Abstract
Varanids are known for conserved sex chromosomes, but there are differences in the size of the W chromosome but not in morphology among species representing varying stages of sex chromosome evolution. We tested for homology of the ZW sex chromosome system with size differences in varanids among four species from two lineages in Australia, the Odatria and the Gouldii. We found that while DNA sequences of the sex chromosomes are conserved in the species we tested, we also identified a homologous region on an enlarged autosomal microchromosome that shares sequences with the W chromosome in some isolated populations of V. acanthurus and V. citrinus from the Odatria lineage. The enlarged microchromosome was unpaired in all individuals tested and is likely an unbalanced segmental duplication translocated between chromosome 1, the W, and another microchromosome. This suggests an ancient balanced duplication homologous to the W and the terminal region of the long arm of chromosome 1. The most parsimonious explanation is that the duplicated region likely originated on chromosome 1. We hypothesised in our reconstruction that genes and related DNA sequences associated with the sex-linkage group have likely originated on an autosome. Subsequently, the sequences may have undergone duplication and translocation to the W chromosome, followed by the accumulation of lineage specific repeat elements and amplifications on the W at different rates in various lineages. Lastly, these sequences are likely to have undergone duplication and translocation to another autosomal microchromosome. Given the role of segmental duplications and translocations as important evolutionary drivers of speciation in other taxa, together with the rapid speciation that has occurred in Australian varanids, our findings provide broader insight into the evolutionary pathway leading to rapid chromosomal and genic divergence of species.
Similar content being viewed by others
Introduction
Chromosomal rearrangements are important structural changes of the genome that have been linked to speciation events1,2,3,4,5. These rearrangements include several classes of rearrangements including segmental or whole chromosome duplications, deletions, inversions or translocations. Chromosomal rearrangements have been linked to speciation because they are associated with different levels of reproductive isolation or species divergence as a result of suppressed recombination of the portion of the altered chromosome1,2,6,7,8,9,10. For example, segmental duplications are long (> 1000 bp) nearly identical regions of the genome that are duplicated and relocated within the genome and considered an important mechanism for the diversification within the primate lineage11,12,13,14. These hosts of large-scale rearrangements provide a genetic substrate for the diversification of genes and opportunities for novel functions to emerge15,16,17. However, determining the class of the rearrangement and teasing out the functional roles they play is not a trivial task and involves multidimensional and interdisciplinary studies18,19,20,21.
Sex chromosomes are among the most rapidly evolving regions of any genome and differences in size and structure arise between sex chromosomes among populations and species22,23. Deletions involving gene loss and repeat motif expansions are usually accompanied by inversions or other structural variants that rearrange the directionality of gene sequences and drive the evolution of the sex chromosomes24,25. Some vertebrates have homologous sex chromosomes while in other vertebrates sex chromosomes are highly variable as a result of rapid sex chromosome turnover26,27,28,29. Sex chromosomes can transition within ZW or XY systems or transition between them30,31,32, and sex determination can even transition between genetic and environmental sex determination systems33,34. It is unknown why rapid sex chromosome turnover occurs in some lineages but not in others35. Most often sex chromosomes are identified by heteromorphy, in which they have diverged due to recombination suppression between the X and the Y or the Z and the W23,27,36,37. However, in other cases, sex chromosomes are cryptic and are less easily identified38. Cryptic sex chromosomes that are homomorphic usually represent early diversification and in rare situations can be indicators of recent turnover and the rapid replacement of sex-determining genes39, because the impacts of recombination suppression have not had time to lead to sex chromosome differentiation or they could be maintained from ongoing recombination for long periods40.
The Varanidae have been proposed to have the oldest conserved ZW sex chromosomes of all vertebrates, estimated to have preceded the varanid radiation in the Jurassic period, 150–180 MYA (Million Years Ago)41,42. Despite this, the Varanid W chromosome displays size variation among closely related species and recently identified within a newly described species V. citrinus (formerly an isolated population of V. acanthurus in the northern distribution of the Northern Territory of Australia43; where the sex chromosomes are cryptic and undifferentiated44. These size differences are likely a result of different rates of evolution of the independent expansion of transposable elements during sex chromosome evolution, and the involved repeat sequences within the sex chromosomes not being conserved between species45,46. For example, the heath goanna (V. rosenbergi) and the sand goanna (V. gouldii) have different sized W chromosomes attributed to CGGn expansions46, while the Komodo dragon (V. komodoensis) does not have any identifiable repeats on the W chromosome and has cryptic sex chromosomes45. The heteromorphic W chromosome in Varanus acanthurus in contrast is highly degraded from gene loss and amplification of (AAT)N repeats46.
Recently two Varanus genomes have been published, V. komodoensis (NCBI accession GCA_004798865.147), and V. acanthurus (NCBI BioProjectID PRJNA737594, Genome Warehouse BioProject accession PRJCA00558325). While the V. komodoensis genome assembly was aided by flow sorting of the chromosomes, the authors were unable to place the sex-linked scaffolds, and instead generated microchromosome pools that likely contain sex chromosome regions47. Zhu et al.25 took an alternative approach by developing a novel pipeline utilising a combination of stLFR (single-tube long fragment linked reads)48, and Illumina reads from both a male and female individual for a de novo assembly. Zhu et al.25 generated putative Z and W-linked scaffolds by read depth ratios between sexes, but so far, these sex-linked regions are only putative without evidence that physically map them to the chromosomes to validate the sequencing patterns of read depth ratios for identifying sex linked reads.
The ridge-tailed goannas are primarily saxicolous lizards, living in rocky outcrops and have a very large distribution across Northern Australia49. It is likely that there are many small, isolated populations separated by marginal habitat which may have been more suitable habitat during the Pliocene which provided a continuous distribution across northern Australia. However, the Holocene Epoch drying effect on the Australian continent may have impacted the current disjunct population distributions. These factors make the ridge-tailed goanna unique model taxa to study higher rates of speciation resulting in the fixation of novel chromosome rearrangements2,5,50 including origin and evolution of sex chromosomes51.
The objective of this study was to characterise the Z and W sex chromosomes in Varanids by testing the pipeline developed in our previous study25, which detected putative sex-linked reads. Herein, we developed FISH (Flourescence in situ Hybridisation) probes from these inferred sex-linked scaffolds and validated the methodology by hybridizing the probes to V. acanthurus individuals of known and validated sex. We then tested for homology of the sex-linked region between isolated V. acanthurus populations which have highly differentiated sex chromosomes25,41,46,49,52 and the newly described sister species V. citrinus43 which has cryptic sex chromosomes44. Cryptic sex chromosomes have also been identified in V. salvator41, V. gouldii46 and V. komodoensis45. We showed that sex chromosomes in the ridge-tailed goannas are highly conserved despite size polymorphisms. To broaden our analysis, we applied the FISH probes developed from V. acanthurus to additional varanid species exhibiting variations in W chromosome size and distinct expansions of repeat elements. Our findings revealed conserved homology in all species tested. Furthermore, we observed a polymorphic enlarged autosomal microchromosome found in both male and female individuals from certain populations of both V. acanthurus and V. citrinus. Remarkably, this microchromosome demonstrated homology with the female-specific W sex chromosome and the terminal region of the long arm of chromosome 1. We discuss our findings in the context of potential segmental duplications and the roles these genome destabilisers play in speciation.
Results
Validation of specificity of sex chromosome probes
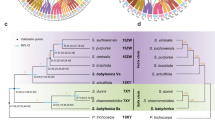
The Z probe consisted of 21,382 polynucleotide sequences from 124 scaffolds and the W probe was 27,392 polynucleotide sequences from 157 scaffolds (Supplementary Tables 1 and 2). A schematic diagram of the probe development process is summarised in methods section Fig. 5. We successfully developed sex-specific probes (Fig. 5) that were hybridised to the chromosomes of 13 V. acanthurus individuals from three different populations (Fig. 1a). After confirming the specificity of these probes for V. acanthurus (n = 13, 8 females, 5 males) within the same species, we proceeded to examine 4 individuals of V. citrinus (3 females, 1 male) with cryptic sex chromosomes. Our analysis revealed that one individual of V. citrinus exhibited a heteromorphic W chromosome, characterised by distinct size differences between the Z and W chromosomes. In contrast, the remaining 3 females displayed cryptic homomorphic sex chromosomes, with the Z and W chromosomes exhibiting similar sizes (Fig. 1b). We then tested our probe against two distantly related species from the Gouldii lineage and further validated the utility of the sex-specific probes in V. rosenbergi (Figs. 2a and 2b) that has a differentiated W (heteromorphic) and V. gouldii with cryptic sex chromosomes (homomorphic).
Summary of FISH results in V. acanthurus and V. citrinus individuals with normal sex chromosomes and individuals with enlarged microchromosomes. The W chromosome is homologous with the telomeric region of the long arm on chromosome 1 as a balanced duplication and with the unpaired enlarged microchromosome as an unbalanced segmental duplication. In V. citrinus the Z chromosome exhibited several segmental duplications with the centromere of a number of microchromosomes. ZW sex chromosomes within yellow box show hybridisation patterns of Z and W probes observed in 3 females of V. citrinus with homomorphic sex chromosomes. ZW sex chromosomes within white box show hybridisation patterns of Z and W probes observed in heteromorphic Z and W chromosome as well as enlarged unpaired microchromosomes from 1 female V. citrinus. EM is an enlarged and unpaired microchromosome first described by Matsubara et al.46; n indicates number of individuals screened.
Hybridisation of the Z and W probes to a male (a) and female (b) V. rosenbergi with heteromorphic sex chromosomes, and to a male (a) and female (b) V. gouldii with homomorphic sex chromosomes. Green arrowheads indicate hybridisation signals of Z chromosomes and pink arrowheads indicate hybridisation signals of W chromosome.
Hybridisation patterns with sex-specific chromosome probes
The sex chromosome-specific probes were hybridised to the chromosomes of 21 individuals from 4 species with karyotype variations (Table 1). There were size differences involving the W chromosome within V. citrinus individuals but all V. acanthurus individuals were highly heteromorphic with respect to the Z, and size variations were observed between V. rosenbergi and V. gouldii. In all males tested, we readily identified ZZ chromosomes and, in all females, we identified a Z and a W, however, both probes showed faint hybridisation signals with other chromosomes Fig. 1, Supplementary Fig. 1). The most striking finding includes hybrisdisation of the W probe to the enlarged unpaired microchromosome in both sexes of V. acanthurus and V. citrinus, in individuals with unpaired enlarged microchromosomes (Figs. 1 and 2, Supplementary Fig. 1). In addition, both the Z and W probes hybridised to the terminal regions of the long arm of chromosome 1 in V. acanthurus and V. citrinus, but there were only weak hybridisation signals of the W probe to the terminal region of the long arm of chromosome 1 in V. rosenbergi and V. gouldii (Fig. 2, Table 1, Supplementary Fig. 1).
In V. acanthurus all females (n = 8) displayed W chromosome heteromorphisms in which the W chromosome was significantly larger than the Z (Fig. 1a). Both Z and W probes showed high level of specificity with ZW sex chromosomes in both V. gouldii and V. rosenbergi which also confirmed ZW sex chromosomes without hybridisation with any other chromosomes (Table 1, Fig. 2). In V. citrinus we identified one female had an enlarged W chromosome that was partially homologous with the enlarged unpaired microchromosome (Fig. 1), and this was mistaken as an enlarged microchromosome pair in a previous study44. The other two females had homomorphic sex chromosomes where the W chromosome was similar in size to the Z (Fig. 1a) and these females did not have enlarged unpaired microchromosome (Fig. 1b).
Discussion
The objective of this study was to test the homology of the Z and W sex chromosomes across species of varanids with cryptic and heteromorphic sex chromosomes using a chromosomics approach. Our analysis identified a novel discovery revealing that an autosomal enlarged microchromosome polymorphism shares homology with the W chromosome in V. acanthurus and V. citrinus indicating a putative segmental duplication and translocation from the sex chromosome to an individual microchromosome. Both the Z and W probes also hybridised to the terminal region of the long arm of chromosome 1 in all individuals implying that the ancestral sequences may have originated there. This finding also provides further support that amniote sex chromosomes may have shared a common ancestry that originated from an autosome23,53. We expanded our analysis with two other species with size and repeat element differences and demonstrated that the genes on the sex chromosomes are conserved despite chromosomal size variation and differences in the repeat element expansions of the highly degraded W chromosome.
The enlarged microchromosome (EM Fig. 1B, Supplementary Fig. 1) is a mutation that has not been observed in any other goanna species and appears to be specific to localised populations of ridge-tailed goannas including individuals from both V. acanthurus and V. citrinus, and likely represent a remnant of ancestral segmental duplication/translocation. It was first reported by Matsubara and colleagues46 from a captive bred lineage but was not identified in wild populations until recently44. EM is not a common mutation and was identified in only 3 out of 34 individuals karyotyped and only found in 2 out of 4 isolated populations44. During our initial cytogenetic analysis, this enlarged microchromosome was mistaken for a W chromosome in individuals that upon dissection were determined to be males. Varanids are notorious for being difficult to sex phenotypically because females can have hemiclitores that resembles hemipenes in males41,45,54,55,56,57,58,59. Therefore, the gonads must be examined (Supplementary Fig. 2) for accurate sex determination.
FISH experiments with our chromosome-specific probes were required to differentiate the sex chromosomes in individuals with cryptic sex chromosomes and to identify the Z chromosome from other microchromosomes. In one individual V. citrinus female, the W chromosome and the enlarged microchromosome were identified as an enlarged pair of autosomal microchromosomes because all other females in that population had cryptic sex chromosomes that were indistinguishable from the Z and other microchromosomes44. However, following FISH experiments with our sex chromosome specific probes, we validated that what was initially considered to be an enlarged pair of microchromosomes, was in fact the unpaired enlarged microchromosome, and a heteromorphic W. This discovery reveals that V. citrinus has two W chromosomes in the population where some females have an enlarged W and other females have a homomorphic W where there is no size difference between the W and the Z. The probe also added clarity that the enlarged microchromosome does not occur in the homozygous form and is present only in some individuals in V. acanthurus and V. citrinus as floating heterokaryotypes. A similar observation was made in the spiny frog (Quasipaa boulengeri) where a number of reciprocal translocations were identified between chromosomes 1 and 6; the result was 5 different karyotypes with 9 possible karyotype combinations60. When two heterokaryotypes breed the expected outcome should produce three karyotypes, homozygous for one morphology, heterokaryotypic individuals, and homozygous for the other morphology. The missing karyotypes could be entirely missing from the population or at an extremely low frequency which could indicate underdominance and/or a lethal mutation could be associated with homozygous individuals with enlarged microchromosomes.
Homology of the enlarged microchromosomes, the W, and the terminal region of chromosome 1 is unlikely to be related to sex determination because it is present in both males and females in isolated populations. The most parsimonious explanation is the homologous sequences originated on chromosome 1 because there was homology to this chromosome in all individuals from four different varanus species. Varanid chromosomes are very similar to pythons, and chromosome 1 in snakes shares genes with the chicken Z23, suggesting that these sequences might be part of an ancestral sex chromosome.
Our FISH probes were designed by masking repeats and targeted single copy regions (including genes) of the sex-linked scaffolds and therefore the shared homology of the W and the enlarged microchromosome is likely a segmental duplication of the W or a portion of the W chromosome that has occurred as a de novo mutation in the common ancestor of V. acanthurus and V. citrinus (Fig. 3, Supplementary Tables 1 and 2). To detect segmental duplications, common repeats of the genome must be removed from the analysis16 which was a critical aspect of our probe design. Segmental duplications are long (> 1 kilo-base pairs) and similar (90–100%) sections of DNA sequences that are located in multiple locations in the genome and are hotbeds for the emergence of new genes and genomic rearrangements13,15,16,61. These genetic changes are major contributors to the divergence between species of primates and are important for understanding species evolution13,14,62,63,64. Segmental duplications were one of the last regions of the human genome to be sequenced due to their nearly identical sequences but are considered critically important for the expansion of the frontal cortex and increased synaptic density since the divergence from other primates14. In Lepidoptera species, segmental duplications often contain transposable elements and the genic content, and the duplicated sequences varied among species ranging from 1.2 to 15.2% of the genome65. Functional analysis indicated the gene families in Lepidoptera may be related to species-specific adaptation. It is unclear what phenotypic consequences are associated with these mutations in goannas, and future research should focus on this topic as they could be significant factors driving the extreme genetic divergence and speciation in the ridge-tailed goannas which was not explained with isolation by distance analysis44.
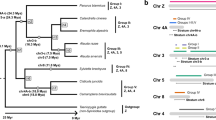
A schematic diagram showing the current hypothetical evolutionary phylogeny for Varanids with chromosome changes indicated, suggesting independent chromosome transition in parallel in several lineages. Arrows indicate nodes of chromosome transitions for both pericentric inversions of autosomes and gain or loss of W chromosome heteromorphisms. The enlarged microchromosome (EM) was likely acquired prior to divergence between V. citrinus and V. acanthurus and was subsequently lost in some populations of V. acanthurus or could be present at a very low frequency in those populations. The phylogenetic relationships of varanus species are based on66.
The probe we developed encompassed regions that align with coding genes, and the documented consequences linked to gene duplications and their implications in speciation are well established67,68. When gene copy number changes due to mutation there are two potential outcomes. In one situation, there could be a positive gene dosage effect where the expression of the gene is increased in response to the copy number69. In the second situation, there is an inverse effect in which gene expression is decreased with the increased dosage correlated to additional gene copies70,71. In the inverse situation, the expression effect can act across the genome and not just impact the chromosome that was modified72. For example, in yeast cells that were tested by the addition of yeast artificial chromosomes containing mostly intronless genes, the mere presence of additional DNA generated a positive dosage response correlated with the number of additional yeast genes, and the imbalance led to a stress response aimed at restoring the wildtype physiology69. In Drosophila the Male Specific Lethal (MSL) complex is assembled on the X chromosomes and can spread to neo-X chromosomes with transposable element activity causing upregulation and disrupting the dosage compensation system72,73,74. Since the duplicated region identified herein contains genes and is homologous with the W, a situation where an additional copy could be lethal or have severe deleterious consequences could explain why homozygous enlarged microchromosomes were not observed in any population, and a similar situation has been demonstrated in the spiny frog60,75. To test this hypothesis additional studies should include gene expression levels associated with the genes on the enlarged microchromosome.
The presence of the enlarged microchromosome in isolated populations of V. acanthurus and V. citrinus is puzzling. We do not have any evidence that it is related to sex chromosome evolution, however, it seems to exhibit the characteristics of a sex chromosome, as it only presents in the heteromorphic situation similar to the W chromosome. We have hypothesised a reconstruction of sex chromosome evolution in Varanidae that includes the appearance of the enlarged microchromosome (Fig. 4). Presumably, an ancient supersex chromosome was repeatedly recruited, or specific gene families that can be traced back to Ambystoma salamanders, have commonly been recruited into a sex related role in other non-avian reptiles, birds, and mammals23,53,76. Autosome-sex chromosome fusions have occurred in several lineages, and these patterns hint at a phylogenetic relationship between fusions of autosomes and sex chromosomes for resolving sexual conflict77. Although the Varanidae exhibits high conservation of sex chromosomes, the expansion of the W chromosome is dynamic and repeat expansions have occurred independently in divergent lineages (Fig. 3)46. We hypothesise that following departure from the ancestral super sex chromosome, the early sex chromosomes were cryptic as observed in V. citrinus, V. salvator, V. komodoensis and V. gouldii. In each of the major Australian lineages and also in the African lineage the W chromosome has undergone independent degradation and accumulated species-specific repeats41,46,78. The W chromosome is in a highly degraded state in V. acanthurus, but in a homomorphic state in V. citrinus which has made it difficult to characterise and study. The chromosome has been expanded with (ATT)N repeats46 and only contains 23 coding genes25.
Theoretical reconstruction of Varanidae sex chromosome evolution. Ancient amniote sex chromosome diverges where different orthologs diverged into sex roles in other lineages. The varanid Z and W chromosomes were likely a homomorphic pair, then independently in the Varius, Gouldii and Odatria lineages the W underwent species independent degradation and accumulation of repeats. Some species in each lineage also maintained the homomorphic pair (V. komodoensis, V. citrinus show no repeat accumulation and V. gouldii exhibit either the earliest stages of repeat accumulation). Other species such as V. acanthurus, V. varius and V. rosenbergi all exhibit heteromorphic W chromosomes, but V. rosenbergi and V. acanthurus do not share repeat content and V. varius still remains to be characterised.
Previous studies have demonstrated different microsatellite repeat accumulations in W chromosomes between the Gouldii (CGG)N and Odatria (ATT)N lineages46. Intriguingly, no significant repeat accumulations were identified in the W chromosomes of V. komodoensis, despite heteromorphisms between Z and W are present in the closest relative of V. komodoensis, V. varius52. However, no additional cytogenetics studies have focused on V. varius to identify the nature of the repeat accumulation in the W chromosome in this species45. Despite the differences in repeat landscapes of Varanid W chromosomes, which have been demonstrated in other species79,80 and not considered significant indicators of orthologs, the genes and single copy regions of these chromosomes are conserved and appear to be homologous in all species investigated in this study. Further work is needed to resolve the functional significance of the enlarged microchromosome and determine if it has a more central role in sex chromosome evolution in the Odatria lineage.
Conclusion
Ridge-tailed goannas are a lineage of dwarf goannas from the subgenus Odatria which has undergone diversification and rapid speciation over the last 5–10 million years. Our research demonstrates that large-scale chromosome rearrangements are actively maintained in this lineage both within and between populations. Segmental duplications of the sex chromosome sequences could be a factor in driving this diversification, but additional work should include V. primordius, V. insulanicus and V. storri as sex chromosomes have not been identified in these other species in the ridge-tailed goanna complex. The pattern here is, however, similar to what occurred in Lepidoptera and Primates over relatively the same amount of time and could add to the broader understanding of the sequential series of speciation events in the other eukaryote lineages. Rapid evolution is often associated with the evolution of sex chromosomes. However, here we show rapid evolution occurring on autosomes with some homology to the sex chromosomes, therefore it could be possible that in Varanidae, sex chromosomes are influencing rapid divergence on the autosomes, but the sex chromosomes remain conserved. In addition, we have identified size differences in W chromosomes across different populations that we have screened with our sex chromosome specific probes (Suppl Fig. 2). Such polymorphisms involving repetitive elements are not unusual and even identified in human Y chromosomes81. Future studies involving in-depth qualitative and quantitative analysis will shed further light, not only on the evolution of sex chromosomes, but also in speciation of varanids.
Materials and methods
Development of sex chromosome specific probes
To develop the sex-specific probes we used the two published Varanus genomes V. komodoensis (NCBI accession GCA_004798865.147), and V. acanthurus (NCBI BioProjectID PRJNA737594, which includes microdissected W chromosome reads from BioSamples: SAMN19718186 and SAMN19718187, Genome Warehouse BioProject accession PRJCA00558325). This study utilized the full pipeline established by Zhu et al.25 to infer both Z and W sex chromosome sequences. A full protocol is detailed in the latter study. The inferred Z-linked sequences were assembled to 134 scaffolds ranging from 1 kb to 3 Mb, and the inferred W-linked sequences were assembled to 441 scaffolds ranging from 5 kb to 230 Kb (Fig. 5, Supplementary Tables 1 and 2). To validate these scaffolds as sex-specific we provided the scaffolds to Arbor Biosciences (Ann Arbor, MI, United States) which used the published V. komodoensis genome47 as a reference to create a hybrid assembly for oligonucleotide probe design. For each probe, highly repetitive sequences were masked from the designs and the resulting myTags® libraries ranged from 45 to 47 nucleotides in length and targeted single copy regions along these scaffolds to avoid non-specific hybridisation. A detailed description and validation of the probe design protocol is described elsewhere82.
Methodology for the development of sex-specific Z and W inferred probes. Step 1. Generate single-tube long fragment reads and Illumina paired-end reads for female and male individuals, assemble and map reads. Step 2. Identify sex-linked reads from read depth coverage. Step 3. BLAST search sex-linked reads against high quality reference genome (V. komodoensis). Step 4. Generate hybrid assembly to identify genomewide repeats and mask repeats regions along sex-linked scaffolds. Step 5. Identify single copy sequences and design myTags® libraries. In step 2, the male to female (M/F) ratio of the Z-specific scaffolds in males is close to 2 whereas the W-specific scaffolds is close to 0. In females, the Z-specific and W-specific scaffolds are close to 1. Steps 4 and 5 were provided by Arbor Biosciences using proprietary methods (Ann Arbor, MI, United States). These probes targeted the single copy regions and functional genes and avoided repetitive elements and were used for FISH (Fluorescence in situ Hybridisation) (Fig modified after).
The probes were hybridised to 21 individuals from 4 species (V. acanthurus, V. citrinus, V. rosenbergi, V. gouldii) with 400 ng probe per slide in a mixture containing both probes (Table 1) with hybridisation buffer (BioCare Medical, Pacheco CA). The cell preparation and FISH protocol is detailed elsewhere44,82. Each slide was freshly prepared with 50 μL cell suspension, dehydrated with ethanol then aged at − 80 °C overnight. Slides were removed from the freezer and immediately immersed in 100% (v/v) ethanol and allowed to dry. 40 μL of buffer and probe mix was added, covered with a coverslip and sealed with rubber cement. The slides were denatured at 68 °C for five minutes and moved to preheated humidity chambers to incubate for 48 h at 37 °C. Following incubation, the coverslips were removed and the slides were washed with 0.4 × SSC: 3 M NaCl, 0.3 M sodium citrate, pH 7, 0.3% (v/v) IGEPAL Sigma-Aldrich at 60 °C for 5 min then a second wash at room temp with 2 × SSC: 3 M NaCl, 0.3% M sodium citrate, pH 7, 0.1% (v/v) IGEPAL Sigma-Aldrich for 3 min. The slides were then immersed in an ethanol series of 70%, 90% and 100% (v/v) for one minute in each. The slides were then stained with Vectashield® antifade mounting medium with DAPI (Vector Laboratories) and observed with a Leica Microsystems Thunder Imaging system. Images were constructed from photographs using Adobe Photoshop 2021.
The W chromosome is in a highly degraded state in all V. acanthurus females, and it is enlarged in V. rosenbergi and in 1 V. citrinus female, and they are cryptic in V. gouldii and 3 V. citrinus females (the enlarged microchromosome is represented by EM).
Data availability
Varanus acanthurus genome scaffolds and sequence data that support the findings of this study are already available via genome warehouse PRJCA005583 and the data is provided in supplementary material (Supplementary Tables 1 and 2).
References
Livingstone, K. & Rieseberg, L. Chromosomal evolution and speciation: A recombination-based approach. New Phytol. 161, 107–112 (2003).
Potter, S. et al. Chromosomal speciation in the genomics era: Disentangling phylogenetic evolution of rock-wallabies. Front. Genet. 8, 1–18 (2017).
Damas, J., Corbo, M. & Lewin, H. A. Vertebrate chromosome evolution. Annu. Rev. Anim. Biosci. 9, 1–27 (2021).
White, M. J. D. chromosomal rearrangements and speciation in animals. Annu. Rev. Genet. 3, 75–98 (1969).
King, M. Species Evolution: The Role of Chromosome Change (Cambridge University Press, New York, 1993). https://doi.org/10.2307/2413666.
Damas, J., O’Connor, R. E., Griffin, D. K. & Larkin, D. M. Avian chromosomal evolution. Avian Genomics Ecol. Evolut. https://doi.org/10.1007/978-3-030-16477-5_4 (2019).
King, M. Speciation and the theoretical approach. Heredity 59, 1–6 (1987).
Murphy, W. J. et al. Evolution: Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 1979(309), 613–617 (2005).
Deakin, J. E. & Ezaz, T. Tracing the evolution of amniote chromosomes. Chromosoma 123, 201–216 (2014).
Kim, J. et al. Reconstruction and evolutionary history of eutherian chromosomes. Proc. Natl. Acad. Sci. U. S. A. 114, E5379–E5388 (2017).
Samonte, R. V. & Eichler, E. E. Evolution of the primate genome. Nat. Rev. Genet. 3, 1–8 (2002).
Locke, D. P. et al. Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol. 4, R50 (2003).
Cantsilieris, S. et al. An evolutionary driver of interspersed segmental duplications in primates. Genome Biol. 21, 1–35 (2020).
Vollger, M. R. et al. Segmental duplications and their variation in a complete human genome. Science (1979) 376 (2022).
Rieseberg, L. H. & Livingstone, K. Evolution: Chromosomal speciation in primates. Science 1979(300), 267–268 (2003).
Pu, L., Lin, Y. & Pevzner, P. A. Detection and analysis of ancient segmental duplications in mammalian genomes Department of Computer Science and Technology, Shandong University Department of Computer Science and Engineering, University of California at San Diego Research School of Com. Genome Res. 28, 901–909 (2018).
Pearse, D. E., Miller, M. R., Abadía-Cardoso, A. & Garza, J. C. Rapid parallel evolution of standing variation in a single, complex, genomic region is associated with life history in steelhead/rainbow trout. Proc. R. Soc. B Biol. Sci 281 (2014).
Iannucci, A. et al. Bridging the gap between vertebrate cytogenetics and genomics with single-chromosome sequencing (Chromseq). Genes 12, 1–13 (2021).
Deakin, J. E. et al. Chromosomics: Bridging the gap between genomes and chromosomes. Genes https://doi.org/10.3390/genes10080627 (2019).
Claussen, U. Chromosomics. Cytogenet. Genome Res. 111, 101–106 (2005).
Liehr, T. Molecular cytogenetics in the era of chromosomics and cytogenomic approaches. Front. Genet. 12, 1–9 (2021).
Matsubara, K. et al. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma 125, 111–123 (2016).
Ezaz, T., Srikulnath, K. & Graves, J. A. M. Origin of amniote sex chromosomes: An ancestral super-sex chromosome, or common requirements?. J. Hered. 108, 94–105 (2017).
Filatov, D. A. The two “rules of speciation” in species with young sex chromosomes. Mol. Ecol. 27, 3799–3810 (2018).
Zhu, Z. X. et al. Diversity of reptile sex chromosome evolution revealed by cytogenetic and linked-read sequencing. Zool. Res. 43, 719–733 (2022).
Furman, B. L. S. et al. Sex chromosome evolution: So many exceptions to the rules. Genome Biol. Evol. 12, 750–763 (2020).
Bachtrog, D. et al. Sex determination: Why so many ways of doing it?. PLoS Biol. 12, 1–13 (2014).
Bachtrog, D. A dynamic view of sex chromosome evolution. Curr. Opin. Genet. Dev. 16, 578–585 (2006).
Ezaz, T., Stiglec, R., Veyrunes, F. & Marshall Graves, J. A. Relationships between vertebrate ZW and XY sex chromosome systems. Curr. Biol. 16, 736–745 (2006).
Van Doorn, G. S. & Kirkpatrick, M. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 186, 629–645 (2010).
Alam, S. M. I., Sarre, S. D., Gleeson, D., Georges, A. & Ezaz, T. Did lizards follow unique pathways in sex chromosome evolution?. Genes 9, 20–28 (2018).
Ezaz, T. et al. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. https://doi.org/10.1007/s10577-005-1010-9 (2005).
Quinn, A. E. et al. Temperature sex reversal implies sex gene dosage in a reptile. Science (1979) 316, 411 (2007).
Holleley, C. E. et al. Sex reversal triggers the rapid transition from genetic to temperature-dependent sex. Nature 523, 79–82 (2015).
Meisel, R. P. et al. Sex chromosome evolution in muscid flies. G3 10, 1341–1352 (2020).
Charlesworth, D., Charlesworth, B. & Marais, G. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95, 118–128 (2005).
Bull, J. J. Evolution of Sex Determining Mechanisms (The Benjamin/Cummings Publishing Company, Inc, 1983).
Palmer, D. H., Rogers, T. F., Dean, R. & Wright, A. E. How to identify sex chromosomes and their turnover. Mol. Ecol. 28, 4709–4724 (2019).
Chalopin, D., Volff, J. N., Galiana, D., Anderson, J. L. & Schartl, M. Transposable elements and early evolution of sex chromosomes in fish. Chromosome Res. 23, 545–560 (2015).
Stöck, M. et al. Ever-young sex chromosomes in European tree frogs. PLoS Biol. 9 (2011).
Iannucci, A. et al. Conserved sex chromosomes and karyotype evolution in monitor lizards (Varanidae). Heredity 123, 215–227 (2019).
Rovatsos, M., Rehák, I., Velenský, P. & Kratochvíl, L. Shared ancient sex chromosomes in varanids, beaded lizards and alligator lizards. Mol. Biol. Evol. 36, 1113–1120 (2019).
Pavón-Vázquez, C. J. et al. Between a rock and a dry place: Phylogenomics, biogeography, and systematics of ridge-tailed monitors (Squamata: Varanidae: Varanus acanthurus complex). Mol. Phylogenet. Evol. 168, 107516 (2022).
Dobry, J. et al. Widespread chromosomal rearrangements preceded genetic divergence in a monitor lizard, Varanus acanthurus (Varanidae). Chromosome Res. 31, 123–135 (2023).
Johnson Pokorná, M. et al. First description of the karyotype and sex chromosomes in the komodo dragon (Varanus komodoensis). Cytogenet. Genome Res. 148, 284–291 (2016).
Matsubara, K. et al. Highly differentiated ZW sex microchromosomes in the Australian varanus species evolved through rapid amplification of repetitive sequences. PLoS One 9 (2014).
Lind, A. et al. A high-resolution, chromosome-assigned Komodo dragon genome reveals adaptations in the cardiovascular, muscular, and chemosensory systems of monitor lizards. Nat. Ecol. Evol. 3, 1241–1252 (2019).
Wang, O. et al. Efficient and unique cobarcoding of second-generation sequencing reads from long DNA molecules enabling cost-effective and accurate sequencing, haplotyping, and de novo assembly. Genome Res. 29, 798–808 (2019).
King, M., Mengden, G. A. & King, D. A pericentric-inversion polymorphism and s ZZ/ZW sex-chromosome system in Varanus acanthurus Boulenger analyzed by G- and C-banding and Ag staining. Genetica 58, 39–45 (1982).
Sites, J. W. J. & Reed, K. M. Chromosomal evolution, speciation, and systematics: Some relevant issues. Herpetologica 50, 237–249 (1994).
Dobry, J. Chromosomal Evolution in the Ridge-Tailed Goannas (2023).
King, M. & King, D. Chromosomal evolution in the lizard genus varanus (reptilia). Aust. J. Biol. Sci. https://doi.org/10.1071/BI9750089 (1975).
Singchat, W. et al. Do sex chromosomes of snakes, monitor lizards, and iguanian lizards result from multiple fission of an “ancestral amniote super-sex chromosome”?. Chromosome Res. 28, 209–228 (2020).
Halverson, J. & Spelman, L. Sex determination and its role in management. In Komodo Dragons: Biology and Conservation (eds Murphy, J. B. et al.) 165–177 (Smithsonian Institution Press, 2002).
Böhme, W. Hemiclitoris discovered: A fully differentiated erectile structure in female monitor lizards (Varanus spp.) (Reptilia: Varanidae). J. Zool. Syst. Evolut. Res. https://doi.org/10.1111/j.1439-0469.1995.tb00220.x (1995).
Morris, P. J., Jackintell, L. A. & Alberts, A. C. Predicting the gender of subadult komodo dragons (Varanus komodoensis) Us i Ng Two-D i Mens i Onal ultrasound imaging and plasma testosterone concentration. Zoo Biol. 15 (1996).
Brown, D. Hemipenal transillumination as a sexing technique in varanids. Biawak 3, 26–29 (2009).
Sulandari, S., Zein, M. S. A., Arida, E. A. & Hamidy, A. Molecular sex determination of captive komodo dragons (Varanus komodoensis) at Gembira Loka Zoo, Surabaya Zoo, and Ragunan Zoo, Indonesia. Hayati 21, 65–75 (2014).
Rovatsos, M. et al. Sexing of Komodo dragons, Varanus komodoensis. Gazella 42, 93–107 (2015).
Qing, L., Xia, Y., Zheng, Y. & Zeng, X. A de novo case of floating chromosomal polymorphisms by translocation in Quasipaa boulengeri (Anura, Dicroglossidae). PLoS One 7 (2012).
Larkin, D. M. et al. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 19, 770–777 (2009).
Bailey, J. A. et al. Recent segmental duplications in the human genome. Science 1979(297), 1003–1007 (2002).
Emanuel, B. S. & Shaikh, T. H. Segmental duplications: An ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet. 2, 791–800 (2001).
Samonte, R. V. & Eichler, E. E. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3, 65–72 (2002).
Zhao, Q., Ma, D., Vasseur, L. & You, M. Segmental duplications: Evolution and impact among the current Lepidoptera genomes. BMC Evol. Biol. 17, 1–11 (2017).
Brennan, I. G. et al. Phylogenomics of monitor lizards and the role of competition in dictating body size disparity. Syst. Biol. 70, 120–132 (2021).
Lynch, M. & Conery, J. S. The evolutionary fate and consequences of duplicate genes. Science 1979(290), 1151–1155 (2000).
Kuzmin, E., Taylor, J. S. & Boone, C. Retention of duplicated genes in evolution. Trends Genet. 38, 59–72 (2022).
Torres, E. M. et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 1979(317), 916–924 (2007).
Zhang, A. et al. Global analysis of gene expression in response to whole-chromosome aneuploidy in hexaploid wheat. Plant Physiol. 175, 828–847 (2017).
Sun, L. et al. Dosage compensation and inverse effects in triple X metafemales of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 110, 7383–7388 (2013).
Zhang, S. et al. Interaction of male specific lethal complex and genomic imbalance on global gene expression in Drosophila. Sci. Rep. 11, 1–18 (2021).
Muyle, A., Bachtrog, D., Marais, G. A. B. & Turner, J. M. A. Epigenetics drive the evolution of sex chromosomes in animals and plants. Philos. Trans. R. Soc. B Biol. Sci. 376 (2021).
Lott, S. E., Villalta, J. E., Zhou, Q., Bachtrog, D. & Eisen, M. B. Sex-specific embryonic gene expression in species with newly evolved sex chromosomes. PLoS Genet. 10 (2014).
Yuan, X., Xia, Y. & Zeng, X. Suppressed recombination of sex chromosomes is not caused by chromosomal reciprocal translocation in spiny frog (Quasipaa boulengeri). Front. Genet. 9, 1–14 (2018).
Smith, J. J. & Voss, S. R. Bird and mammal sex-chromosome orthologs map to the same autosomal region in a salamander (Ambystoma). Genetics 177, 607–613 (2007).
Sigeman, H. et al. Repeated sex chromosome evolution in vertebrates supported by expanded avian sex chromosomes. Proc. R. Soc. B Biol. Sci. 286 (2019).
Srikulnath, K., Uno, Y., Nishida, C. & Matsuda, Y. Karyotype evolution in monitor lizards: Cross-species chromosome mapping of cDNA reveals highly conserved synteny and gene order in the Toxicofera clade. Chromosome Res. 21, 805–819 (2013).
Suwala, G. et al. Evolutionary variability of w-linked repetitive content in lacertid lizards. Genes 11 (2020).
Rovatsos, M., Farkačová, K., Altmanová, M., Johnson Pokorná, M. & Kratochvíl, L. The rise and fall of differentiated sex chromosomes in geckos. Mol. Ecol. 28, 3042–3052 (2019).
Harris, P., Boyd, E., Young, B. & Ferguson-Smith, M. Determination of the DNA content of human chromosomes by flow cytometry. Cytogenet. Cell. Genet. 41, 14–21 (1986).
Dobry, J. et al. Fixed allele differences associated with the centromere reveal chromosome morphology and rearrangements in a reptile (Varanus acanthurus BOULENGER). Mol. Biol. Evol. 40 (2023).
Acknowledgements
This work was funded by the Australian Government Research Training Program (RTP) stipend scholarship awarded to Jason Dobry. We would like to thank Frank Retes, Gavin Bedford, Max King, Brian Green and Fiorenzo Guarino for their insights for fieldwork; Tristan Ford, Sara, Adelynn and Jarrett Dobry for their help with field collections; Lee Donald and Graeme Adlam at Ross River Resort, Wally Klein, Joella Klein and Billy Nelson at Orange Creek Station, and Peter “Spud” Murphy at Stewarts Well for permission to collect specimens on their properties. We also thank Arthur Georges, Emily Stringer, Bernd Gruber, Luis Mijangos Araujo, for helpful comments. Finally, we would like to thank Steve Sarre and Jacqui Richardson for their support with the captive colony.
Funding
This work was funded by the Australian Government Research Training Program (RTP) stipend scholarship awarded to Jason Dobry.
Author information
Authors and Affiliations
Contributions
J.D., T.E., E.W., and J.E.D. conceptualised the study. J.D. conducted fieldwork, laboratory work, data analysis and wrote the first draft of the manuscript. Z.Z and Q.Z provided sequence data and analysis support. All authors provided insight on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This work was approved by the University of Canberra Animal Ethics Committee under project number 20180306 as required by the ACT Animal Welfare Act 1992 for the authorization of research using animals to conduct experiments. The animals in this project were collected under Northern Territory Parks and Wildlife Commission permit number 63414, and Queensland Government Department of Environment and Science Permit number WA0010049, and they were imported into the ACT under the License number LT201829. All methods reported in this study were conducted in accordance with the relevant guidelines and regulations. The study is reported in accordance with ARRIVE guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dobry, J., Zhu, Z., Zhou, Q. et al. The role of unbalanced segmental duplication in sex chromosome evolution in Australian ridge-tailed goannas. Sci Rep 15, 8545 (2025). https://doi.org/10.1038/s41598-025-93574-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-93574-5