Abstract
Current nucleic acid amplification techniques for the diagnosis of pulmonary tuberculosis (TB) lack the simplicity and affordability of achieving the goals envisioned in the WHO END-TB strategy, particularly for low- and middle-income countries (LMICs). Here we report a cost-effective real-time LAMP (rt-LAMP) assay with high sensitivity for the early diagnosis of pulmonary TB. A sample size of 350 was calculated using Buderer’s formula. The assay was validated in the laboratory using mpt64 cloned gene targets and clinical patient samples. The lower limit of detection for the rt-LAMP assay was observed as 10 copies/µl. Out of 350 suspected TB patient samples, 47 were positive for MGIT culture (microbiological reference standard, MRS), 42 were positive for Xpert MTB/RIF and 41 were positive for rt-LAMP assay. Compared to MRS, rt-LAMP showed a sensitivity of 89.36% (95% CI 76.9–96.45%) and a specificity of 94.06% (95% CI 90.77–96.44%). When compared to Xpert MTB/RIF, rt-LAMP showed a sensitivity of 93.33% (95% CI 83.80–98.15%), a specificity of 98.62% (95% CI 96.51–99.62%), negative predictive value of 98.62% (95% CI 96.52–99.47%), positive predictive value of 93.33% (95% CI 84.07–97.38%). Youden index value of rt-LAMP was 0.92 when compared with Xpert MTB/RIF, indicating a significantly low false positive rate. As the technology has been developed in an open platform, the assay will be useful in early diagnosis of pulmonary TB, particularly in screening large susceptible populations in LMICs.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is a highly contagious disease caused by the bacterium Mycobacterium tuberculosis. Nearly 1.8 billion people globally, close to one-quarter of the world’s population, are infected with TB, making it one of the leading communicable diseases1. The likelihood of contracting TB is closely related to social determinants and various health-related risk factors. Certain populations, particularly those who are vulnerable or with low socio-economic status, are disproportionately affected by TB2.
Unlike other infectious diseases like HIV and malaria, tuberculosis currently lacks a simple, fast and affordable test. Implementation of an accurate tuberculosis diagnostic test, readily available in clinical centres and hospitals, has the potential to prevent approximately 200,000 deaths annually3.
WHO suggests that due to inadequate access to rapid diagnostic tests, only 71% of new infections were reported. According to the recently released WHO 2023 report, the COVID-19 pandemic has had a detrimental impact on tuberculosis diagnosis and care, resulting in a surge of cases and fatalities. In 2021, there were 10.6 million new tuberculosis cases, compared to 10.1 million in 2020, with 1.6 million deaths, up from 1.5 million the previous year. Furthermore, tuberculosis incidence rose by 3.6% in 2021, which is a reversal of the nearly two-decade-long trend of a 2% annual decrease3.
In recent years, global initiatives have been implemented to "find the missing cases" of TB4. To achieve accurate diagnosis and early treatment of TB, it is crucial to strengthen TB diagnosis activities by prioritising microbiological detection. Clinical examination methods for TB usually have low specificity, which can result in incorrect diagnosis and unnecessary treatment. The WHO 2021 guideline has endorsed various diagnostic technologies, including amplifying and detecting M. tuberculosis complex (MTBC) nucleic acids, which have proven to be highly sensitive and specific1.
Among the nucleic acid amplification tests (NAATs), Xpert MTB/RIF and Xpert Ultra (Cepheid, Sunnyvale, CA, United States) emerged as reliable solutions because of their ability to detect low levels of bacterial DNA, thereby providing a more accurate diagnosis along with rifampicin susceptibility5,6,7,8.
Delays in diagnosis and underdiagnosis pose significant challenges to the elimination of TB. To address this, efforts have been made to use Xpert MTB/RIF and Xpert Ultra as screening tools by pooling patient samples9,10. However, implementing large-scale TB screening in resource-limited countries necessitates the development of new, accurate, low-cost, high-throughput screening tools.
Isothermal nucleic acid amplification techniques are gaining popularity for pathogen detection due to their affordability. Techniques such as rolling circle amplification (RCA)11, recombinase polymerase amplification (RPA)12, strand displacement amplification (SDA)13, Ladder-shape melting temperature isothermal amplification of nucleic acids (LMTIA)14, and loop-mediated amplification of DNA (LAMP)15 allow for simple and cost-effective detection. These methods are also faster than the qPCR but have shown lower sensitivity and specificity than the qPCR technique16. For example, a large amount of evidence on LAMP assay supports its robustness but is found to have a lower sensitivity in disease diagnosis17,18,19.
Here, we report a novel real-time LAMP (rt-LAMP) assay for the diagnosis of pulmonary tuberculosis in adults having a high sensitivity and specificity comparable to the Xpert MTB/RIF.
Materials and methods
Study settings
In this study, we collected sputum samples at the State Tuberculosis Demonstration and Training Centre (State TB Cell) in Thiruvananthapuram, Kerala, India. The State TB Cell has TB testing facilities for culture, Xpert MTB/RIF, and smear test methods. The study was conducted from October 2019 to March 2020 and January 2023 to March 2024, during which we collected sputum samples from presumptive TB patients. The gap in the study was due to the SARS-CoV-2 pandemic. The samples were tested using MGIT Culture as the microbiological reference standard (hereafter named MRS), sputum smear microscopy, Xpert MTB/RIF, and rt-LAMP assays.
Sample size calculation
Buderer’s formula was utilized to determine the required sample size to evaluate the assay’s diagnostic parameters to achieve the desired level of accuracy in sensitivity and specificity20. Assuming a sensitivity and specificity of 90% (0.90) for rt-LAMP and a 10% prevalence of TB among suspected cases being tested at the State TB cell in Trivandrum21 we calculated a sample size of 346. Therefore, we included 350 patients in the study to ensure a representative sample.
Recruitment and inclusion criteria
All patients suspected of having TB and visiting the State TB Cell were referred for CB-NAAT or culture tests by a chest physician at peripheral, secondary, or tertiary hospitals. The State TB Center offers free TB diagnostic testing for suspected patients. This study included patients over 15 years old who were suspected of pulmonary TB.
Real-time LAMP (rt-LAMP) kit
rt-LAMP primers for the mpt64 gene (GenBank accession number NC_000962.3) were designed based on melting temperature (Tm) critical for the LAMP assay using LAMP designer 1.16 software (Premier Biosoft Interpairs, USA). The primer details, including its positions in the mpt64 gene and specificity to the M. tuberculosis complex, are provided in Supplementary Table S1.
The rt-LAMP kit consisted of a 200 µL master mix containing 1.6 µM Forward and Backward Inner Primers (FIP and BIP), 0.4 µM Forward and Backward outer primers (F3 and B3), 0.2 µM Loop primers (LF and LB), 1X WarmStart LAMP Master Mix (NEB, USA), and 2 µM SYTO 16, with nuclease-free water added to reach a final volume for 10 reactions. For each reaction, 20 µL of the master mix was aliquoted into a PCR tube, and 5 µL (1.5 ng/µL) of Mycobacterium tuberculosis H37Rv genomic DNA (ATCC 27294) was added, making the final reaction volume to 25 µL. The rt-LAMP assay was carried out at 65⁰C for 40 min in a QuantStudio 5 qPCR machine (ThermoFisher Scientific, USA) programmed in isothermal cycles. The fluorescence levels were recorded every minute for 40 min using QuantStudio Design and Analysis Software v1.5.1.
Development of proof of concept of the rt-LAMP
Recombinant mpt64 plasmid as positive control
For rt-LAMP, we selected the mpt64 gene, which is a single-copy gene per genome of M. tuberculosis. The primary reason for this selection was to make the assay more quantitative and to avoid false negatives, especially for the South Asian population, where Mycobacterium tuberculosis complex (MTB complex) may lack the IS6110 gene22.
The mpt64 gene construct was cloned using the Gibson assembly protocol23. The primers were designed to amplify the mpt64 gene fragment and 20 base pairs of homology to the linearized (digested with Xba1 and HindIII) cloning vector pL4440 (Addgene #1654) using the software NEBuilder Assembly tool, v 2.7.1. The primer sequences are given below:
Forward 5′ cgaggtcgacggtatcgatATCGTTTTGCTCTGTTGTTC 3′.
Reverse 5′ ccaccgcggtggcggccgctACGTGGGACCAATACCTG 3′.
The amplified mpt64 gene fragment (666 bp) was cloned into the XbaI and HindIII digested pL4440 (2712 bp) vector. The recombinant plasmid was further transformed into DH5α E.coli strain and isolated on a large scale. The plasmid containing mpt64 was named pL-mpt64 and was used as the positive control for both assay standardisation and for the limit of detection assay.
Limit of detection
To assess the quantitative sensitivity of the rt-LAMP assay, serial titration of pL-mpt64 DNA in deionized water was prepared and used as templates. The pL-mpt64 plasmid concentration measured was converted into copy numbers by following the equation24: Number of DNA copies/µl = (M x 6.022x1023x10-9)/ (n x660)
M is the amount of DNA in ng/µl, n is the total base pair of DNA target (3334 bp for pLmpt-64) plasmid, Avogadro’s number 6.022 × 1023 and 660 is the average molecular weight of the DNA base pair.
The amplification time of rt-LAMP was plotted against the log values of copies of DNA/µl for the analysis. The assays were performed with DNA copies ranging from 1.91 × 10⁸ to 1.91 × 10⁰, with each dilution assessed in triplicate.
Validation of the rt-LAMP using clinical samples
In this study, 350 presumptive pulmonary TB sputum samples were collected and subjected to smear microscopy, Xpert MTB/RIF, MRS, and rt-LAMP assays. Patients’ written informed consent was obtained to collect the epidemiological, demographic and clinical data (Table S2).
Sample collection
For the study, the morning and spot sputum samples were collected from 350 presumptive TB patients in sterile sputum collection tubes. The morning sputum sample was used for MRS and the spot sputum sample was further divided into 1 ml each for smear microscopy testing, Xpert MTB/RIF and rt-LAMP assay.
The flow chart below illustrates the collection and allocation of samples for the specified tests (Fig. 1).
Sputum Smear test
The smear test was carried out, using NALC-NaOH processed sputum samples, following the Ziehl–Neelsen (ZN) staining procedure. The results were graded according to RNTCP guidelines25.
MRS: MGIT culture
The sputum samples were decontaminated, adding an equal volume of NALC-NaOH solution (containing 2% NaOH).
The culturing of MTB was performed using MGIT protocol in tubes containing Middle brook 7H9 broth26. The tubes were incubated at 37 °C for 42 days and monitored for fluorescence using the MGIT 960 instrument (BD, USA) to detect mycobacterial growth. All MGIT-positive cultures were confirmed as Mycobacterium tuberculosis complex (MTBc) using the MPT64 antigen-based test (SD Bioline TB Ag MPT64 Rapid, Abbott, USA)27.
MGIT culture results are taken as microbiological research standard (MRS).
Xpert MTB/RIF
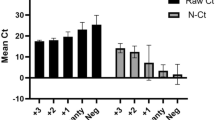
The Xpert MTB/RIF assay was performed by following the kit protocol (Cepheid, CA, USA)28. The results were interpreted semi-quantitatively using cycle threshold (Ct) values as follows: very low (Ct above 28), low (Ct 22–28), medium (Ct 16–22), and high (Ct below 16) using GeneXpert software (see Supplementary Table S2).
DNA extraction from sputum samples for rt-LAMP
From sputum samples treated with NALC-NaOH, the DNA was extracted using the standard protocol with modifications29. Briefly, around 2 ml of patients’ sputum samples were treated with an equal volume of NALC-NaOH solution. The sample was centrifuged at 800 × g for 10 min, and the supernatant was discarded. 100 µl Lysis buffer (0.4 M NaCl, 2 mM EDTA, 10 mM Tris–Cl, and 0.5% Triton X-100, pH 8) was added to the pellet and resuspended by vortexing. The tubes were incubated at 95 °C for 20 min before adding 100 µl neutralization buffer (10 mM Tris–Cl, pH 7.5). The tubes were sealed and stored at 4 °C till use. The samples were used for rt-LAMP assay without further purification.
Evaluation of rt-LAMP assay
A volume of 5 µl of the extracted DNA sample was added to a 20 µL single-tube rt-LAMP kit containing 1X WarmStart LAMP mix, six primers, and SYTO 16 dye. The rt-LAMP assay was performed at 65 °C for 40 min using a QuantStudio™ 5 qPCR machine (ThermoFisher Scientific, USA). Fluorescence readings were recorded every minute for 40 min using QuantStudio™ Design and Analysis Software v1.5.1.
Quality control (QC)
QC was ensured in the form of sterility and performance testing of each batch of rt-LAMP. Performance testing of reagents was carried out using positive (pL-mpt64 DNA) and negative control (without DNA) in each set of tests.
Data analysis
rt-LAMP kit was evaluated against the reference Standards, MRS, Xpert MTB/RIF, and smear test. The discriminative ability of the rt-LAMP test was quantified by measures of diagnostic accuracy, including sensitivity and specificity, positive and negative predictive values (PPV, NPV), likelihood ratio, Youden index, ROC curve, and accuracy compared to culture, Xpert MTB/RIF and smear tests.
Ethical considerations
Ethical clearance for the research study was obtained from the SCTIMST Institutional Ethics Committee (IEC/1230). Patients provided written informed consent to collect their epidemiological, demographic, and clinical data, which was available at the State TB Center when the sample was submitted. The study was conducted in accordance with the Declaration of Helsinki.
Results
Optimization of rt-LAMP
The study evaluated the effectiveness of the rt-LAMP assay and showed a robust exponential amplification within 10–20 min with a high ΔRn value (Fig. 2A).
(A) Limit of detection analysis of rt-LAMP for the mpt64 gene A. l to 9 serially diluted pL-mpt64 DNA with a concentration of 1.91 × 108, 1.91 × 107, 1.91 × 106, 1.91 × 105, 1.91 × 104, 1.91 × 103, 1.91 × 102, 1.91 × 101, 1.91 × 100 copies/µl and 10 : No Template Control. (B) The standard curve of the rt–LAMP assay shows a linear correlation of concentration DNA with amplification time R2 = 0.97. n = 3.
pL-mpt64 plasmid was serially diluted from an initial concentration of 1.91 × 10⁸ copies/µl to a final concentration of 1.91 × 100 copies/µl. rt-LAMP assays were performed to compare the detection limit. The results indicated that the rt-LAMP assay with SYTO16 could detect DNA having 10 copy numbers/µl, (Fig. 2B) (R2 = 0.97), maintaining a concentration-dependent linearity.
Analytical sensitivity and specificity of rt-LAMP
Among the 350 participants recruited for the study, the majority (63%) were male, while 37% were female. The participants ranged in age from 15 to 85 years, with a median age of 53 years. Among them, 16 (2.51%) reported a history of contact with known TB cases and 9 (2.51%) were under anti-TB treatment (Table 1).
Out of 350 suspected TB samples, 47 were MRS positive, 42 were smear (SS) positive and 60 were positive for Xpert MTB/RIF and rt-LAMP assay, respectively.
Compared to MRS, rt-LAMP showed a sensitivity of 89.36% (95% CI 76.9–96.45%) and a specificity of 94.06% (95% CI 90.77–96.44%) (Table 2). When compared to Xpert MTB/RIF, rt-LAMP showed a very high sensitivity of 93.33% (95% CI 83.80–98.15%) and specificity of 98.62% (95% CI 96.51–99.62%) (Table 2). We observed rt-LAMP assay compared to Xpert MTB/RIF showing a higher negative predictive value (NPV) of 98.62% (95% CI 96.51–99.62%), positive predictive value (PPV) of 93.33% (95% CI 83.80–98.15%) (Table 2). Youden index value was 0.91, suggesting significantly low false positives. The positive likelihood ratio (LR+) was 67.33 and the negative likelihood ratio (LR−) was 0.07. Kappa value was 0.92 (0.86 to 0.97) (Table 2). Two-way comparison of the rt-LAMP with MRS positive, Xpert MTB/RIF positive and sputum smear (SS+) test-positive samples showed a 100% sensitivity and 95.85% specificity (Table 2). A similar result was observed for the two-way comparison of rt-LAMP with MRS-positive and sputum smear-positive (SS+) samples (Table 2). In MRS-positive and sputum smear-negative (SS-) groups, the sensitivity and specificity were 73.68% and 95.8%, respectively (Table 2).
Analyzing logistic regression using MRS as the gold standard, it was observed that Xpert MTB/RIF and rt-LAMP had overlapping plots with an AUC of 0.905 and 0.917 respectively, suggesting that rt-LAMP has a robust detection accuracy (Fig. 3).
We compared the performance of the Xpert MTB/RIF and rt-LAMP assays with MRS. The rt-LAMP assay demonstrated slightly higher sensitivity and specificity than the Xpert MTB/RIF assay, with both tests showing high accuracy (Table 3). The Youden Index values were 0.81 and 0.83 for the Xpert MTB/RIF and rt-LAMP assays, respectively. The sensitivity and specificity of the SS test were 59.97% (95% CI 44.27–73.63%) and 95.38% (95% CI 92.37–97.45%) respectively compared to MRS (Table 3). Both Xpert MTB/RIF and rt-LAMP assays showed higher specificity of detection compared to the SS test (Table 3). Among the 42 SS + cases, 7 patients were negative for MRS, Xpert and rt-LAMP assay, indicating that there are false positives reported in the smear tests (see Supplementary Fig. S1). Among the TB treatment group (9 cases; 2.5% of the study population), no patient showed positive for any of the TB tests, including rt-LAMP.
Discussion
This study reports a newly developed rt-LAMP assay is highly specific and sensitive for early diagnosis of pulmonary tuberculosis. Youden index of 0.92 suggests a high discriminative ability of the test. Ideally, a good diagnostic test should have a LR+ of > 10 and an LR- of 0.130. The rt-LAMP assay showed a LR+ of 67.63 and 15.04 and LR- of 0.07 and 0.11, with Xpert MTB/RIF and MRS respectively, meeting the requirements.
TB-LAMP by Eikon, Japan, is the predicate assay for the newly developed rt-LAMP assay and has been recommended by WHO as an alternate assay to sputum smear1. TB-LAMP is an affordable and reliable endpoint reading tool for amplifying nucleic acids without requiring sophisticated equipment. However, compared to MRS, TB-LAMP has shown a pooled sensitivity of 81.1% (95% CI 70.6–88.5)1 (see Supplementary Table S3). Our newly developed rt-LAMP assay kit has an improved sensitivity of 89.36% (95% CI 76.9–96.45%) compared to MRS. Besides, endpoint reading TB-LAMP assays give only one data point, hence, it becomes a challenge to differentiate non-specific amplifications in the assay. The newly developed rt-LAMP kit gives 40 data points for each sample and shows exponential amplification signals in positive samples within 10 min.
The rt-LAMP assay kit has been developed as an open platform system that can run in an isothermal fluorescent reader or any qPCR machine programmed to run under isothermal cycles. During the COVID-19 pandemic, qPCR equipment has become an integral component of most healthcare and diagnostic testing centres which can be used to run the rt-LAMP assay. We compared the rt-LAMP assay with the recommendations by the Treatment Action Group for TB point of care test31 and found it to meet the requirements for all major points, including high throughput screening capabilities (see Supplementary Table S4). When utilizing a quantitative polymerase chain reaction (qPCR) machine for the rt-LAMP assay, it is possible to process either 48, 96, or 384 tests in a single run. This throughput allows for the completion of more than four runs per day, depending on the specific machine configuration and operational efficiency. This capacity significantly increases testing efficiency and facilitates timely diagnosis and treatment for patients. On the other hand, commercially available LAMP PCR machines can handle 16 tests in each run. Similar to qPCR, these machines can also conduct more than four runs per day, depending on the laboratory workflow and testing demand. The flexibility and efficiency of the open rt-LAMP assay present a notable advancement over traditional pooled testing methods, such as those using the Xpert MTB/RIF or Xpert Ultra systems. This improvement is crucial for enhancing the speed and reliability of TB diagnostics, ultimately contributing to better patient outcomes.
Implementing the rt-LAMP assay in diagnostic centres, however, presents a few challenges. One of the main issues is that it requires a separate DNA extraction procedure, which must be performed either manually or through automation. This necessitates a designated extraction area equipped with tools such as centrifuges, vortex mixers, pipettes, and more. Additionally, trained personnel with expertise in DNA extraction from sputum and clinical specimens are essential. These competencies are typically available in COVID-19 testing centres, making it a viable option to repurpose these facilities for TB diagnosis. Since the rt-LAMP assay is a cost-effective (see table S4) and accurate diagnostic method, it can reduce the burden on Xpert MTB/RIF as the first set of tests, which is mainly used to detect rifampicin resistance—a crucial marker for drug-resistant (DR) and multidrug-resistant (MDR) TB. By incorporating rt-LAMP, we can enhance the screening process for suspected cases, reserving more expensive tests, such as Xpert MTB/RIF and line probe assay, for follow-up analysis for DR and MDR in positive cases. This approach could lead to a better, cost-effective disease control programme.
Our study also indicates that the sputum smear test, one of the most affordable test in LMICs, shows high false positivity. Several factors could contribute to this, including improper staining techniques, cross-contamination, the experience of the reader, non-tuberculous mycobacteria (NTMs), and environmental contamination32. Many of these issues stem from manual errors or procedural practices. These can be reduced with corrective actions and by following good laboratory practices. However, an affordable nucleic acid-based test may be a better alternative to the smear test, as it could enhance the specificity of the diagnosis.
The rt-LAMP assay reported here would be ideal for catering for a large population at an affordable cost and could be placed as an alternative for smear test. The rt-LAMP assay could be deployed in low-income countries, contributing significantly to the early detection of TB and in the disease eradication program.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
References
WHO Consolidated Guidelines on Tuberculosis. Module 3: Diagnosis—Rapid Diagnostics for Tuberculosis Detection, 2021 Update. (World Health Organization; 2021. Licence: CC BY-NC-SA 3.0 IGO, 2021).
Litvinjenko, S., Magwood, O., Wu, S. & Wei, X. Burden of tuberculosis among vulnerable populations worldwide: an overview of systematic reviews. Lancet Infect. Dis. https://doi.org/10.1016/s1473-3099(23)00372-9 (2023).
Global Tuberculosis Report 2023. (World Health Organization; 2023. Licence: CC BY-NC-SA 3.0 IGO, 2023).
Pande, T. et al. Finding the missing millions: lessons from 10 active case finding interventions in high tuberculosis burden countries. BMJ Glob. Health. 5, e003835 (2020).
Boehme, C. C. et al. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363, 1005–1015 (2010).
Dorman, S. E. et al. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect. Dis. 18, 76–84 (2018).
Geleta, D. A. et al. Xpert MTB/RIF assay for diagnosis of pulmonary tuberculosis in sputum specimens in remote health care facility. BMC Microbiol. 15, 220 (2015).
Lawn, S. D. et al. Advances in tuberculosis diagnostics: the Xpert MTB/RIF assay and future prospects for a point-of-care test. Lancet Infect. Dis. 13, 349–361 (2013).
Cuevas, L. E. et al. Systematic review of pooling sputum as an efficient method for Xpert MTB/RIF tuberculosis testing during the COVID-19 pandemic—volume 27, number 3—March 2021—Emerging infectious diseases journal—CDC. Emerg. Infect. Dis. 27, 719–727 (2021).
Quan, Z. et al. Pooling sputum samples for the Xpert MTB/RIF Ultra assay: A sensitive and effective screening strategy. Tuberculosis 149, 102575 (2024).
Fire, A. & Xu, S. Q. Rolling replication of short DNA circles. Proc. Natl. Acad. Sci. 92, 4641–4645 (1995).
Lutz, S. et al. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA). Lab a Chip 10, 887–893 (2010).
Seckinger, D. Strand displacement amplification and fluorescence polarization. Clin. Chem. 42, 1720–1720 (1996).
Wang, D. et al. Ladder-shape melting temperature isothermal amplification of nucleic acids. BioTechniques 71, 358–369 (2021).
Notomi, T. et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, e63–e63 (2000).
Özay, B. & McCalla, S. E. A review of reaction enhancement strategies for isothermal nucleic acid amplification reactions. Sens. Actuators Rep. 3, 100033 (2021).
Shete, P. B., Farr, K., Strnad, L., Gray, C. M. & Cattamanchi, A. Diagnostic accuracy of TB-LAMP for pulmonary tuberculosis: a systematic review and meta-analysis. BMC Infect. Dis. 19, 268 (2019).
Garg, N., Ahmad, F. J. & Kar, S. Recent advances in loop-mediated isothermal amplification (LAMP) for rapid and efficient detection of pathogens. Curr. Res. Microb. Sci 3, 100120 (2022).
Kim, S.-H., Lee, S.-Y., Kim, U. & Oh, S.-W. Diverse methods of reducing and confirming false-positive results of loop-mediated isothermal amplification assays: A review. Anal. Chim. Acta 1280, 341693 (2023).
Buderer, N. M. F. Statistical methodology: I. Incorporating the prevalence of disease into the sample size calculation for sensitivity and specificity. Acad. Emerg. Med. 3, 895–900 (1996).
Mohan, A. et al. Prevalence of tuberculosis infection among adults of Thiruvananthapuram district of Kerala as measured by interferon gamma release assay—A cross-sectional study. Indian J. Community Med. Off. Publ. Indian Assoc. Prev. Soc. Med. 47, 501–505 (2022).
Howard, S. T., Oughton, M. T., Haddad, A. & Johnson, W. M. Absence of the genetic marker IS6110 from a strain of Mycobacterium tuberculosis isolated in Ontario. Can. J. Infect. Dis. Méd. Microbiol. 9, 48–53 (1998).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Ma, B., Yu, H., Fang, J., Sun, C. & Zhang, M. Employing DNA binding dye to improve detection of Enterocytozoon hepatopenaei in real-time LAMP. Sci. Rep. 9, 15860 (2019).
Revised National Tuberculosis Control Programme Laboratory Network: Guidelines for Quality Assurance of smear microscopy for diagnosing tuberculosis. https://tbcindia.mohfw.gov.in/wp-content/uploads/2023/05/4234099618RNTCP-Lab-Network-Guidelines.pdf (2005).
Tortoli, E. et al. Use of BACTEC MGIT 960 for recovery of mycobacteria from clinical specimens: Multicenter study. J. Clin. Microbiol. 37, 3578–3582 (1999).
Armstrong, D., Pretty, L., D’Agostino, K., Redhead-Harper, R. & Parrish, N. Diagnostic Accuracy of the Abbott SD Bioline MPT64 Antigen Test for Identification of MTB Complex in a U.S. Clinical Mycobacteriology Laboratory. (2024) https://doi.org/10.2139/ssrn.4681916.
Xpert® MTB/RIF Assay. https://www.cepheid.com/content/dam/www-cepheid-com/documents/package-insert-files/Xpert-MTB-RIF-ENGLISH-Package-Insert-301-1404-Rev-G.pdf.
Crudu, V. et al. First evaluation of an improved assay for molecular genetic detection of tuberculosis as well as rifampin and isoniazid resistances. J. Clin. Microbiol. 50, 1264–1269 (2012).
Deeks, J. J. & Altman, D. G. Diagnostic tests 4: likelihood ratios. BMJ 329, 168 (2004).
Treatment Action Group experts meeting report on defining specifications for TB Point of Care test. https://www.msfaccess.org/sites/default/files/MSF_assets/TB/Docs/TB_event_POC_fullsurveyreport_ENG_2008.pdf (2009).
Zaporojan, N. et al. Evolution of laboratory diagnosis of tuberculosis. Clin. Pr. 14, 388–416 (2024).
Acknowledgements
We would like to acknowledge the following individuals for their valuable contributions towards the conduct of the study: Dr. R. Ajay Kumar from Rajiv Gandhi Centre for Biotechnology, Trivandrum; Dr. Praveen Sanker Shivasankaran, Ms Renu and Ms Remya from State Tuberculosis Demonstration and Training Centre, Trivandrum; Dr. Sanjeev Nair, Professor at Trivandrum Medical College; Dr. M Sunil Kumar, Honorary Secretary of TB Association of Kerala, Trivandrum; Dr. Ranjani Ramachandran and Dr. Anand from WHO India Country Office; and Dr. Nishant Kumar, Deputy Director of Central TB Division, Ministry of Health and Family, Government of India. Dr. Dinoop K.P, Associate Professor, Department of Microbiology, SCTIMST for critically evaluating the manuscript. The study was funded by TRC programme, Department of Science and Technology, Government of India (AT).
Author information
Authors and Affiliations
Contributions
A.T. conceived the idea and planned the experiments; S.S.N., R.E.V. and A.S. conducted the experiments; A.T. and S.S.N. wrote the main manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nair, S.S., Varghese, R.E., Saji, A. et al. Validation study of a novel, rapid, open platform real-time LAMP assay for early diagnosis of pulmonary tuberculosis. Sci Rep 15, 10069 (2025). https://doi.org/10.1038/s41598-025-93565-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-93565-6