Abstract
Applying deep learning algorithms to mine ultrasound features of breast cancer and construct a machine learning model that accurately predicts Ki-67 expression level. This multi-center retrospective study analyzed clinical and ultrasound data from 929 breast cancer patients. We integrated deep features from the tumor and peritumoral areas to build a fusion model for predicting Ki-67 expression. The model underwent performance validation on both internal and external test datasets. Its accuracy as well as clinical usefulness were evaluated by diverse statistical metrics. In the ultrasound depth feature model for the tumor area, the Support Vector Machine (SVM) algorithm achieved the highest performance, with an accuracy of 0.782, ROAUC of 0.771 (95% CI 0.704–0.838), sensitivity of 0.905, specificity of 0.543, and F1 score of 0.846. In the depth feature model for the peritumoral area, the Light Gradient Boosting Machine (LightGBM) algorithm demonstrated superior performance, achieving an accuracy of 0.728, ROAUC of 0.623 (95% CI 0.545–0.702), sensitivity of 0.892, specificity of 0.407, and F1 score of 0.813. The SVM algorithm exhibited superior performance in both internal and external test sets when validated the fusion model integrating depth features from tumor and peritumoral area. Internal test set validation in clinical application indicated significantly lower disease-free survival in the high Ki-67 expression group compared to the low expression group (P = 0.005). Through comprehensive analysis of breast cancer ultrasound images and the application of machine learning techniques, we developed a highly accurate model for predicting Ki-67 expression levels.
Similar content being viewed by others
Introduction
Breast cancer is the most prevalent malignant tumor among women, with increasing global incidence and mortality rates1,2. In 2020, there were 2.26 million new cases of breast cancer globally3. Despite advancements in breast cancer treatment over the past 30 years, it continues to be a major cause of mortality among women worldwide4. Enhancing the accuracy of early breast cancer diagnosis is a pressing challenge in contemporary medical research.
Accurately assessing tumor cell proliferation is essential in breast cancer research and treatment. When highly expressed, Ki-67, a cell proliferation marker, indicates faster cell division and worse prognosis5. Research indicates a significant elevation of Ki-67 expression in breast cancer tissues, which correlates with clinical features6,7. However, current Ki-67 detection mainly relies on invasive tissue biopsy. It imposes additional burdens on patients and delays treatment decisions. Thus, finding an efficient, fast, non-invasive Ki-67 detection method is crucial. Ultrasound is an important non-invasive diagnostic tool for breast cancer8. Cui, et al.'s study suggests that ultrasound features, such as breast cancer lesions’ shape, margin, and internal echogenicity, significantly correlate with the Ki-67 expression levels9. Huang’s research indicates that ultrasound image features can effectively predict Ki-67 expression10. The correlation between ultrasound and Ki-67 expression levels indicates that clinicians can use ultrasound to predict breast cancer malignancy and prognosis. However, the conventional ultrasound features used to predict Ki-67 expression exhibit certain limitations. For instance, Cheng et al. investigated the feasibility of predicting Ki-67 expression in breast cancer using ultrasonography and clinical features, but their approach faced issues like incomplete feature extraction and subjective variable selection due to the direct use of radiological parameters11. Consequently, there remains a significant disparity between this approach and optimal diagnostic strategies.
As an essential technical approach in artificial intelligence, deep learning can automatically learn complex features from data by constructing neural network models. It has shown outstanding performance and broad application potential in many tasks12. Deep learning combined with ultrasonic technology exhibits promising clinical applications in cancer diagnosis. Xiang et al. developed the OvcaFinder model to differentiate benign and malignant ovarian tumors13. Xie et al. utilized ultrasonic radiofrequency signals for rapid intraoperative molecular diagnosis of glioma biomarkers14. Li et al. predicted cervical lymph node metastasis risk in papillary thyroid cancer patients using deep learning and ultrasound images15. Roongruedee Chaiteerakij et al. enhanced liver cancer screening efficiency with an AI model based on YOLOv5 and ultrasound results16. Significant advancements have also been made in breast cancer research using deep learning.
Deep learning can effectively mitigate the subjective bias introduced by manual feature extraction, providing powerful technical support for medical diagnosis, accuracy of Dynamic Contrast-Enhanced Magnetic Resonance Imaging (DCE-MRI) diagnosis in breast cancer17. The successful development of an AI system assisting in breast cancer detection has notably improved the efficiency and accuracy of early screening18. A research team accurately predicted the expression states of molecular markers, including Estrogen Receptor (ER), Progesterone Receptor (PR), and Human Epidermal Growth Factor Receptor 2 (Her-2), using pathological slide images of breast cancer patients. There are pivotal for assessing the hormone sensitivity of breast cancer and devising personalized treatment plans19. In predicting Ki-67, deep learning has also exhibited significant advantages. Researchers used deep learning algorithms and find that quantitative imaging features from DCE-MRI of breast tumors correlated with Ki-67 expression in breast cancer20. We aim to explore deep learning technology further to ascertain whether it is possible to construct a predictive model based on ultrasound imaging data. Our goal is to predict pre-surgical Ki-67 levels in breast cancer patients with a non-invasive and efficient manner.
In summary, we aim to extract deep features from breast cancer ultrasound images to build a machine-learning model that predicts Ki-67 expression. The study conducted a retrospective analysis. From multiple centers, we recruited breast cancer patients meeting the inclusion criteria between 2019 and 2023 and collected their clinical and ultrasound data. After preprocessing, the data was randomly split into a training set, an internal and external testing set. We standardized the data and reduced the noise, then proceeded to precisely delineated the ultrasound images. Our study employed the pre-trained Densenet169 model various machine-learning algorithms to create predictive models. We combined features and used LASSO feature selection to build a fusion predictive model. It was validated on internal and external test sets. We anticipate that predicting Ki-67 expression will provide a more accurate, convenient, and non-invasive tool for diagnosing and treating breast cancer, ultimately improving patients’ quality of life and prognosis.
Materials and methods
Selection of research subjects and data preprocessing techniques
A retrospective study of 929 breast cancer patients (2019–2023) from two centers investigated outcomes post-radical mastectomy and lymph node dissection. Comprehensive clinical data collection (age, body mass index (BMI), TNM staging, disease-free survival (DFS), etc.) and biomarker reassessment (epidermal growth factor receptor (EGFR), ER, Her-2, PR, Ki67) facilitated model development. Data were randomly split into training (N = 557), internal testing (N = 239), and external test sets (N = 133). Baseline consistency among cohorts was verified using t-tests, underpinning subsequent analyses. This retrospective study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Guangxi Medical University and the need for informed consent was waived due to the retrospective nature of the study and the use of anonymized data. This study adhered to scientific ethical standards and was approved by the Medical Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (Approval number 2024-S822-01). All methods were performed in accordance with the relevant guidelines and regulations.
Detection using IHC technique
The expression level of Ki-67 was assessed using IHC techniques, following these specific steps: Tumor tissue specimens were fixed in 10% neutral formalin, subsequently embedded in paraffin, and sectioned into 4 μm-thick slices. These slices underwent heat-induced antigen retrieval and were then incubated with an anti-Ki-67 polyclonal antibody (Proteintech, 27309-1-AP) for 60 min. After DAB staining, the nuclei were counterstained with hematoxylin, and the slides were mounted. The expression level of Ki-67 was calculated as the percentage of nuclear staining-positive cells. Ten high-power fields (400 ×) were randomly selected, with at least 100 cells counted in each field, to determine the percentage of positive cells. An expression level of Ki-67 greater than 20% was defined as high expression, while a level of 20% or less was defined as low expression.
Ultrasonic image data acquisition and depth feature extraction
Among all the patients, we collected one cross-sectional ultrasound image of each breast tumor. All images were sourced from our Picture Archiving and Communication System and patients’ breast Static ultrasound images (JPG format) were obtained from ultrasound equipment by manufacturers including Philips and Toshiba. We removed annotations and markings from the image by minimally cropping the edges for anonymization. Then, to eliminate variations introduced by different physicians and equipment, we utilized interpolation to standardize all ultrasound images to a resolution of 960 × 720. This standardization process ensured that all images possessed a uniform resolution for subsequent analysis, thereby providing a more consistent data foundation for model training and validation. Furthermore, to improve image quality and enhance the accuracy of subsequent analysis, we processed the images using an average filter to reduce noise and artifacts within them. Experienced ultrasound physicians manually delineated the regions of interest (ROI) using ITK-SNAP (version 4.0.0, open-source software http://itksnap.org/). We implemented a comprehensive quality control protocol, which included independent sketching by multiple experts and comparison of their results, to ensure the accuracy and reliability of the sketch data.
Feature extraction from the peritumoral area (PTA)
Imaging features of the PTA, including texture, morphology, and functional information, have been proven to significantly improve the diagnostic accuracy and prognostic prediction capabilities for breast cancer21. Based on the biological characteristics of the tumor microenvironment, and considering the balance between imaging resolution and computational efficiency22, studies typically define the PTA by expanding the tumor ROI outward by 10 voxels (approximately 5–10 mm)23. Although this range selection has limitations, such as the neglect of biological heterogeneity, it can more effectively capture imaging features of the PTA compared to other voxel expansions, while avoiding the introduction of excessive background noise due to an overly large expansion. Therefore, in this study, we used Python programming to expand each patient’s tumor ROI outward by 10 voxels to form the PTA of breast tumors. Subsequently, we conducted precise segmentation of the ROI for both the tumor area (TA) and the PTA.
Transfer learning
DenseNet169, as a sophisticated deep convolutional neural network, exhibits exceptional capabilities in accurately localizing and identifying complex lesions with intricate morphological characteristics24. Gao et al. successfully utilized DenseNet169 to extract deep features from ultrasound images, achieving precise diagnosis of ovarian cancer22. Furthermore, Zhou et al.'s research also demonstrated that DenseNet169 is effective in extracting features of tumors and their surrounding areas in breast cancer imaging analysis21. Based on the successful experience of these studies, this study utilized Densenet169 as the feature extraction framework, enhancing its performance via transfer learning implemented in Python 3.7 (Anaconda). Deep learning models were developed using TensorFlow 2.0 and Keras 2.2.4, with ReLU activation, binary cross-entropy loss, Adam optimizer, and sigmoid classification. Model parameters were set at a learning rate of 0.001, Alpha 0.500, and 100 epochs. Breast TA and PTA ultrasound images were split 2:1 into training and test sets. Comprehensive training and parameter optimization were performed, tracking loss and accuracy per epoch. The CNN model with optimal accuracy and minimal loss was selected for deep feature extraction.
Extraction and selection of deep features
In this study, we extract the deep features of ROI through a pre-trained DenseNet169 model. Then, the penultimate layer (avgpool layer) features are extracted as the basis for subsequent machine learning model development. Due to the high-dimensional nature of deep feature data, we utilized Principal Component Analysis (PCA) for initial dimensionality reduction and converted the original dateset into independent and linearly uncorrelated representations via a linear transformation (Supplementary File 1). Post-PCA application, each deep feature matrix is reduced to 256 dimensions. Following feature extraction, we used Spearman’s rank correlation coefficient to exclude features with correlation coefficients greater than 0.9. The LASSO was then applied to further reduce the number of features. We selected those deep features most correlated with Ki-67 expression.
Developing machine learning models and multi-dimensional validation
Our study employed 6 machine learning algorithms to develop multiple predictive models based on the ultrasound depth features extracted from the TA and PTA. Firstly, these models were validated for their performance in an internal test set. Additionally, we investigated and executed feature fusion strategies by integrating the depth features of the tumor with its surrounding area, thereby acquiring a more comprehensive set of features. To improve the model’s predictive accuracy, we used the LASSO method for feature selection and identified key features that significantly influenced Ki-67 expression prediction. The model’s performance was rigorously validated on both internal and external test datasets.
Statistical analysis and performance assessment
This study leveraged Python 3.7.2 for rigorous data analysis and model validation. We evaluated model performance using metrics such as accuracy, AUC, sensitivity, specificity, and F1 score. Clinical decision curve analysis determined net benefit across Ki-67 expression probabilities. Results follow normal distribution (mean ± standard deviation). Independent t-test or Wilcoxon test for continuous variables and Pearson’s chi-square for categorical variables were employed. All analyses were two-sided, with P < 0.050 indicating significance.
Survival analysis methodology
Disease-free survival (DFS) is defined as the time interval from the date of surgery to the date of first recurrence or death from any cause. All patients underwent regular follow-up at intervals of every 3 months for a duration of 5 years. Survival curves were plotted using the Kaplan–Meier method to compare DFS between groups with different Ki-67 expression levels (high vs. low). Patients were stratified into high-risk (high Ki-67 expression) and low-risk (low Ki-67 expression) groups based on Ki-67 expression levels to assess its impact on patient prognosis. Patients in the high-risk group were expected to have shorter DFS, whereas those in the low-risk group were expected to have longer DFS.
Results
Clinical pathological results
A total of 929 breast cancer patients, with an average age of 53.0 ± 10.2 years, were recruited from two research centers. All patients underwent radical mastectomy with concurrent central and axillary lymph node dissection on the same side. We obtained all patients’ pathological results postoperatively. Among the cases analyzed, Ki-67 exhibited low expression in 267 instances and high expression in 662. A univariate analysis was conducted to evaluate the clinical-pathological data. The patient’s age, height, weight, BMI, and TMN stage showed no significant association with the level of Ki-67 expression (P > 0.050) (Table 1). In our center, 796 cases were identified and randomized into a training set (N = 557) and an internal testing set (N = 239) in a 7:3 ratio. Samples from additional centers (N = 133) served as an external test set, and the clinical baseline data were compared across the training, internal test, and external test sets. No significant differences were observed among the three groups (P > 0.050) (Table 2).
Performance of ultrasonic depth feature models for tumor and peritumoral area
We extracted 81,536 ultrasonographic features from the TA and the PTA using the pre-trained Densenet169 model. These features were then compressed into 256 deep features through PCA. After feature dimension reduction, we constructed deep feature models for both tumor and peritumoral ultrasonographic images using 6 machine learning algorithms. The efficacy of these models was validated using an internal test set. As presented in Table 3, the SVM algorithm demonstrated the best performance in the tumor deep feature model, with accuracy, ROAUC, sensitivity, specificity, and F1 values of 0.782, 0.771 (95% CI 0.704–0.838), 0.905, 0.543, and 0.846, respectively. In contrast, the LightGBM algorithm exhibited optimal performance in the peritumoral deep feature model, with accuracy, ROAUC, sensitivity, specificity, and F1 values of 0.728, 0.623 (95% CI 0.545–0.702), 0.892, 0.407, and 0.813, respectively.
Evaluation of a deep feature fusion model for ultrasound imaging of tumor peripheries
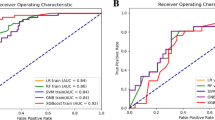
By integrating the tumor and peritumoral deep features using a pre-fusion approach, we used 512 deep features to construct a machine-learning model. Utilizing LASSO feature selection, we have identified 22 deep features significantly associated with Ki-67 expression, including 8 depth features specific to the TA and 14 depth features about the PTA (Fig. 1A–C). Similarly, 6 machine learning algorithms were employed to construct the fusion model (Supplementary File 2). The ROAUC is shown in Fig. 1D in the training set, with Logistic Regression and SVM performing the best. The models were validated with internal and external test sets (Table 4, Supplementary File 3). The SVM algorithm demonstrated the optimal performance in the internal test data, with an accuracy of 0.778, ROAUC of 0.811 (95% CI 0.752–0.870), sensitivity of 0.823, specificity of 0.691, and F1 score of 0.831 (Fig. 2A,B). The prediction scores obtained for each internal test data sample through the SVM algorithm model demonstrate a significant distinction in Ki-67 expression (Fig. 2C). The clinical decision curve showed that significant net benefit could be obtained using the model with a Ki-67 high expression probability ranging from 0.200 to 0.800 (Fig. 2D).
Selection of ultrasound depth features and performance of the model training set. (A) Through LASSO regression analysis, 22 deep features significantly correlated with Ki-67 expression were selected, with 8 originating from TA and 14 from PTA. (B) The optimal λ value in LASSO regression was determined by minimizing cross-validation error. (C) The weight distribution of the 22 selected deep features in TA and PTA is illustrated, with PTA features having slightly higher weights than those of TA. (D) A comparison of the ROAUC performance of six machine learning algorithms in the training set revealed that LR and SVM algorithms exhibited the best performance.
Performance of the fusion model in the internal test set. (A) When comparing the ROAUC performance of six machine learning algorithms within the internal test set, the SVM algorithm significantly outperformed the others. (B) The confusion matrix of the SVM model demonstrates the classification results within the internal test set, with a sensitivity of 0.823 and a specificity of 0.691. (C) There is a significant difference in the predictive probability distribution between high and low Ki-67 expression samples using the SVM model. (D) The clinical decision curve suggests that when the probability of predicting high Ki-67 expression by the SVM model falls between 0.200 and 0.800, the model curve lies above the reference lines for “Treat all” and "Treat none," indicating that within this range, the net benefit of the model is higher than these two simple strategies.
Similarly, the validation of the fusion model using an external test set yielded consistent results, with SVM accuracy, ROAUC, sensitivity, specificity, and F1 values of 0.827, 0.817 (95% CI 0.720–0.915), 0.884, 0.684, and 0.880, respectively (Fig. 3A–C). The model demonstrates significant net benefits for predicting elevated Ki-67 expression within the 0.100–0.900 range (Fig. 3D). These findings confirmed that the SVM machine learning algorithm performs best in a fusion model of breast cancer tumors and peritumoral depth features.
Performance of the fusion model in the external test set. (A) A comparison of six machine learning algorithms based on their ROAUC performance in an external test set revealed that the SVM algorithm performed best. (B) The confusion matrix of the SVM model demonstrates the classification results in the external test set, with a sensitivity of 0.884 and a specificity of 0.684. (C) The SVM model exhibits significant differences in predictive probability distributions between samples with high and low Ki-67 expression. (D) The clinical decision curve suggests that when the probability predicted by the SVM model for high Ki-67 expression falls between 0.100 and 0.900, the model curve lies above the reference lines for “Treat all” and "Treat none," indicating that the net benefit of the model within this range is higher than those of these two simple strategies.
The practical significance of the model
We evaluated the clinical applicability of the model by calculating the predicted probability of high Ki-67 expression for each sample and assessing the patients’ DFS status. Patients were categorized into high-scoring and low-scoring groups based on mean predicted probability values. The internal test set showed a significantly lower DFS in the high-scoring group compared to the low-scoring group (P = 0.005) (Fig. 4A). In contrast, no significant difference in DFS was observed between these groups in the external test set (P = 0.058) (Fig. 4B).
Survival analysis of DFS in internal and external test sets. (A) The KM survival curve for the internal test set suggests that the DFS in the high-score group is significantly lower than that in the low-score group (P = 0.005). (B) The KM survival curve for the external test set shows no significant difference in DFS between the two groups (P = 0.058).
Discussion
In the in-depth exploration of breast cancer, accurately assessing the proliferation state of tumor cells is crucial25. Ki-67, a key biomarker for evaluating breast cancer cell proliferation, has been increasingly recognized for its precise predictive value26. Our study conducted a multi-center retrospective analysis of clinical and ultrasound data from 929 breast cancer patients, extracted intra-tumoral and peritumoral features, and successfully developed a predictive model by deep learning technology. This model demonstrated exceptional accuracy and reliability in predicting Ki-67 expression levels. Furthermore, it achieved highly accurate predictions for patients’ DFS, providing clinicians with precise tools for personalizing treatment and highlighting the model’s potential value in prognosis assessment.
Ki-67 serves as a proliferation marker in breast cancer, closely associated with tumor progression and prognosis, with its expression modulated by various factors. Li et al. confirm that the high expression of EGFR and Her-2 is significantly associated with elevated Ki-67 proliferation27. Mayara Bocchi et al. found that Ki-67 expression correlates with ER and PR levels28. Our study found that Ki-67 expression is not associated with patient age, height, weight, BMI, or TMN stage; however, it correlates with key biomarkers’ expression such as EGFR, ER, Her-2, and PR. Some of our findings are consistent with prior studies, broadly supporting the potential role of these molecules in the development of breast cancer. However, Nishit et al. observed that Ki-67 expression was related to patient age, tumor size, and lymph nodes29. Zhu et al. found that Ki-67 expression was associated with BMI and tumor stage30. The differences may stem from varying criteria for evaluating Ki-67 positivity, so future research must further standardize the Ki-67 cutoff value. Besides, it is essential to investigate the specific effects of varying Ki-67 expression levels on tumor classification, prognostic evaluation, and treatment response. These advancements will enhance our understanding of breast cancer’s biological characteristics and offer clinicians more personalized treatment recommendations. Then, we performed rigorous data segmentation and validation. Comparative analysis revealed no significant differences in clinical baseline data among the training, internal validation, and external test sets. This confirms the homogeneity and representativeness of the study sample, ensuring the validity and reliability of the results.
Machine learning algorithms show great potential in enhancing medical diagnosis and treatment performance. For instance, machine learning is widely used in skin cancer screening31. Wei et al. employed machine learning algorithms to accurately classify colorectal cancer32, and Chen et al. improved their prediction of prostate cancer utilizing a machine learning-based model33. Meanwhile, using deep learning technology to extract depth features from ultrasound images and build models has become a critical factor in improving the accuracy of tumor diagnosis. Gao et al. utilized deep learning to develop a model capable of automatically evaluating ultrasound images, achieving greater accuracy in ovarian cancer detection than existing methods22. Xu et al. constructed a radiomics model to predict the expression levels of breast cancer molecular biomarkers, including Ki-67, by selecting and analyzing the largest ultrasound slice from patient lesions34. However, these studies primarily focused on the characteristics of the tumor itself, without adequately incorporating the features of the peritumoral tissue. In fact, cells in the peritumoral region can significantly influence tumor cell proliferation, invasion, and metastasis. Neglecting peritumoral features may impact the accurate assessment of tumor biological behavior and treatment response. To address this gap, the present study not only conducted an in-depth analysis of tumor imaging characteristics but also carefully examined peritumoral features, providing a more reliable basis for clinical decision-making. Ma’s model uses DCE-MRI imaging phenotypic features to predict the Ki-67 expression in breast cancer (AUC = 0.77320. Yasemin Kayadibi employed an MRI-based radiogenomics model to predict Ki-67 expression levels. With a Ki-67 threshold of 14%, the model attained an AUC of 0.849 on the test set35. In our study, SVM outperformed 6 mainstream machine learning algorithms, becoming the optimal model for predicting Ki-67 expression level (AUC = 0.811). Besides, the model performed stably in both internal testing and external validation. This proved its reliability in predicting Ki-67 and providing technical support for precision medical breast cancer treatment. Although some MRI models have reported high AUC values, the ultrasound technology that we employed is cost-effective and easily implementable, rendering it capable of offering comprehensive and precise treatment recommendations in resource-limited settings36. It is more likely to become a valuable tool for breast cancer screening management than MRI. Therefore, the ultrasound-based Ki-67 expression level prediction model presents advantages such as convenience, economy, and real-time monitoring for clinical applications.
This study also demonstrates significant advantages in multiple aspects beyond MRI. Firstly, with a multicenter dataset comprising 929 cases, the sample representativeness is enhanced, conferring greater reliability compared to single-center studies37. Secondly, the adoption of the LASSO feature selection algorithm effectively removes redundant and noisy features, thereby improving the prediction accuracy and performance of the model, which outperforms traditional image smoothing processing methods38. Finally, we explored the clinical application potential of this multi-center fusion model. DFS, a critical indicator for assessing breast cancer prognosis, assists clinicians in accurately evaluating patient conditions and predicting survival. Our study concentrated on the model’s effectiveness in predicting DFS. We stratified patients into high- and low-risk groups based on model-predicted Ki-67 expression probabilities. Internal validation confirmed model accuracy in DFS assessment, revealing significantly reduced DFS in high-risk patients.
Limited
The ultrasound-based Ki-67 expression prediction model developed in this study demonstrates potential in clinical application, but it still exhibits the following limitations: Despite utilizing a multicenter dataset, geographical bias and patient diversity issues may still exist, affecting the model’s accuracy and generalization ability. Additionally, biases may be introduced during manual delineation of the ROI and image denoising processes. These factors may slightly diminish the model’s predictive accuracy and clinical reliability. To mitigate these limitations, future studies could further expand the scale and diversity of the dataset to include patients from more regions, ethnicities, and pathological types. Furthermore, semi-automatic or fully automatic ROI delineation algorithms and adaptive denoising algorithms based on deep learning could be trained to enhance model accuracy.
Conclusions
The study utilizes deep features from multi-center breast cancer ultrasound imaging to develop and validate a Ki-67 expression prediction model. Future developments may include the development of clinical decision support tools based on this model and its integration into hospital information systems for automated analysis, both of which can aid physicians in formulating personalized treatment plans, thereby reducing their workload while enhancing treatment efficacy and minimizing resource waste.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DCE-MRI:
-
Dynamic contrast-enhanced magnetic resonance imaging
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- Her-2:
-
Human epidermal growth factor receptor 2
- BMI:
-
Body mass index
- EGFR:
-
Epidermal growth factor receptor
- ROI:
-
Region of interest
- TA:
-
Tumor area
- PTA:
-
Peritumoral area
- PCA:
-
Principal component analysis
- SVM:
-
Support vector machine
- LightGBM:
-
Light gradient boosting machine
- DFS:
-
Disease-free survival
- Binary cross-entropy:
-
A metric used to measure the difference between the probability distribution predicted by the model and the true labels
- ROI:
-
The target area within an image used for analysis
- ReLU:
-
An activation function that enhances the model’s ability to fit non-linear relationships
- PCA:
-
A dimensionality reduction technique used for initial dimensionality reduction of extracted deep features, thereby reducing computational complexity
- LASSO:
-
A linear regression analysis method used to further reduce the number of features
- AUC:
-
A metric used to evaluate the performance of classification models. A higher AUC indicates better model performance
- F1 Score:
-
A metric that balances precision and recall, used to evaluate the balanced performance of a model
References
Grabinski, V. F. & Brawley, O. W. Disparities in breast cancer. Obstet. Gynecol. Clin. N. Am. 49(1), 149–165 (2022).
Wang, W. et al. Spatial pattern and environmental drivers of breast cancer incidence in Chinese women. Environ. Sci. Pollut. Res. Int. 30(34), 82506–82516 (2023).
Wilkinson, L. & Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 95(1130), 20211033 (2022).
Katsura, C. et al. Breast cancer: Presentation, investigation and management. Br. J. Hosp. Med. (Lond) 83(2), 1–7 (2022).
Ge, Z. et al. Compared with high-intensity interval exercise, moderate intensity constant load exercise is more effective in curbing the growth and metastasis of lung cancer. J. Cancer 13(5), 1468–1479 (2022).
Song, Y. et al. The potential value of serum chemerin in patients with breast cancer. Sci. Rep. 11(1), 6564 (2021).
Yoshida, M. et al. An antisense amido-bridged nucleic acid gapmer oligonucleotide targeting SRRM4 alters REST splicing and exhibits anti-tumor effects in small cell lung cancer and prostate cancer cells. Cancer Cell Int. 23(1), 8 (2023).
Hu, X. et al. Preoperative nomogram for predicting sentinel lymph node metastasis risk in breast cancer: A potential application on omitting sentinel lymph node biopsy. Front. Oncol. 11, 665240 (2021).
Cui, H. et al. Identifying ultrasound features of positive expression of Ki-67 and P53 in breast cancer using radiomics. Asia Pac. J. Clin. Oncol. 17(5), e176–e184 (2021).
Huang, Z. et al. Molecular markers, pathology, and ultrasound features of invasive breast cancer. Clin. Imaging 79, 85–93 (2021).
Cheng, C. et al. Predicting the expression level of Ki-67 in breast cancer using multi-modal ultrasound parameters. BMC Med. Imaging 21(1), 150 (2021).
Xia, D. et al. Research progress and hotspot of the artificial intelligence application in the ultrasound during 2011–2021: A bibliometric analysis. Front. Public Health 10, 990708 (2022).
Xiang, H. et al. Development and validation of an interpretable model integrating multimodal information for improving ovarian cancer diagnosis. Nat. Commun. 15(1), 2681 (2024).
Xie, X. et al. Rapid intraoperative multi-molecular diagnosis of glioma with ultrasound radio frequency signals and deep learning. EBioMedicine 98, 104899 (2023).
Zhou, L. Q. et al. Deep learning predicts cervical lymph node metastasis in clinically node-negative papillary thyroid carcinoma. Insights Imaging 14(1), 222 (2023).
Chaiteerakij, R. et al. Artificial intelligence for ultrasonographic detection and diagnosis of hepatocellular carcinoma and cholangiocarcinoma. Sci. Rep. 14(1), 20617 (2024).
Witowski, J. et al. Improving breast cancer diagnostics with deep learning for MRI. Sci. Transl. Med. 14(664), eabo4802 (2022).
Suh, Y. J., Jung, J., & Cho, B. J. Automated breast cancer detection in digital mammograms of various densities via deep learning. J. Pers. Med. 10(4) (2020).
Niyas, S. et al. Automated molecular subtyping of breast carcinoma using deep learning techniques. IEEE J. Transl. Eng. Health Med. 11, 161–169 (2023).
Ma, W. et al. Breast cancer Ki67 expression prediction by DCE-MRI radiomics features. Clin. Radiol. 73(10), 909.e1-909.e5 (2018).
Zhou, J. et al. Diagnosis of benign and malignant breast lesions on DCE-MRI by using radiomics and deep learning with consideration of peritumor tissue. J. Magn. Reson. Imaging 51(3), 798–809 (2020).
Gao, Y. et al. Deep learning-enabled pelvic ultrasound images for accurate diagnosis of ovarian cancer in China: Aa retrospective, multicentre, diagnostic study. Lancet Digit. Health 4(3), e179–e187 (2022).
Chen, S. et al. Machine learning-based models enhance the prediction of prostate cancer. Front. Oncol. 12, 941349 (2022).
Al-Rimy, B.A.S., et al. An adaptive early stopping technique for DenseNet169-based knee osteoarthritis detection model. Diagnostics (Basel) 13(11) (2023).
Tong, Y. et al. Ki-67 increase after core needle biopsy associated with worse disease outcome in HER2-negative breast cancer patients. Sci. Rep. 13(1), 2489 (2023).
Zhang, A. et al. The role of Ki-67 in evaluating neoadjuvant endocrine therapy of hormone receptor-positive breast cancer. Front. Endocrinol. (Lausanne) 12, 687244 (2021).
Li, F. Y. et al. Prognostic value of Ki-67 in breast cancer patients with positive axillary lymph nodes: A retrospective cohort study. PLoS One 9(2), e87264 (2014).
Bocchi, M. et al. Expression of Ki-67 and p53 proteins: Breast cancer aggressivity markers in Brazilian young patients. J. Adolesc. Young Adult Oncol. 10(4), 379–388 (2021).
Nishit, A. et al. Association of Ki-67 with clinicopathological factors in breast cancer. Cureus 13(6), 15621 (2021).
Zhu, X. et al. The prognostic and predictive potential of Ki-67 in triple-negative breast cancer. Sci. Rep. 10(1), 225 (2020).
Das, K. et al. Machine learning and its application in skin cancer. Int. J. Environ. Res. Public Health 18(24) (2021).
Wei, W., Li, Y., & Huang, T. Using machine learning methods to study colorectal cancer tumor micro-environment and its biomarkers. Int. J. Mol. Sci. 24(13) (2023).
Chen, S., et al. Machine learning-based models enhance the prediction of prostate cancer. Front Oncol. 12, 941349. https://doi.org/10.3389/fonc.2022.941349 (2022).
Xu, R. et al. Ultrasound-based radiomics model for predicting molecular biomarkers in breast cancer. Front. Oncol. 13, 1216446 (2023).
Ma, W. et al. Breast cancer Ki-67 expression prediction by DCE-MRI radiomics features. Clin. Radiol. 73(10), 909.e1-909.e5 (2018).
Kayadibi, Y. et al. Radioproteomics in breast cancer: Prediction of Ki-67 expression with MRI-based radiomic models. Acad. Radiol. 29(Suppl 1), S116-s125 (2022).
Kurepa, D. et al. Neonatal lung ultrasound exam guidelines. J. Perinatol. 38(1), 11–22 (2018).
Yang, H. et al. Radiomic machine learning in invasive ductal breast cancer: Prediction of Ki-67 expression level based on radiomics of DCE-MRI. Technol. Cancer Res. Treat 23, 15330338241288752 (2024).
Li, Y. Z. et al. Breast MRI segmentation and Ki-67 high- and low-expression prediction algorithm based on deep learning. Comput. Math. Methods Med. 2022, 1770531 (2022).
Acknowledgements
We thank the Key Laboratory of Ultrasonic Molecular Imaging and Artificial Intelligence, Guangxi Zhuang Autonomous Region Engineering Research Center for Artificial Intelligence Analysis of Multimodal Tumor Images, and Guangxi Key Laboratory of Early Prevention and Treatment for Regional High Frequency Tumor/Key Laboratory of Early Prevention and Treatment for Regional High Frequency Tumor (Guangxi Medical University), Ministry of Education for their support of this study.
Funding
This study was supported by the Youth Science Foundation of Guangxi Medical University (grant number GXMUYSF202327); the National Natural Science Foundation of China (grant number 82160336); the Natural Science Foundation of Guangxi (grant number 2023GXNSFDA026013, 2020GXNSFDA238005); Self-financed scientific research project of the Health Commission of Guangxi Zhuang Autonomous Region (grant number Z-A20240410).
Author information
Authors and Affiliations
Contributions
Y.W., H.Y. M.W. and QS.C. are responsible for study conceptualization. M.W. generated most of the data, assisted by QS.C.. M.W. and SY.Z. checked the statistical method. M.W., Y.W. and SY.Z. prepared the figures. QS.C. and M.W. wrote the manuscript. Y.W. and H.Y. provided resources, supervision, and editing. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cen, Q., Wang, M., Zhou, S. et al. Multi-center study: ultrasound-based deep learning features for predicting Ki-67 expression in breast cancer. Sci Rep 15, 10279 (2025). https://doi.org/10.1038/s41598-025-94741-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-94741-4