the altered foliar fungal community composition (Fig. 2) and the significant reduction in foliar fungal richness across all fungal categories, except for epiphytes, in P. contorta plantations in Patagonia, compared to those in the USA (Fig. 3), indicate that the introduction process has caused large changes in the fungal community. Moreover, the observed decrease in both the richness and relative abundance of plant pathogens in Patagonia compared to the USA (Fig. 3b,h) indicates that pathogen release occurred during P. contorta’s introduction from the USA to Patagonia, and the co-invasion of fungal taxa from the USA to Patagonia is a primary assembly mechanism for Patagonian foliar fungal communities19. In support of this, we identified several crucial plant pathogens, including the pathogens P. fissurella and L. baculiferum, both of which were found to be more abundant in the USA than in Patagonia (Table S4). Notably, the literature indicates these pathogens are native to coniferous forests in the Northern Hemisphere and are absent from the Nothofagus forests of the Southern Hemisphere58,63, indicating that they have followed a co-introduction trajectory into Patagonian P. contorta plantations. These data suggest that relatively few taxa from local Patagonian Nothofagus forests, with notable exceptions such as the generalist saprotroph Hormonema macrosporum64, can colonize P. contorta, and that co-invasion from the Northern Hemisphere is a key process that has led to significant changes in community composition, reduced species richness, and pathogen release following the introduction of P. contorta from the USA to Patagonia. The local randomness of co-invasion may also explain why pathogens in Patagonia exhibit higher beta-dispersion than the native range (Fig. S1), which could occur if different co-invading taxa initially dominate at different sites.
Taken together, we observed significant differences in foliar fungal community composition, along with reduced richness and relative abundance of plant pathogens, following the introduction of P. contorta across a broad geographic range—particularly in the southern NIRP (Figs. 3 and S2; Tables S2, S3, and S4). Consequently, our first hypothesis was supported that P. contorta would associate with distinct foliar fungal communities and undergo pathogen release during its large-scale introduction. The reduction in fungal richness appeared to be especially pronounced in Patagonia (Fig. 3), where phylogenetically related tree species are absent, whereas changes in composition appeared to be strongest in the northern NIRP (Fig. 2), where a phylogenetically similar tree species was present. These findings suggest that distinct ecological processes operated in introduction contexts with varying phylogenetic similarity. In Sweden, the presence of the phylogenetically related P. sylvestris appears to significantly influence the colonization and composition of foliar fungal communities in P. contorta plantations. The relatively greater dissimilarity between Swedish and Canadian P. contorta fungal communities can thus plausibly be explained by a distinct yet partially suitable foliar fungal community associated with P. sylvestris in Sweden. In contrast, in Patagonia, the partial co-invasion of fungal taxa accompanying P. contorta from North America appears to play a role, albeit gradually, in reducing fungal community richness and abundance, thereby facilitating pathogen release within the southern NIRP. Thus, community assembly via co-invasion in Patagonia may help explain the relatively greater community similarity in the southern (93.3%) than northern (87.9%) NIRP, the strong reduction in richness (Fig. S2), and greater pathogen community dispersion in Patagonia compared to North America (Fig. S1).
Foliar fungal communities of Patagonian plantations versus invaders
At the local scale, microbe-climate differences between source plantations and invasion fronts can lead to fungal dispersal limitations, which subsequently influence microbial co-invasion and invasion success; however, as the invasion progresses, micro-environments are likely to converge65. For example, in the same stand network, Núñez, et al.39 demonstrated that the key belowground fungal mutualists were restricted to areas close to source plantations, while lower levels of mycorrhizal colonization and fewer fungal species were observed at greater distances from the original plantings. In our study, the presence of common fungal species across plantations and invasion fronts (Table S6), along with the lack of significant differences in multivariate dispersion across nearly all fungal categories, suggests that dispersal limitation is not a major factor influencing foliar fungal communities over distances spanning from plantations to invasion fronts. Although the differences in fungal composition between source plantations and invasion fronts were relatively modest (R² ranging from 0.01 to 0.06 for all fungi, pathogens, endophytes, saprotrophs, and unknown ecological groups; Fig. 4), and fungal richness differences were also limited (Fig. 5), our results suggest that fungal community divergence was primarily driven by differences in species relative abundances rather than species presence.
Factors such as differences in tree age or degree of canopy heterogeneity may lead to distinct microenvironments that influence the relative abundances of fungal taxa25,66,67. This pattern, consistent with Gundale, et al.18, contrasts the larger inter-continental scale comparison we made between native stands in the USA and Patagonia plantations (Fig. 3; Tables S2 andS4), where differences in species presence-absence were a larger contributor to the difference in community composition, which corresponded with a reduction in richness in Patagonia. In brief, while significant alterations in fungal community composition were observed between P. contorta plantations and invasion fronts in Patagonia, there was insufficient evidence to suggest dispersal limitation or pathogen release occurs at this scale during the spread of P. contorta from source plantations.
To date, no studies have investigated the variation in foliar fungal communities of P. contorta across such an extensive geographical range, incorporating replicated native and introduced sites along with the phylogenetic similarity to the resident plant community. Our study reveals that assembly mechanisms shaping foliar fungal communities differ significantly across various environmental contexts and also highlights the critical role that foliar fungal communities can have in intercontinental introductions, where pathogen release can contribute to a species’ establishment and invasion success.
Data availability
The datasets generated and/or analyzed during the current study are available in the European Nucleotide Archive (ENA), accession number is PRJEB82887.
References
Lombardero, M. J., Alonso-Rodríguez, M. & Roca-Posada, E. P. Tree insects and pathogens display opposite tendencies to attack native vs. non-native pines. For. Ecol. Manag. 281, 121–129. https://doi.org/10.1016/j.foreco.2012.06.036 (2012).
Nuñez, M. A. & Medley, K. A. Pine invasions: climate predicts invasion success; something else predicts failure. Divers. Distrib. 17, 703–713. https://doi.org/10.1111/j.1472-4642.2011.00772.x (2011).
Otsing, E., Barantal, S., Anslan, S., Koricheva, J. & Tedersoo, L. Litter species richness and composition effects on fungal richness and community structure in decomposing foliar and root litter. Soil Biol. Biochem. 125, 328–339. https://doi.org/10.1016/j.soilbio.2018.08.006 (2018).
Gaytan, A. et al. Changes in the foliar fungal community between oak leaf flushes along a latitudinal gradient in Europe. J. Biogeogr. 49, 2269–2280. https://doi.org/10.1111/jbi.14508 (2022).
Ridout, M. & Newcombe, G. The frequency of modification of Dothistroma pine needle blight severity by fungi within the native range. For. Ecol. Manag. 337, 153–160. https://doi.org/10.1016/j.foreco.2014.11.010 (2015).
Steel, G., Dickie, I. & Sapsford, S. A risk to the forestry industry? Invasive pines as hosts of foliar fungi and potential pathogens. New Z. J. Ecol. 46, 3741. https://doi.org/10.20417/nzjecol.46.13 (2022).
Morrison, E. W., Kasson, M. T., Heath, J. J. & Garnas, J. R. Pathogen and endophyte assemblages co-vary with Beech bark disease progression, tree decline, and regional climate. Front. Forests Glob. Change 4, 673099. https://doi.org/10.3389/ffgc.2021.673099 (2021).
Hierro, J. L., Maron, J. L. & Callaway, R. M. A biogeographical approach to plant invasions: the importance of studying exotics in their introduced and native range. J. Ecol. 93, 5–15. https://doi.org/10.1111/j.0022-0477.2004.00953.x (2004).
Downing, J. L. et al. Generalized mycorrhizal interactions and fungal enemy release drive range expansion of orchids in Southern Florida. Ecosphere 11, e03228. https://doi.org/10.1002/ecs2.3228 (2020).
Blaisdell, G. K. & Roy, B. A. Two tests of enemy release of commonly co-occurring bunchgrasses native in Europe and introduced in the united States. Biol. Inv. 16, 833–842. https://doi.org/10.1007/s10530-013-0541-9 (2013).
Roy, B. A. et al. Population regulation by enemies of the grass Brachypodium sylvaticum: demography in native and invaded ranges. Ecology 92, 665–675. https://doi.org/10.1890/09-2006.1 (2011).
Mitchell, C. E. & Power, A. G. Release of invasive plants from fungal and viral pathogens. Nature 421, 6923. https://doi.org/10.1038/nature01317 (2003).
Parker, I. M. & Gilbert, G. S. The evolutionary ecology of novel plant-pathogen interactions. Annu. Rev. Ecol. Evol. Syst. 35, 675–700. https://doi.org/10.1146/annurev.ecolsys.34.011802.132339 (2004).
Fahey, C., Koyama, A. & Antunes, P. M. Vulnerability of non-native invasive plants to novel pathogen attack: do plant traits matter? Biol. Inv. 24, 3349–3379. https://doi.org/10.1007/s10530-022-02853-z (2022).
Phillips, B. L. et al. Parasites and pathogens lag behind their host during periods of host range advance. Ecology 91, 872–881. https://doi.org/10.1890/09-0530.1 (2010).
Colautti, R. I., Ricciardi, A., Grigorovich, I. A. & MacIsaac, H. J. Is invasion success explained by the enemy release hypothesis? Ecol. Lett. 7, 721–733. https://doi.org/10.1111/j.1461-0248.2004.00616.x (2004).
Taylor, K. T. et al. Drivers of plant invasion vary globally: evidence from pine invasions within six ecoregions. Glob. Ecol. Biogeogr. 25, 96–106. https://doi.org/10.1111/geb.12391 (2015).
Gundale, M. J. et al. Differences in endophyte communities of introduced trees depend on the phylogenetic relatedness of the receiving forest. J. Ecol. 104, 1219–1232. https://doi.org/10.1111/1365-2745.12595 (2016).
Tierney, P. A., Caffrey, J. M., Matthews, S. M., Costantini, E. & Holland, C. V. Evidence for enemy release in invasive common Dace Leuciscus leuciscus in Ireland: a helminth community survey and systematic review. J. Helminthol. 94, e191. https://doi.org/10.1017/S0022149X20000759 (2020).
Mårald, E., Jönsson, J., Kardell, Ö., Sjögren, J. & Tunlid, A. An exotic tree in a foreign country: A cultural biography of the lodgepole pine in Sweden. Environ. History 30, 483–506. https://doi.org/10.3197/096734023x16869924234822 (2024).
Elfving, B., Ericsson, T. & Rosvall, O. The introduction of lodgepole pine for wood production in Sweden- a review. For. Ecol. Manag. 141, 15–29. https://doi.org/10.1016/S0378-1127(00)00485-0 (2001).
Ledgard, N. The spread of lodgepole pine (Pinus contorta, Dougl.) in New Zealand. For. Ecol. Manag. 141, 43–57. https://doi.org/10.1016/S0378-1127(00)00488-6 (2001).
Simberloff, D. et al. Spread and impact of introduced conifers in South America: lessons from other Southern hemisphere regions. Austral Ecol. 35, 489–504. https://doi.org/10.1111/j.1442-9993.2009.02058.x (2010).
McIntosh, A. C. S., Macdonald, S. E. & Gundale, M. J. Tree species versus regional controls on ecosystem properties and processes: an example using introduced Pinus contorta in Swedish boreal forests. Can. J. For. Res. 42, 1228–1238. https://doi.org/10.1139/x2012-049 (2012).
Langdon, B., Pauchard, A. & Aguayo, M. Pinus contorta invasion in the Chilean Patagonia: local patterns in a global context. Biol. Inv. 12, 3961–3971. https://doi.org/10.1007/s10530-010-9817-5 (2010).
Richardson, D. M., Williams, P. A. & Hobbs, R. J. Pine invasions in the Southern hemisphere: determinants of spread and invadability. J. Biogeogr. 21, 511–527. https://doi.org/10.2307/2845655 (1994).
Rejmánek, M. & Richardson, D. M. What attributes make some plant species more invasive? Ecology 77, 1655–1661 (1996).
Gundale, M. J. et al. Can model species be used to advance the field of invasion ecology? Biol. Inv. 16, 591–607. https://doi.org/10.1007/s10530-013-0610-0 (2013).
Zeb, U. et al. Comparative plastid genomics of Pinus species: insights into sequence variations and phylogenetic relationships. J. Syst. Evol. 58, 118–132. https://doi.org/10.1111/jse.12492 (2019).
Allen, R. B. & Lee, W. G. Seedling establishment microsites of exotic conifers in Chionochloa rigida tussock grassland, Otago, new Zealand. N. Z. J. Bot. 27, 491–498. https://doi.org/10.1080/0028825x.1989.10414133 (1989).
Varmola, M., Salminen, H., Rikala, R. & Kerkelä, M. Survival and early development of lodgepole pine. Scand. J. For. Res. 15, 410–423. https://doi.org/10.1080/028275800750172619 (2010).
Liziniewicz, M., Ekö, P. M. & Agestam, E. Effect of spacing on 23-year-old lodgepole pine (Pinus contorta Dougl. Var. latifolia) in Southern Sweden. Scand. J. For. Res. 27, 361–371. https://doi.org/10.1080/02827581.2011.639798 (2012).
Simberloff, D., Relva, M. A. & Nuñez, M. Gringos En El Bosque: introduced tree invasion in a native Nothofagus/Austrocedrus forest. Biol. Inv. 4, 35–53. https://doi.org/10.1023/a:1020576408884 (2002).
Ennos, R. A. The introduction of lodgepole pine as a major forest crop in Sweden: implications for host ± pathogen evolution. For. Ecol. Manag. 141, 85–96. https://doi.org/10.1016/S0378-1127(00)00491-6 (2001).
Andersson, B., Rosvall, O., Engelmark, O. & Sjoeberg, K. Environmental Impact Analysis (EIA) Concerning Lodgepole Pine Forestry in Sweden; Miljoekonsekvensbeskrivning (MKB) Av Skogsbruk Med Contortatall I Sverige. (Forestry Research Institute of Sweden, Uppsala, 1999).
Davis, K. T. et al. Severity of impacts of an introduced species corresponds with regional eco-evolutionary experience. Ecography 42, 12–22. https://doi.org/10.1111/ecog.04014 (2018).
Nuske, S. J. et al. Soil biotic and abiotic effects on seedling growth exhibit context-dependent interactions: evidence from a multi-country experiment on Pinus contorta invasion. New Phytol. 232, 303–317. https://doi.org/10.1111/nph.17449 (2021).
Gundale, M. J. et al. Interactions with soil biota shift from negative to positive when a tree species is moved outside its native range. New Phytol. 202, 415–421. https://doi.org/10.1111/nph.12699 (2014).
Núñez, M. A., Horton, T. R. & Simberloff, D. Lack of belowground mutualisms hinders Pinaceae invasions. Ecology 90, 2352–2359. https://doi.org/10.1890/08-2139.1 (2009).
Hayward, J., Horton, T. R. & Núñez, M. A. Ectomycorrhizal fungal communities coinvading with Pinaceae host plants in Argentina: Gringos Bajo El Bosque. New Phytol. 208, 497–506. https://doi.org/10.1111/nph.13453 (2015).
Gundale, M. J. et al. Functional traits differ across an invasive tree species’ native, introduced, and invasive populations. Biol. Inv. 26, 1–17. https://doi.org/10.1007/s10530-024-03316-3 (2024).
Straub, D. et al. Interpretations of environmental microbial community studies are biased by the selected 16S rRNA (Gene) amplicon sequencing pipeline. Front. Microbiol. 11, 550420. https://doi.org/10.3389/fmicb.2020.550420 (2020).
Baylis, F. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 271. https://doi.org/10.1038/s41587-020-0435-1 (2020).
Grüning, B. et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods 15, 475–476. https://doi.org/10.1038/s41592-018-0046-7 (2018).
da Leprevost, V. BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics 33, 2580–2582. https://doi.org/10.1093/bioinformatics/btx192 (2017).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. https://doi.org/10.1093/bioinformatics/btw354 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. https://doi.org/10.14806/ej.17.1.200 (2011).
Callahan, B. J. et al. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. https://doi.org/10.1038/nmeth.3869 (2016).
Abarenkov, K. UNITE general FASTA release for fungi. Unite Community (2020).
R: A language and environment for statistical computing. (R Foundation for Statistical Computing, Vienna, Austria, 2024).
Oksanen, J. et al. Community Ecology Package. (2018).
Nguyen, N. H. et al. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. https://doi.org/10.1016/j.funeco.2015.06.006 (2016).
Kruse, J., Pautasso, M. & Aas, G. A test of the enemy release hypothesis for plants in the Ecological-Botanical gardens, Bayreuth, using data on plant parasitic microfungi. Nova Hedwigia 103, 239–249. https://doi.org/10.1127/nova_hedwigia/2016/0348 (2016).
Liu, Z., Wang, X., Jia, G., Jiang, J. & Liao, B. Introduction of broadleaf tree species can promote the resource use efficiency and gross primary productivity of pure forests. Plant Cell Environ. https://doi.org/10.1111/pce.15096 (2024).
Crous, P. W. et al. Fungal planet description sheets: 785–867. Persoonia 41, 238–417. https://doi.org/10.3767/persoonia.2018.41.12 (2018).
Reignoux, S. N., Green, S. & Ennos, R. A. Molecular identification and relative abundance of cryptic Lophodermium species in natural populations of Scots pine, Pinus sylvestris L. Fungal Biol. 118, 835–845. https://doi.org/10.1016/j.funbio.2014.07.002 (2014).
Baral, H. O. et al. Venturioscypha Nigropila (Hyphodiscaceae, Helotiales) – a new genus and species from xeric Pinus bark. Karstenia 28-48 https://doi.org/10.29203/ka.2022.516 (2023).
Cannon, P. & Minter, D. Descriptions Fungi Bacteria Sheet 795 (1984).
Minter, D. & Millar, C. Descriptions of Fungi and Bacteria Sheet 1146 (1993).
Lane, B. R. et al. Fungicide-mediated shifts in the foliar fungal community of an invasive grass. Phytobiomes J. 7, 198–207. https://doi.org/10.1094/pbiomes-03-22-0018-r (2023).
Torchin, M. E. & Mitchell, C. E. Parasites, pathogens, and invasions by plants and animals. Front. Ecol. Environ. 2, 183–190. https://doi.org/10.2307/3868313 (2004).
Kraus, C., Damm, U., Bien, S., Voegele, R. T. & Fischer, M. New species of Phaeomoniellales from a German vineyard and their potential threat to grapevine (Vitis vinifera) health. Fungal Syst. Evol. 6, 139–155. https://doi.org/10.3114/fuse.2020.06.08 (2020).
Bogomolova, E. V. & Minter, D. W. Descriptions Fungi Bacteria Sheet 1557 (2003).
Bergmann, G. E. & Busby, P. E. The core seed mycobiome of Pseudotsuga menziesii Var. menziesii across provenances of the Pacific Northwest. USA Mycol. 113, 1169–1180. https://doi.org/10.1080/00275514.2021.1952830 (2021).
García, R. A. et al. From Biocultural Homogenization to Biocultural Conservation Ecology and Ethics Ch. Chapter 15 245–263 (2018).
Wang, J. et al. Darwin’s naturalization conundrum reconciled by changes of species interactions. Ecology 104, e3850. https://doi.org/10.1002/ecy.3850 (2023).
Philibert, A. et al. Predicting invasion success of forest pathogenic fungi from species traits. J. Appl. Ecol. 48, 1381–1390. https://doi.org/10.1111/j.1365-2664.2011.02039.x (2011).
Acknowledgements
We acknowledge the financial support from ANID PIA/BASAL FB210006 (awarded to AF) and UofA PER funding (awarded to AM). Special thanks to Chantal Ricard, funded by the Augustana Summer Research Award, and Lara Canovai, funded by a Mitacs Globalink internship, for their field sampling in Canada. We also thank Mark Bonner and Meredith Blackburn for their field sampling assistance in Patagonia and Sweden, respectively, and Ilse Van Duuren for laboratory support.
Funding
Open access funding provided by Swedish University of Agricultural Sciences.
This project was supported by the Swedish Research Council (2016 − 01055 awarded to MJG).
Author information
Authors and Affiliations
Contributions
Ruirui Zhao: Data curation, methodology, writing and drafting; Susan J. Nuske: Conceptualization, investigation, methodology, editing; Martín A. Nuñez: Conceptualization, investigation, editing; Alex Fajardo: Conceptualization, investigation, editing; Jaime Moyano: Conceptualization, investigation, editing; Anne C. S. McIntosh: Conceptualization, investigation, editing; Marie-Charlotte Nilsson: Conceptualization, editing; Michael J. Gundale: Supervision, conceptualization, investigation, editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhao, R., Nuske, S.J., Nuñez, M.A. et al. Distinct foliar fungal communities in Pinus contorta across native and introduced ranges: evidence for context dependency of pathogen release. Sci Rep 15, 7273 (2025). https://doi.org/10.1038/s41598-025-91639-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-91639-z