Abstract
Accessory ostium [AO] is one of the important anatomical variations in the maxillary sinus. AO is often associated with sinus pathology. Radiographic imaging plays a very important role in the detection of AO. Deep learning models have been used in maxillofacial imaging for interpretation and segmentation. However, there have been no research papers investigating the effectiveness of CNN in detecting AO in radiographs. To fill this gap of knowledge, we conducted a study to determine the accuracy of deep learning models in detecting AO in coronal CBCT images. Two examiners collected 454 coronal section images (227 with AO and 227 without AO) from 856 large field of view [FOV] cone beam tomography [CBCT] scans in the dental radiology archives of a teaching hospital. The collected images were then pre-processed and augmented to obtain 1260 images. Three pre-trained models, the Visual Geometry Group of the University of Oxford-16 layers [VGG16], MobileNetV2, and ResNet101V2, were used as base models. The performance of all the models was analyzed, and ResNet101v2 was selected for classification of images. Fine-tuning approach was employed with L1 (Lasso regression) regularization to avoid overfitting. The test accuracy and loss of the ResNet-101V2 classification model was 0.81 and 0.51, respectively. The precision, recall, F1-score, and AUC of the classification model were 0.82, 0.81, 0.81, and 0.87 respectively. ResNet-101V2 showed good accuracy in the detection of AO from coronal CBCT images. The present study used cropped two-dimensional images of CBCT scans. Future work can be carried out to determine the accuracy of deep learning models in the detection of AO in three-dimensional CBCT scans.
Similar content being viewed by others
Introduction
The close proximity of the maxillary posterior teeth with the maxillary sinus and the application of sinus lift procedures for dental implants made maxillary sinus an important anatomical site1,2. However, the primary ostium, which is the main pathway for the drainage of the maxillary sinus, is located in an unfavorable location3. In addition to the difficult location, the primary ostium is also susceptible to blockage during inflammation4. Accessory ostium [AO], also called the Girade’s orifice, is one of the important anatomical variations in the maxillary sinus5. The AOs may be located unilaterally or bilaterally, either as solitary or multiple apertures, between the uncinate process and inferior turbinate6,7. AO tends to occur more frequently on the posterior fontanelle, which is part of the lateral nasal wall covered only by mucoperiosteum8.
Studies using computed tomography [CT], cone beam computed tomography [CBCT], endoscopy and cadaveric analysis have shown a wide range of variations in the prevalence of AO9,10. Some studies have reported the association between AO and sinus pathologies3,10. The presence of AO leads to an increase in the ventilation of the sinus; however, it also leads to reverse drainage into the sinus from the middle meatus into the sinus11. The reverse drainage a causes reduction in the level of nitrous oxide and a buildup of mucous in the maxillary sinus, leading to pathologies like retention cyst, mucosal thickening, and maxillary sinusitis12,13. Recent studies have shown that CBCT can be effectively used for imaging the anatomy of the sinonasal structures and AO with precision and lower radiation dose10,14.
Artificial intelligence models have been explored for detection and segmentation anatomical structures in the craniofacial region15,16. Deep learning models which, are a subset of machine learning and artificial intelligence, have shown promising results in interpreting medical images when combined with residual neural networks17,18,19. Experts suggest that the use of AI makes radiology workflow efficient by reducing image reading time, speeding disease detection, and improving diagnostic accuracy20.
Tasks such as segmentation of the maxillary sinus, upper airway space, and detection of sinus pathology have been achieved using deep learning models with high accuracy21,22,23. Deep learning models have shown promising results in the detection of nasal septal deviation and fracture of the nasal bones16,24.
However, there is no research exploring the use of neural networks in the detection of AO using radiographic images. To fill this gap of knowledge, we conducted a study to determine the accuracy of the deep learning model in the detection of AO in coronal CBCT images.
Materials and methods
We conducted a retrospective cross-sectional study using the radiology archives of University Dental Hospital, Sharjah, United Arab Emirates between 1st January 2024 and 30th June 2024. The CBCT scans were made for various diagnostic purposes using Planmeca Viso 7 (Finland) at 95 kilovoltage peak (kVp), 5 milliampere (mA), and 0.2-millimeter (mm) resolution. Two examiners with 10 years of clinical experience screened 3278 CBCT scans to obtain 856 scans, which were obtained with a large field of view (FOV) with a region of coverage extending from the base of the mandible to the cranial base (20 × 17 cm).
CBCT scans of patients below the age of 18 years and scans of patients with a history of trauma and tumors in the sinonasal region were excluded from the study. CBCT scans of patients with congenital deformities of the sinonasal region and cleft palate were excluded from the study. CBCT scans of patients with history of nasal polyps, choanal atresia, acute sinusitis, and severe nasal septal deviation (deviated septum contacting the lateral nasal wall) were excluded from the study.
The two examiners analyzed 856 CBCT scans for AO. In case of a disagreement between the examiners, a third examiner with equal experience was called in to detect the presence of AO. The inter-rater reliability was calculated, and 10% of the scans were re-evaluated by each of the examiners after 2 weeks to obtain intra-rater reliability.
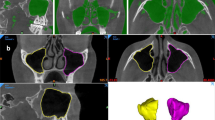
The examiners scrolled the coronal CBCT sections from the mesial side of the maxillary first premolar to the distal side of the maxillary second molar. The examiners then cropped the image from coronal CBCT sections at the site of AO. To maintain uniformity in cropping the images, the medial boundary was set at the nasal septum to distally the vertical line crossing the middle of the sinus. Inferiorly one centimeter below the level of the hard palate and superiorly at the level of the cribriform plate (Fig. 1).
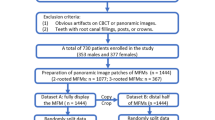
The images were saved in the Joint Photographic Experts Group (JPEG) format and labeled with the letter ‘A’ preceding the patient’s hospital registration number. Example: 706788RA (R/L implies right or left side). The examiners obtained 227 images from the 856 CBCT scans. Since these were the highest possible number of images we could get from our radiology archives, we followed the convenience sampling. We then obtained 227 coronal images from the CBCT images without AO following the same boundaries and labelled them as ‘N’ after the registration number. Example: 65097LN. To maintain uniformity, all the ‘N’ images were cropped from the coronal section coinciding with the medial aspect of the maxillary first molar. The outline of the methodology (data reprocessing, image classification, and image classification) followed in our study is shown in Fig. 2.
Flowchart of the steps followed in the present study. In the first step, coronal CBCT images were preprocessed. This was followed by image augmentation (rotation, shift width, zoom). In the next step, fine-tuning of the base model and custom layers was carried out. In the last step the classification output (normal = without AO, abnormal = with AO) of the model was obtained.
The images were first segregated into two separate folders (Data cleaning) based on the presence and absence of AO. As a part of preprocessing, the images were resized and subjected to a sharpening filter using ‘ImageFilter’, a package of Python Imaging Library (PIL) available in python Fig. 3.
The custom dataset had a total 454 images of two classes (227 normal images and 227 accessory ostium images). Among the 227 images, 118 were from the right side and 109 from the left side. Whereas 114 normal images were obtained from the right side and 113 from the right side. To avoid overfitting of the model, a data augmentation technique was used to create 1260 (630 normal and 630 accessory ostium) images by using 420 images from the training data set. The rest of the 34 images were kept for testing of the model. The overall distribution of images for training, validation, and testing is presented in Table 1.
The “ImageDataGenerator from tensorflow.keras.preprocessing.image” package was used to increase the multiplicity of data for training models like rotating, shifting, zooming, and width shift of the images Fig. 4.
The parameters of “ImageDataGenerator” are also shown below in Fig. 5. Rotation range was set at 7 (this parameter specifies the range of degrees [0-180] within which the images can be rotated). Rotation augments the model’s robustness to orientation changes25,26. The width shift range was set at 0.2, and the height shift range was set at 0.2 (these parameters specify the range of horizontal and vertical shifts [as a fraction of the image size] that can be applied to the images). Width shift enhances the model’s robustness to object positioning and reduces overfitting to specific object locations25,26. The zoom range was set at 0.2 (this parameter specifies the range of zoom factors [as a fraction of the original image size] that can be applied to the images). Object size variations reduce overfitting of the model to specific object sizes25,26. Horizontal flip was set at false, meaning disabled. (This parameter specifies whether the images should be flipped horizontally [mirrored]). Fill mode was set at nearest, (this parameter specifies how to fill the newly created pixels when applying transformations [rotation, shifting]. In our study, the nearest neighbour interpolation method is used.
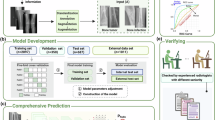
Three pre-trained models: Visual Geometry Group of the University of Oxford-16 layers [VGG16], MobileNetV2, and ResNet101V2, were used as base models. They were chosen as the base models due to their performance in previous deep learning studies in the sinonasal region, well-established architectures, strong feature extraction capabilities, availability of pre-trained weights, and balance between accuracy and computational costs16. The performance of all models was analyzed (Table 2and Fig. 6), and ResNet101v2 was selected as a base model.
We used the fine-tuning approach in our study (Fig. 7). In the initial steps, pre-processing and augmentation of the input data. The base model was then frozen, and a classification layer was added over it. Training and evaluation of the base model was then carried, out followed by unfreezing some of the top layers. Retraining of the whole model (lower training rate) was carried out, followed by evaluation. The fine-tuning was repeated if an improvement in performance was observed. This cycle was repeated till a pause in the improvement of performance metrics was noticed. The main idea of the proposed fine-tuning framework was to achieve a gradual increase in the level of layers that are to be unfrozen and tuned. To avoid overfitting, L1 regularization, also known as Lasso regularization, was used. It added a penalty term to the model’s loss function, which encourages the model to reduce the magnitude of its weights27. The model was trained with the following hyperparameters: 20 epochs, batch size 32, Adam optimizer with a learning rate of 1e-5, binary-crossentropy loss function, and sigmoid activation function for the top layer classification.
The analysis was performed on a local workstation running Ubuntu with Intel(R) Core (TM) M-5Y71 CPU @ 1.20 GHz, 1.40 GHz, and 8GB RAM, using Python to build the system using the deep learning frameworks Keras with TensorFlow as a back end.
Flow chart showing steps in the training and fine-tuning of the model used in the study. Training and evaluation of the base model was then carried out, followed by unfreezing some of the top layers. Retraining of the whole model was carried out, followed by evaluation. If performance metrics improved, the cycle was continued. The cycle was stopped till no further improvement was exhibited by the model.
Statistical analysis
The inter-rater and intra-rater reliability was evaluated using the Kappa Cohen test. The performance metrics of the model was evaluated in terms of accuracy, precision, F1-score, and area under curve (AUC).
Results
In the present study, the examiners analyzed 856 CBCTs and found AOs in 207 scans with an estimated prevalence of 24.18%. Among the 207 CBCT scans, 20 showed bilateral AOs [40 AOs], 98 showed right unilateral AOs, and 89 showed left unilateral AOs. Therefore, the total number of AOs was 227.
The inter-rater reliability between the two examiners for the detection of AO was 0.87, indicating almost perfect agreement. The intra-rater reliability for examiners 1 and 2 was 0.91 and 0.95, respectively.
The evaluation of performance metrics of the classification model revealed a training accuracy of 99%, and a valid accuracy of 81% (Fig. 8). The training and valid loss is shown in Fig. 9. The formula used for calculating accuracy = (TN + TP)/(TP + FP + TN + FN) [TN = True negative, True Positive, FP = False positive, FN = False negative].
The test accuracy and test loss of the unseen dataset was 0.81 and 0.51 respectively (Fig. 10).
The classification report of model (in terms of accuracy, precision, F1-score) and confusion matrix is shown in Figs. 11 and 12. The AUC value found to be at 0.87 are shown in Fig. 13.
Discussion
Studies have revealed that the prevalence of AOs is as high as 30% in patients with chronic sinusitis and 10–20% in healthy individuals, suggesting a strong link between the existence of AM and sinus pathologies6,28,29. In the present study, the prevalence of AOs in the CBCT scans of healthy individuals was estimated at 24.15%.
Some researchers believe that AO leads to the re-entry of the mucous that is drained out of the maxillary sinus through the primary ostium30. This complication associated with AO is known as “two-hole syndrome”31. The AO-linked mucous recirculation has been associated with chronic sinusitis32. The close proximity of the sinus to the posterior teeth makes the sinus pathology important to dental professionals.
In the present study, the inter-rater agreement for the detection was 0.87 for the detection of AO. A previous study in the same region for the detection of AO in CBCT scans using two observers has reported similar (0.83) inter-rater agreement values10. However, slightly lower (0.67) inter-rater agreement was reported in a study conducted at the University of Hong Kong1.
In the present study, we used the ImageDataGenerator to increase the multiplicity of data for training models, like rotating, shifting, zooming, and width shifting of the images. A recently published study using CBCT images in different planes of the maxillofacial region also used ImageDataGenerator with settings: rotation of 15 degrees, height and width shift of 0.1, and zoom by a factor of 0.533.
In general radiology studies, ImageDataGenerator has been used to generate a large number of chest x-ray images for the detection of COVID-19 using deep convolutional neural networks DCNN34. Flipping, rotation, and translation are the common methods used for augmentation of CT images35. Similar augmentation methods were used by the ImageDataGenerator used in our study to increase the data pool.
Recently, studies have revealed that deep learning models exhibit good performance metrics in image classification and segmentation36,37. In the present study, we used the ResNet-101V2 classification of model with a test accuracy of 81%. Recently published studies revealed that ResNet-101 showed higher accuracy compared to ResNet-50 and ResNet-152 in the classification chest X-rays for COVID related changes38. In another study ResNet showed best performance in classification of dental radiographs39. Another recent study using ResNet-101V2 for the detection of furcal bone loss showed a test accuracy of 91%40. The valid accuracy of the classification model used in our study was 81%, and the probability of misclassification is 19% (Fig. 14).
The probable factors for misclassification in our model could be due to a relatively smaller data set. The other reason could be due to wide variations in the anatomical position of the lateral nasal wall and the AO in the coronal CBCT images41,42,43.
Some recently published studies have used ResNet in the radiographic evaluation of the sino-nasal region16,44. A pretrained ResNet al.ong with a Swin transformer showed 99% accuracy in detecting boundaries of maxillary sinus pathologies in CBCT scans44. Similarly, another study on the classification of sinus pathologies in CT scans using ResNet showed an accuracy of 95%45.
In the present study, the ResNet-101V2 classifier showed an AUC value of 0.87 in the detection of AO. There is pre-activation of weights in version 2.00 of ResNet101, thus leading to better generalization compared to version 1.0046. Version 2.00 also produces a more normalized and regularized output signal leading to reduced overfitting46.
Though there are no studies exploring the accuracy of deep learning models in the detection of AO, one recent study has used ResNet-101V2 for the detection of nasal septum deviation in coronal CBCT images16. The AUC value of the classifier model used in that study (0.83) was slightly lower than in our study16. Slightly higher AUC values (0.92) were reported when ResNet-101 was used to detect sinusitis in paranasal sinus [PNS] radiographs47. The mild variation in the AUC could be due to the difference in the region of interest [ROI] and quantity of training datasets in these studies.
We carried out fine-tuning of the classification model in our study. Fine tuning improves the speed and computing efficiency of the AI model48. Since our dataset was comparatively smaller, we used the transfer learning technique. In this technique, the model is initially trained on a smaller dataset, and the features that have been grasped are readapted for use on training a different dataset48.
In the present study, we used L1 regularization, also known as Least Absolute Shrinkage and Selection Operator (LASSO) regularization to reduce overfitting. A recently published study used L1 regularization in ResNet to construct a compact model for reading ECG signals49.
Most of the recently published research papers on the application of deep learning models in the sino-nasal region focus on the classification of sinus pathologies, detection of deviated nasal septum, and detection of concha15,16,50. We have made an attempt to pioneer a study in the AI-based detection of AO in CBCT scans. We were able to develop an AI model for the detection of AO with an accuracy of 0.81 and an AUC of 0.87. We can further develop our model to detect AO in three-dimensional CT scans using the present study as the foundation.
However, there are some limitations in our study. Firstly, we have used two-dimensional cropped coronal sections from the CBCT scan and not 3D CBCT scans. The major challenges in developing a classification model for 3D CBCT scans are (1) complex 3D anatomical representations. (2) requires higher computing resources and (3) higher computational costs51.
The other limitation is a smaller dataset because of the lack of availability of large FOV CBCT scans, which are not frequently made in a dental imaging setup. The main disadvantage of using a smaller dataset is overfitting52. Overfitting leads to reduced generalizability and transferability of the classification model, causing poor performance when used on newer datasets52. The generalizability of our model is further affected because our data was obtained from one hospital setup. Future studies involving three-dimensional CBCT scans and larger dataset from different hospitals can be carried out with different deep learning models to further support our findings.
Conclusion
ResNet-101V2 showed good accuracy in detection of AO from coronal CBCT images. The findings of the present study can provide a base for future AI-based imaging studies on AO and other sino-nasal variations.
Data availability
The datasets generated and/or analysed during the current study are available in the Figshare repository [https://doi.org/10.6084/m9.figshare.28094912].
References
Hung, K., Montalvao, C., Yeung, A. K., Li, G. & Bornstein, M. M. Frequency, location, and morphology of accessory maxillary sinus Ostia: a retrospective study using cone beam computed tomography (CBCT). Surg. Radiol. Anat. 42, 219–228 (2020).
Shetty, S. R. et al. Application of a Cone-Beam Computed Tomography-Based Index for Evaluating Surgical Sites Prior to Sinus Lift Procedures-A Pilot Study. Biomed. Res. Int. 9601968 (2021). (2021).
Serindere, G., Gunduz, K. & Avsever, H. The relationship between an accessory maxillary ostium and variations in structures adjacent to the maxillary sinus without polyps. Int. Arch. Otorhinolaryngol. 26, e548–e555 (2022).
Prasanna, L. C. & Mamatha, H. The location of maxillary sinus ostium and its clinical application. Indian J. Otolaryngol. Head Neck Surg. 62, 335–337 (2010).
Rajendiran, D., Nagabooshanam, M. & Venugopal, R. Cross-sectional observational study on accessory ostium of the maxillary sinus. J. Clin. Sci. Res. 12, 88–92 (2023).
Yenigun, A. et al. The effect of the presence of the accessory maxillary ostium on the maxillary sinus. Eur. Arch. Otorhinolaryngol. 273, 4315–4319 (2016).
Mahajan, A., Mahajan, A., Gupta, K. & &Verma, P. Anatomical variations of accessory maxillary sinus Ostium: an endoscopic study. Int. J. Anat. Res. 5, 3484–3490 (2017).
Do, J. & Han, J. J. Anatomical characteristics of the accessory maxillary ostium in Three-Dimensional analysis. Medicina 58, 1243 (2022).
Bani-Ata, M. et al. Accessory maxillary Ostia: Prevalence of an anatomical variant and association with chronic sinusitis. Int. J. Gen. Med. 13, 163–168 (2020).
Shetty, S. et al. A study on the association between accessory maxillary ostium and maxillary sinus mucosal thickening using cone beam computed tomography. Head Face Med. 17, 28 (2021).
Soikkonen, K. & Ainamo, A. Radiographic maxillary sinus findings in the elderly. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 80, 487–491 (1995).
Swamy, A. & Sarumathi, T. Evaluating the prevalence, location, morphology of accessory maxillary sinus Ostia: A retrospective, cross sectional study using cone beam computed tomography. J. Indian Acad. Oral Med. Radiol. 35, 241–245 (2023).
Aksoy, O. & Orhan, K. Association between odontogenic conditions and maxillary sinus mucosal thickening: A retrospective CBCT study. Clin. Oral Investig. 23, 123–131 (2019).
Ali, I. K. et al. Cone-beam computed tomography analysis of accessory maxillary ostium and Haller cells: Prevalence and clinical significance. Imaging Sci. Dent. 47, 33–37 (2017).
Zou, C. et al. Preliminary study on AI-assisted diagnosis of bone remodeling in chronic maxillary sinusitis. BMC Med. Imaging. 24, 140 (2024).
Shetty, S. et al. The application of mask Region-Based convolutional neural networks in the detection of nasal septal deviation using cone beam computed tomography images: Proof-of-Concept study. JMIR Form. Res. 8, e57335 (2024).
Wanni, X., You-Lei, F. & Dongmei, Z. ResNet and its application to medical image processing: Research progress and challenges. Computer Methods and Programs in Biomedicine. 240, 107660 (2023).
Showkat, S. & Qureshi, S. Efficacy of transfer Learning-based ResNet models in chest X-ray image classification for detecting COVID-19 pneumonia. Chemometr Intell. Lab. Syst. 224, 104534 (2022).
Hasanah, S. A., Pravitasari, A. A., Abdullah, A. S., Yulita, I. N. & Asnawi, M. H. A deep learning review of ResNet architecture for lung disease identification in CXR image. Appl. Sci. 13, 13111 (2023).
van Leeuwen, K. G., de Rooij, M., Schalekamp, S., van Ginneken, B. & Rutten, M. M. How does artificial intelligence in radiology improve efficiency and health outcomes? Pediatr. Radiol. 52, 2087–2093 (2022).
Bayrakdar, I. S. et al. Artificial intelligence system for automatic maxillary sinus segmentation on cone beam computed tomography images. Dentomaxillofac Radiol. 53, 256–266 (2024).
Altun, O. et al. Automatic maxillary sinus segmentation and pathology classification on cone-beam computed tomographic images using deep learning. BMC Oral Health. 24, 1208 (2024).
Sin, Ç. et al. A deep learning algorithm proposal to automatic pharyngeal airway detection and segmentation on CBCT images. Orthod. Craniofac. Res. 24 Suppl 2, 117–123 (2021). (2021).
Wang, S., Fei, J., Liu, Y., Huang, Y. & Li, L. Study on the application of deep learning artificial intelligence techniques in the diagnosis of nasal bone fracture. Int. J. Burns Trauma. 14, 125–132 (2024).
Elgendi, M. et al. The effectiveness of image augmentation in deep learning networks for detecting COVID-19: A geometric transformation perspective. Front. Med. (Lausanne). 28, 629134 (2021).
Prezja, F., Annala, L., Kiiskinen, S. & Ojala, T. Exploring the efficacy of base data augmentation methods in deep Learning-Based radiograph classification of knee joint osteoarthritis. Algorithms 17, 8 (2024).
Safi, S. K. Empowering deep learning for images: A comparative analysis of regularization techniques in CNNs. J. Pub Int. Res. Eng. Manag. 5, 1–13 (2021).
Soylemez, U. O. & Atalay, B. Investigation of the accessory maxillary Ostium: A congenital variation or acquired defect? Dentomaxillofac Radiol. 50, 20200575 (2021).
Jones, N. S. CT of the paranasal sinuses: A review of the correlation with clinical, surgical and histopathological findings. Clin. Otolaryngol. Allied Sci. 27, 11–17 (2002).
Gutman, M. & Houser, S. Iatrogenic maxillary sinus recirculation and beyond. Ear Nose Throat J. 82, 61–63 (2003).
Mladina, R., Vuković, K. & Poje, G. The two holes syndrome. Am. J. Rhinol Allergy. 23, 602–604 (2006).
Chung, S. K., Dhong, H. J. & Na, D. G. Mucus circulation between accessory ostium and natural ostium of maxillary sinus. J. Laryngol Otol. 113, 865–867 (1999).
Kats, L., Goldman, Y. & Kahn, A. Automatic detection of image sharpening in maxillofacial radiology. BMC Oral Health. 21, 411 (2021).
Mezzoudj, S., Belkessa, I., Bouras, F. & Meriem, K. A novel distributed deep learning approach for large-scale chest X-ray covid-19 images detection, 07 February 2023, PREPRINT (Version 1) available at Research Square [https://doi.org/10.21203/rs.3.rs-2534755/v1]
Hu, R. et al. Automated diagnosis of covid-19 using deep learning and data augmentation on chest CT. 1–11 https://doi.org/10.1101/2020.04.24.20078998(2020).
Magat, G. et al. Automatic deep learning detection of overhanging restorations in bitewing radiographs. Dentomaxillofac Radiol. 53, 468–477 (2024).
Orhan, K., Bayrakdar, I. S. & Yakin, E. Second mesio-buccal Canal segmentation with YOLOv5 using CBCT images. Int. Dent. J. 74, S13 (2024).
Abdrakhmanov, R., Viderman, D., Wong, K. S. & Lee, M. Few-Shot Learning based on Residual Neural Networks for X-ray Image Classification. IEEE International Conference on Systems, Man, and Cybernetics (SMC), Prague, Czech Republic 2022, 117–1821, (2022). https://doi.org/10.1109/SMC53654.2022.9945469
Cejudo, J. E., Chaurasia, A., Feldberg, B., Krois, J. & Schwendicke, F. Classification of dental radiographs using deep learning. J. Clin. Med. 10, 1496 (2021).
Shetty, S. et al. Application of artificial intelligence-based detection of furcation involvement in mandibular first molar using cone beam tomography images- a preliminary study. BMC Oral Health. 24, 1476 (2024).
AlQabbani, A., Aldhahri, R. & Alhumaizi, A. Rare variation of accessory maxillary ostium. Cureus 12, e11921 (2020).
Okumus, O. & Şalli, G. A. The relationship between accessory maxillary Ostium, maxillary sinus pathologies, and sinonasal region variations. J. Stoma. 76, 182–190 (2023).
Zahedi, F. D., Yaacob, N. M., Wang, Y. & Abdullah, B. Radiological anatomical variations of the lateral nasal wall and anterior skull base amongst different populations: A systematic review and meta-analysis. Clin. Otolaryngol. 48, 271–285 (2023).
Çelebi, A. et al. Maxillary sinus detection on cone beam computed tomography images using ResNet and Swin Transformer-based UNet. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 138, 149–161 (2024).
Alhumaid, M. & Fayoumi, A. G. Transfer Learning-Based classification of maxillary sinus using generative adversarial networks. Appl. Sci. 14, 3083 (2024).
He, K., Zhang, X., Ren, S. & Sun, J. Deep Residual Learning for Image Recognition. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 2016, 770–778. (2016). https://doi.org/10.1109/CVPR.2016.90 (2016).
Kim, H. G., Lee, K. M., Kim, E. J. & Lee, J. S. Improvement diagnostic accuracy of sinusitis recognition in paranasal sinus X-ray using multiple deep learning models. Quant. Imaging Med. Surg. 9, 942–951 (2019).
Yosinski, J., Jeff Clune, J., Bengio, Y. & Lipson, H. How transferable are features in deep neural networks? NIPS’14: Proceedings of the 27th International Conference on Neural Information Processing Systems. 2, 3320–3332 (2014).
Ukil, A., Marin, L. & Jara, A. J. L1 and L2 Regularized Deep Residual Network Model for Automated Detection of Myocardial Infarction (Heart Attack) Using Electrocardiogram Signals. Proceedings of the CIKM 2021 Workshops, November 01–05, (2021). https://ceur-ws.org/Vol-3052/paper9.pdf
Parmar, P. et al. An artificial intelligence algorithm that identifies middle turbinate pneumatisation (concha bullosa) on sinus computed tomography scans. J. Laryngol Otol. 134, 328–331 (2020).
Muzahid, A. M. et al. Deep learning for 3D object recognition: A survey. Neurocomputing, 608, (2024). https://doi.org/10.1016/j.neucom.2024.128436
Bailly, A. et al. Effects of dataset size and interactions on the prediction performance of logistic regression and deep learning models. Comput. Methods Programs Biomed. 213, 106504 (2022).
Acknowledgements
Not applicable.
Author information
Authors and Affiliations
Contributions
S.S: Protocol/project development, Data analysis, Manuscript writing/editing. T. W: data collection or management. A.N: data collection or management. A.S: data collection or management. S.M: data collection or management. E.M: data collection or management. G. K: data collection or management8) N.S: data collection or management9) O.I: data collection or management10) O. U: Protocol/project development, Data analysis, Manuscript writing/editing11) D. L: Protocol/project development, Data analysis, Manuscript writing/editingAll authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods were carried out in accordance with relevant guidelines and regulations. All experimental protocols were approved by the Research ethics committe e Ref. no. REC-24-12-07-01-F, (University of Sharjah).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shetty, S., Talaat, W., Al-Rawi, N. et al. Accuracy of deep learning models in the detection of accessory ostium in coronal cone beam computed tomographic images. Sci Rep 15, 8324 (2025). https://doi.org/10.1038/s41598-025-93250-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-93250-8
Keywords
This article is cited by
-
Convolutional neural network for maxillary sinus segmentation based on the U-Net architecture at different planes in the Chinese population: a semantic segmentation study
BMC Oral Health (2025)
-
Deep learning-assisted CBCT segmentation provides reliable volumetric assessment of mandibular defects compared with micro-CT for 3D printing and surgical planning
Scientific Reports (2025)